Introduction
Breast cancer (BC) is a prevalent cancer type among
women, with an annual rate of 30 to 40/100,000 individuals in
general, with a heterogeneous nature with marked differences
between patients in terms of incidence, clinical outcome, response
to treatment and prognosis (1,2). BC is
classified according to a variety of factors, including expression
of estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor 2 (HER2), and according to
histological classification (3,4). BC
has become the most common cancer worldwide, surpassing lung
cancer, according to Global Cancer Data 2020 (5). Although the management strategies in
place for BC have rapidly advanced, including early detection and
effective therapies, metastatic disease continues to be a major
contributor to poor prognosis (6,7). Given
the limited availability of reliable biomarkers to accurately
predict BC prognosis due to tumor heterogeneity, there is a need to
identify new prognostic indicators and to design personalized
treatments. Progress has been made with surgical techniques,
chemotherapy, radiation therapy, hormone therapy and targeted
therapies with respect to the diagnosis and treatment of
early-stage BC, although the outlook for patients with advanced BC
remains poor (6). Furthermore,
recent data suggest that the majority of patients newly diagnosed
with BC are already at an advanced stage. BC is recognized as a
systemic disease by researchers around the world, and surgery
remains the mainstay of treatment (8). Although surgery is the main treatment
method for BC, its efficacy in the treatment of distant metastasis
is limited. Endocrine therapy is only effective for patients with
ER-positive and PR-positive BC; moreover, the rapid drug resistance
that develops to chemotherapy and endocrine therapy reduces the
benefits available to BC patients (9). This has led to the emergence of
immunotherapy as a potentially effective systemic treatment for BC.
However, despite decades of research, only a few immunotherapy
strategies have been implemented in a clinical setting, and even
fewer have shown promising therapeutic results (10).
Atypical chemokine receptors (ACKRs) have been
reported to fulfill a crucial role in the regulation of chemokine
availability via scavenging chemokines, and also have the potential
to elicit downstream signaling via coupling to β-arrestin (11). However, conventional chemokine
receptors have been reported to induce immune cell migration
directly via G-protein-coupled signaling (12). The following have been reported as
members of the ACKR family: ACKR1 [also known as Duffy antigen
receptor for chemokines (DARC)], ACKR2 (D6), ACKR3 [C-X-C chemokine
receptor type 7 (CXCR-7)], ACKR4 [C-C chemokine receptor type 11
(CCRL1)], ACKR5 (CCRL2) and ACKR6 [phosphatidylinositol transfer
protein, membrane-associated, 3 (PITPNM3)]. Additionally, a recent
study has reported that other receptors, including GPR182, GPR1 and
C5aR2, that display some, or all, of the abnormal biological
characteristics of ACKRs (13).
These receptors, however, have not received as much attention as
potential therapeutic targets in cancer due to a general lack of
understanding of their biological function (14).
ACKRs have conventionally been understood to act
solely as decoy or scavenger receptors, as they bind chemokines
without triggering signaling coupled to downstream G-proteins
(7). Tumor growth and metastasis
are both mediated by the tumor cell expression of ACKRs, whereas
antitumor immunity is mediated by the stromal cell expression of
ACKRs (15). Certain receptors that
are related to ACKRs, such as CCL2 and CXCL14, which serve
important roles in tumor biology, have also been studied in recent
years (16,17). Recent discoveries of additional
ACKRs that have atypical properties, and an expanding understanding
of the biological functions of ACKRs in cancer, both point to the
need for further investigation of this subgroup of receptors as
therapeutic targets in cancer (18–21).
However, the specific functions and prognostic significance of
individual ACKR family members in breast cancer (BC), remain both
poorly defined and poorly understood. Therefore, the present study
assessed the expression levels of ACKRs in breast carcinoma (BRCA)
tissues compared with normal breast tissues through a series of
database analyses, and the specific functions and prognostic
significance of certain individual ACKR family members in BC were
also explored.
Materials and methods
RNA-sequencing data and bioinformatics
analysis
Normalized RNA sequencing data and associated
clinical features were downloaded from The Cancer Genome Atlas
(TCGA) in the Breast Invasive Cancer (TCGA BRCA) dataset,
TCGA.BRCA.sampleMap/HiSeqV2_PANCAN (https://tcga.xenahubs.net). A total of 1,097 samples
of BC tissue and 114 samples of healthy breast tissue were
obtained. Transcript expression levels were estimated using the
FPKM (fragments per kilobase per million fragments mapped) method
for the high throughput sequencing data. Finally, the expression of
ACKRs in adjacent benign tissues that were case-matched to BCs in
TCGA database were paired for comparison using the Xiantao platform
(https://www.xiantaozi.com/).
Cell lines and cell culture
The human MCF7, SK-BR3, BT549 and MDA-MB-231 BC cell
lines, and the normal MCF10A mammary epithelial cell line were
purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. SK-BR3, MCF-7, BT549 and MDA-MB-231
cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% (v/v) fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) and 1% (v/v) penicillin/streptomycin
solution (Beyotime Institute of Biotechnology). MCF10A cells were
grown in MCF-10A cell medium (Procell Life Science & Technology
Co., Ltd.). Standard cell culture techniques were used to grow and
passage all cell lines in an incubator at 37°C in a 5%
CO2 atmosphere.
University of ALabama at Birmingham
CANcer data analysis Portal (UALCAN) database
Based on TCGA database, UALCAN (http://ualcan.path.uab.edu) is a comprehensive cancer
data analysis website (22,23). Data on the mRNA expression of ACKRs
and promoter methylation levels in BRCA and normal tissues (from
healthy controls; TCGA-BRCA), as well as the associations between
ACKR expression and clinicopathological parameters were assessed
using UALCAN. The differences between ACKR mRNA expression and
promoter methylation levels in BC and the matched controls (healthy
controls) were assessed using Welch's t-test. Differences in ACKR
expression among the different tumor substages were compared using
one-way ANOVA with Dunnett's multiple-comparison test as the
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference
ACKR-related gene function enrichment
analyses
Differentially expressed genes (DEGs) were analyzed
using R package (DESeq2, version 1.26.0). The threshold of
log2FC >2 and adjusted P-value of <0.05 were
chosen to consider genes as differentially expressed. Gene Ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses
were performed to evaluate potential gene functions associated with
TPMs based on the TCGA database with R package (org.Hs.eg.db,
version 3.10.0 and clusterProfiler, version 3.14.3).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from the cell lines using
the Total RNA Extraction Kit (Qiagen GmbH). cDNA was prepared using
a HiScript®III First-Strand cDNA Synthesis Kit (+gDNA
wiper) (Vazyme Biotech Co., Ltd.) according to the manufacturer's
instructions. Subsequently, qPCR was performed using ChamQ
Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.). Primers
were synthesized by GenScript. The reaction steps were as follows:
i) pre-denaturation at 95°C for 30 sec; ii) PCR reaction of 40
cycles at 95°C for 5 sec, followed by incubation at 60°C for 30
sec. The sequences of the β-actin and the ACKR primers are
presented in Table I. Cycle
thresholds were recorded and the relative expression of target
genes was calculated and quantified using the 2−∆∆Cq
method with β-actin as the reference gene (24).
 | Table I.Primer sequences used for reverse
transcription- quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription- quantitative PCR.
| Gene | Species | Sequence
(5′-3′) |
|---|
| Β-actin | Human | F:
CTCCATCCTGGCCTCGCTGT |
|
|
| R:
GCTGTCACCTTCACCGTTCC |
| ACKR1 | Human | F:
CCCTCAACTGAGAACTCAAGTC |
|
|
| R:
AGGTTGGCACCATAGTCTCCA |
| ACKR2 | Human | F:
CTTGCTCCGTTACGTGCCT |
|
|
| R:
GAAACTCCCGAAGACCCAATG |
| ACKR3 | Human | F:
TCTGCATCTCTTCGACTACTCA |
|
|
| R:
GTAGAGCAGGACGCTTTTGTT |
| ACKR4 | Human | F:
GTTTTCGTCATTGGACTTGCAG |
|
|
| R:
GCTACAGCCAAATTCAGGATGT |
| ACKR5 | Human | F:
AGCGATGAGGCAGAGCAATG |
|
|
| R:
GGACACCGATCACAAACACAG |
| ACKR6 | Human | F:
TCGCTTGTCTCTCACCTGAAC |
|
|
| R:
CAGGAACTCTCTGTAGACCTGG |
Tumor immune estimation resource
(TIMER)
TIMER 2.0 (https://cistrome.shinyapps.io/timer/) is an intuitive
web interface with six basic analysis modules for the systematic
assessment of numerous types of immune cell infiltration and their
clinical impact (25). ACKRs were
selected as the input using the gene module and scatterplots were
generated for the purposes of visualizing the correlation between
their expression levels and the level of immune infiltration in
BC.
Gene expression profiling interactive
analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/index.html, accessed
December 21, 2017) is an analytical tool that uses standard
processing procedures which incorporate data from thousands of
normal and cancer tissue samples (26). In the present study, GEPIA was used
to analyze differential gene expression between tumor and normal
tissues. Pathological staging analysis and associated prognosis
analysis were also performed. Statistical analysis of expression
levels or pathological stage analysis were performed using Welch's
t-test. Patient survival was also analyzed using Kaplan-Meier
curves for further validation.
cBioPortal
The cBio Cancer Portal (cBioPortal) is a
comprehensive web resource which provides multidimensional
visualization and access to cancer genomics data (www.cbioportal.org) (27). In the present study, gene mutations
of ACKRs in BRCA were analyzed using this resource.
GeneMANIA
GeneMANIA (http://www.genemania.org) is a resource-rich website
for gene information, gene list analysis and high-precision
prediction algorithm-prioritized gene function analysis. This
website was used to establish the ACKR interaction networks.
STRING
The STRING online database (https://string-db.org/) is a website for the
investigation of protein-protein interactions (PPIs). Through the
PPI network analysis of STRING, the different expression levels and
potential PPIs of ACKRs were identified and assessed.
Statistical analysis
GraphPad Prism software (version 8.0; Dotmatics) and
R (https://www.r-project.org; version
3.6.3) were used for statistical analysis. Transcriptional data and
clinical data for BRCA were also downloaded from TCGA database,
subjected to one-way Cox regression analysis, and analyzed using
the R software package ‘Forestplot’ (version 2.0.1; http://CRAN.R-project.org/package=forestplot).
Receiver operating characteristic (ROC) curves were drawn using
‘pROC’ package (version 1.18.0; http://CRAN.R-project.org/package=pROC) in R. Finally,
multivariate analysis of overall survival (OS), distant
metastasis-free survival (DMFS), post-progression survival (PPS)
and recurrence free survival (RFS) were performed using the
‘survival’ package (version 3.2, year 13; http://CRAN.R-project.org/package=survival) in R.
The correlation between ACKRs expression and immune infiltration
was evaluated by Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
ACKR expression and promoter
methylation in patients with BRCA
The expression of ACKRs in patients with BRCA was
assessed using the UALCAN and GEPIA databases. The mRNA expression
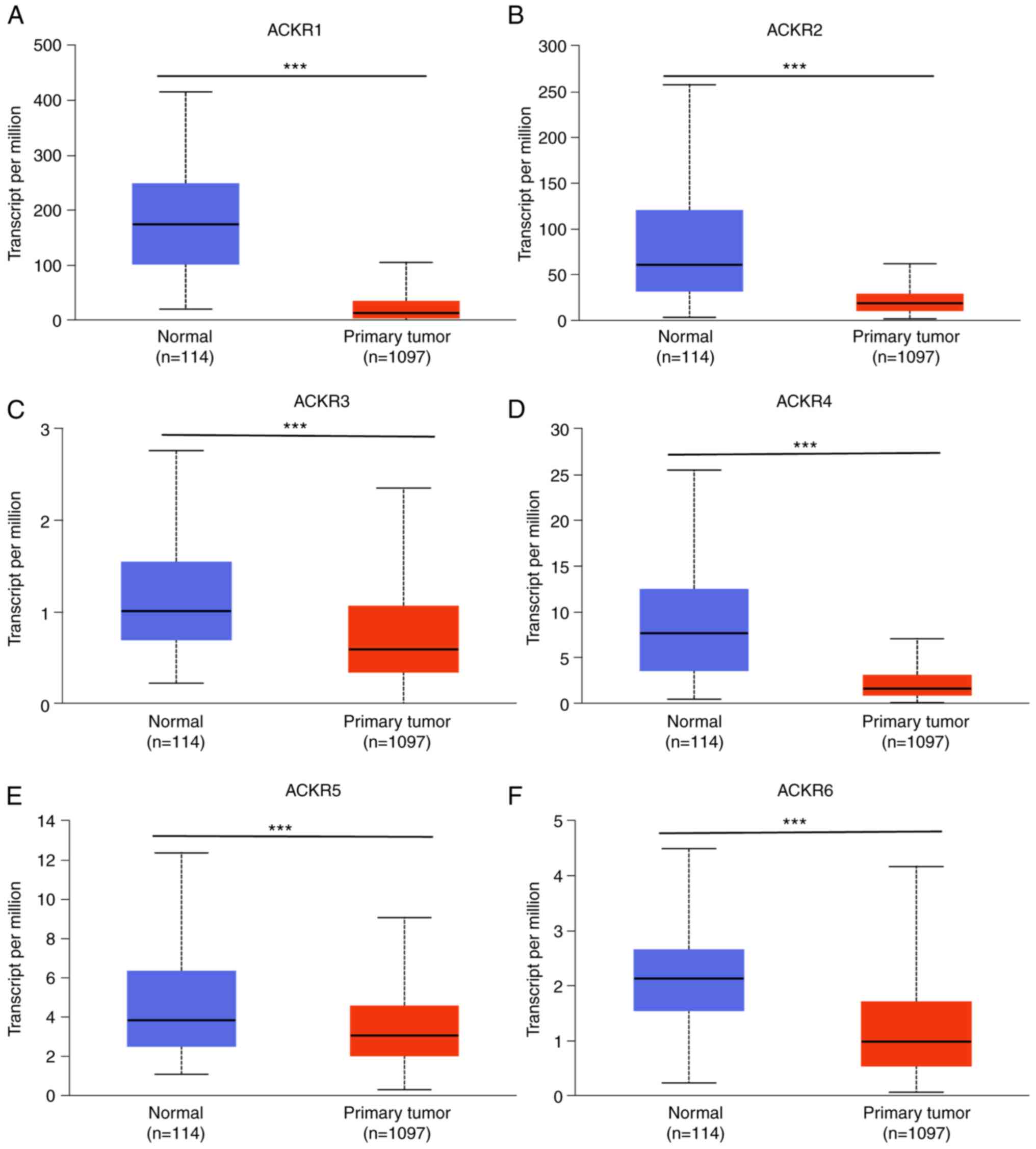
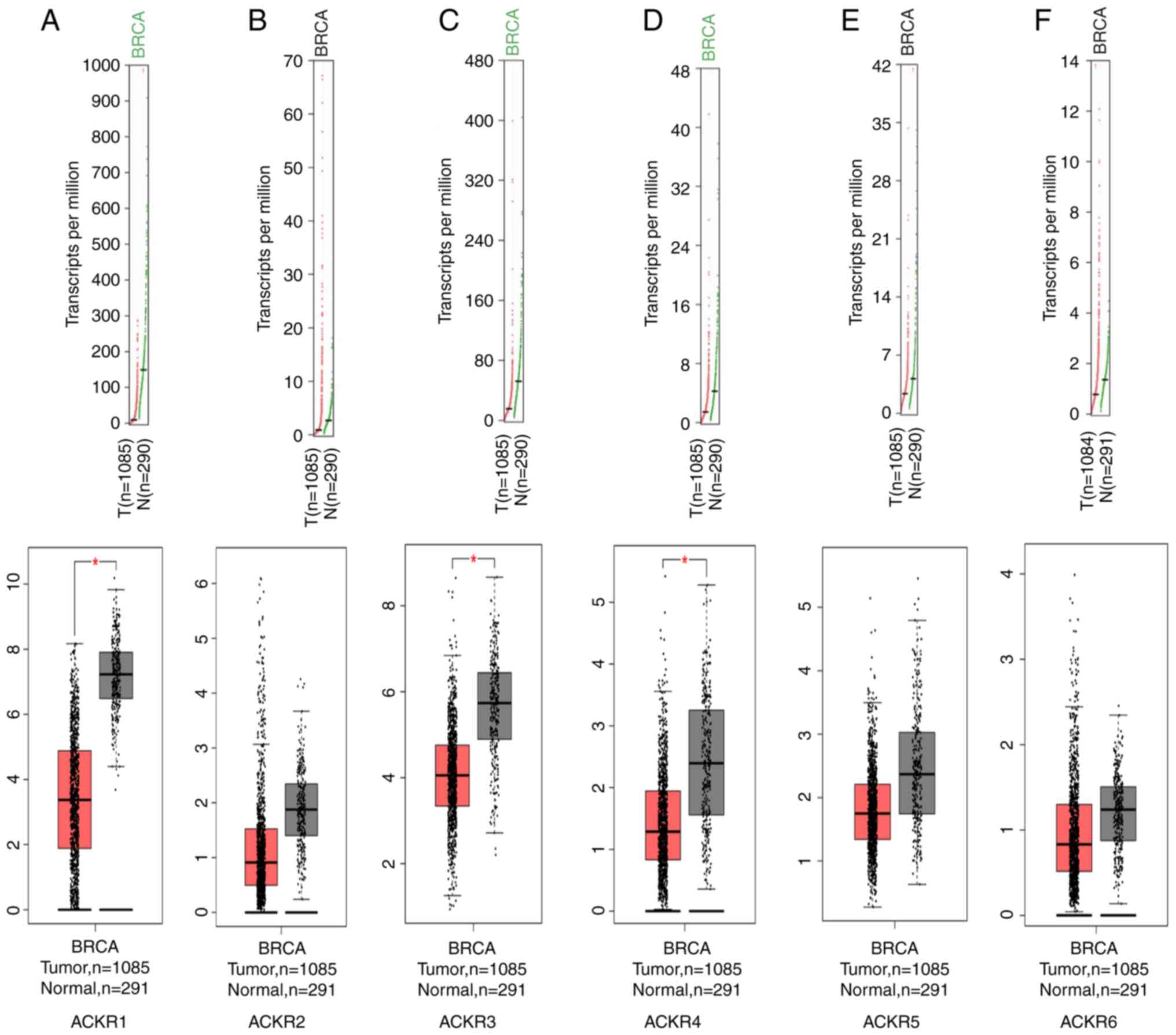
levels of ACKR1 (Figs. 1A and
2A), ACKR3 (Figs. 1C and 2C) and ACKR4 (Figs. 1D and 2D) were demonstrated to be significantly
upregulated in BRCA compared with normal tissues. The expression of
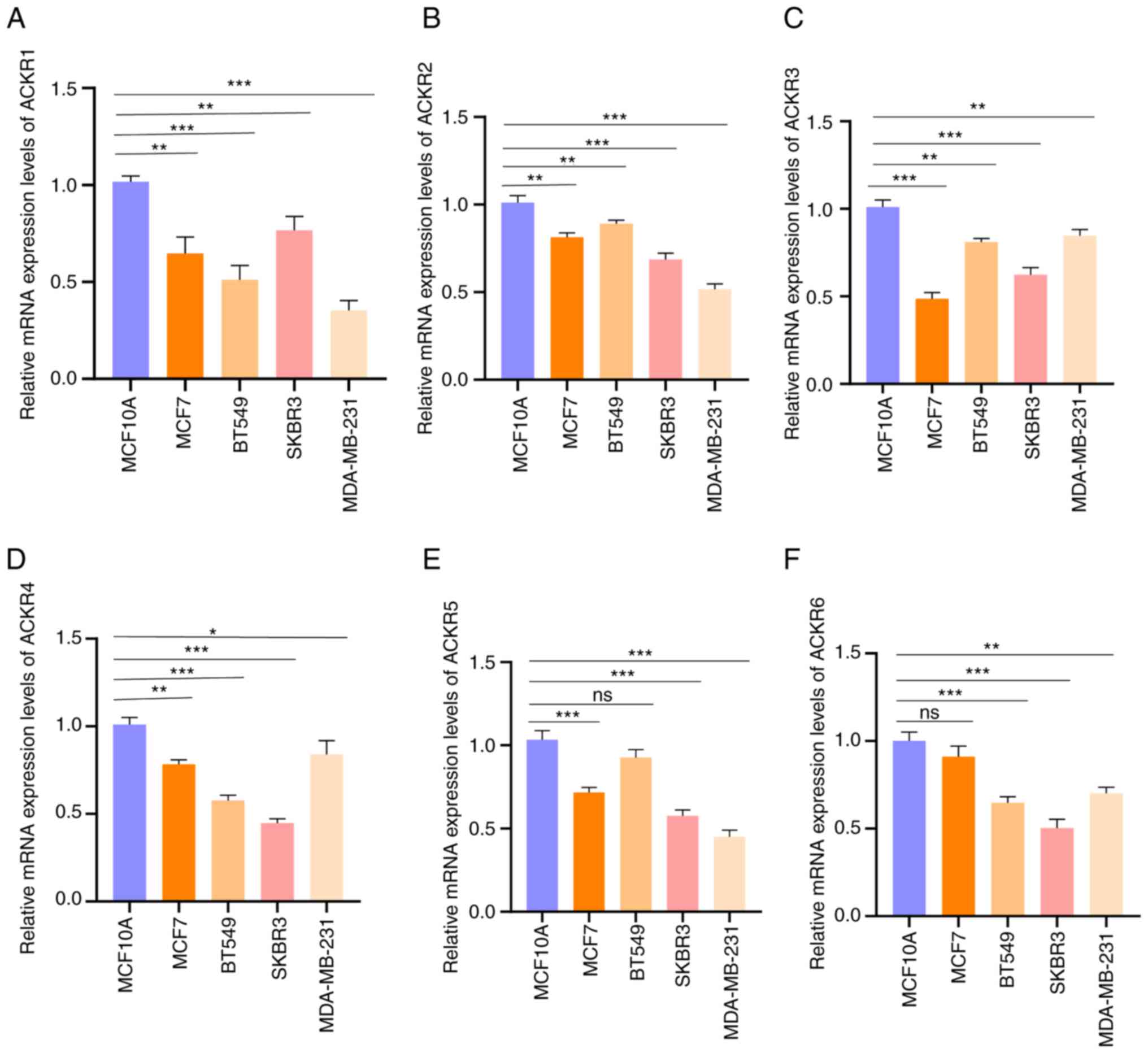
levels of ACKRs in the normal BC cells (MCF-10A, MCF7, SKBR3 and
MDA-MB-231) were assessed using RT-qPCR. Consistent with the
aforementioned results, markedly higher expression levels of the
ACKRs were detected in BRCA cells compared with normal MCF10A
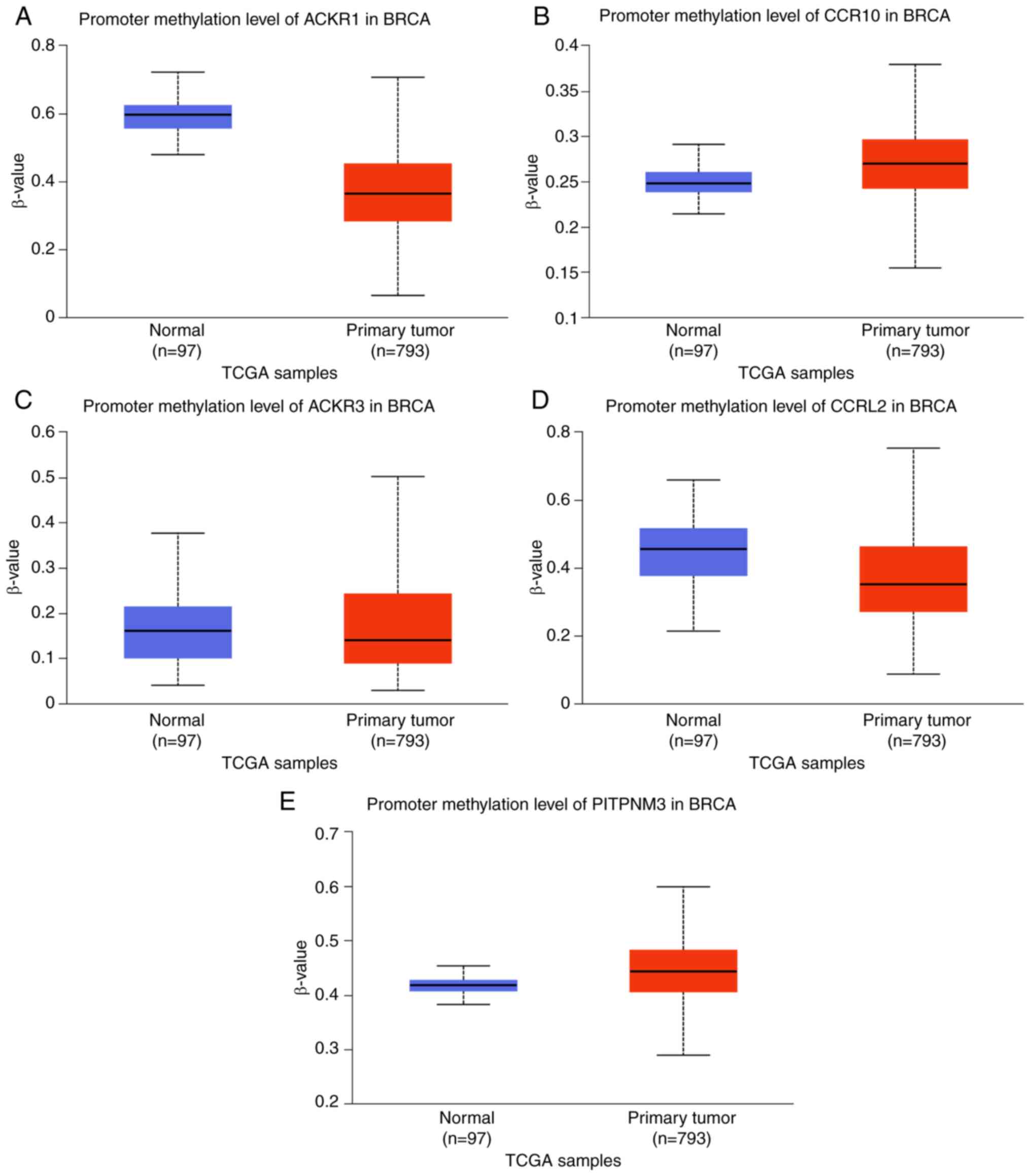
cells, except ACKR5 in BT549 and ACKR6 in MCF7 (Fig. 3). Furthermore, promoter methylation
was demonstrated to be notably correlated with BC progression.
Marked hypermethylation was demonstrated in the ACKR2
promoter (Fig. 4B) and in the
ACKR6 promoter (Fig. 4E).
However, marked hypomethylation was demonstrated in the
ACKR1 (Fig. 4A),
ACKR3 (Fig. 4C) and
ACKR5 (Fig. 4D)
promoters.
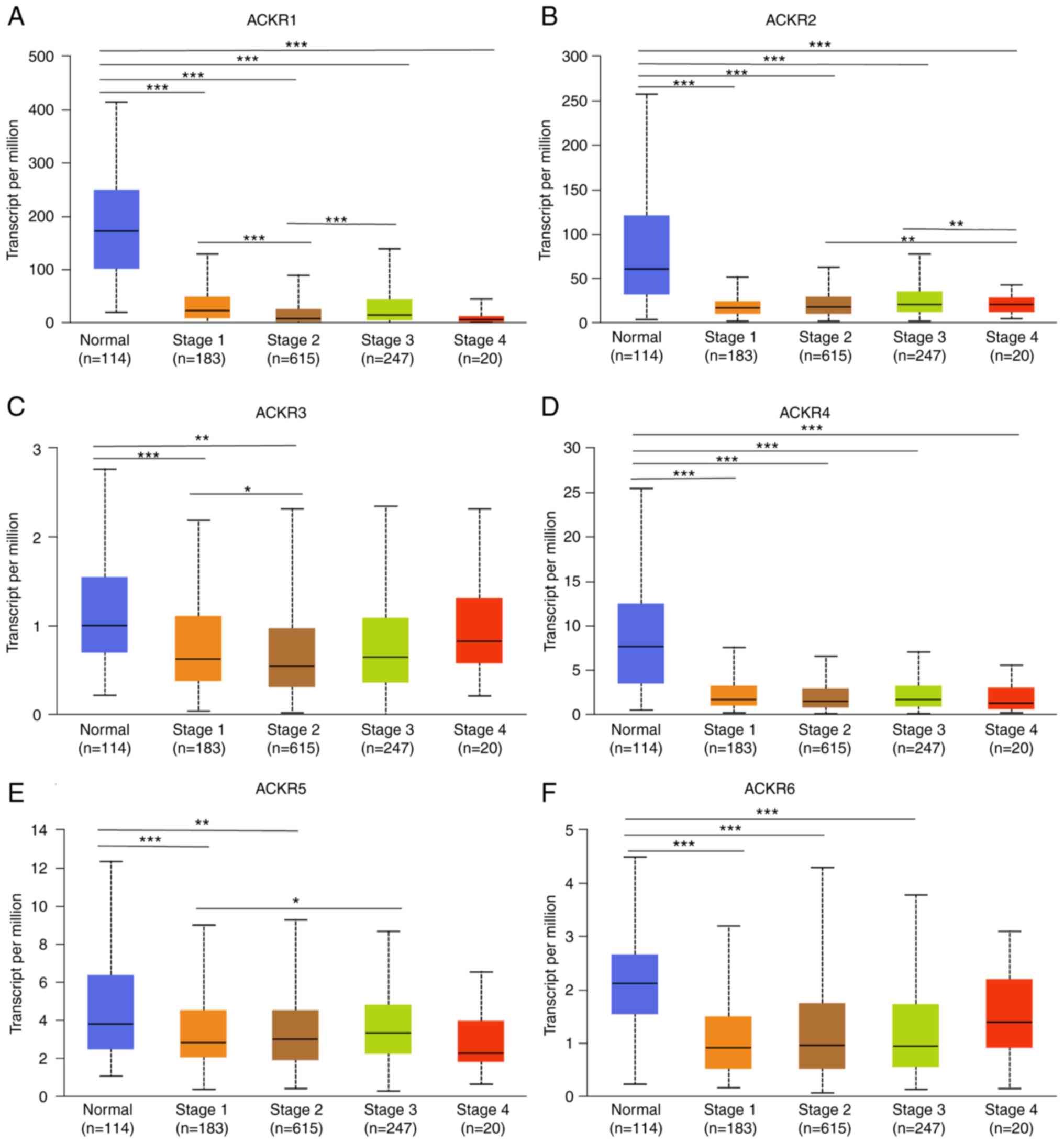
Association between ACKRs and the
cancer stages of patients with BRCA
Data from the UALCAN database demonstrated that BC
stage was markedly negatively correlated with the expression of
ACKR1 (Fig. 5A), ACKR2 (Fig. 5B), ACKR3 (Fig. 5C), ACKR4 (Fig. 5D), ACKR5 (Fig. 5E) and ACKR6 (Fig. 5F). These findings suggested that the
expression levels of ACKRs were closely associated with the staging
of patients with BRCA.
Prognostic and diagnostic value of
ACKR mRNA expression in patients with BC
To assess the value of differently expressed ACKRs
in determining BC progression, the correlation between different
ACKRs and clinical outcomes was analyzed using Kaplan-Meier plots.
A low expression of ACKR1 (Fig. 6A)
was significantly correlated with a shorter DMFS in patients with
BC. Furthermore, low mRNA expression levels of ACKR1 (Fig. 6B and C), ACKR4 (Fig. 6N and O) and ACKR6 (Fig. 6W) were significantly correlated with
shorter recurrence-free survival (RFS) times and shorter OS times
in patients with BC. In addition, a high mRNA expression level of
ACKR3 (Fig. 6I-K) was demonstrated
to be significantly correlated with shorter RFS, OS and DMFS times
in patients with BC. A low mRNA expression level of ACKR4 (Fig. 6P) was a predictor of shorter PPS in
patients with BC, while other effects of ACKRs were not significant
(Fig. 6). Furthermore, it was
demonstrated that ACKRs had high accuracy in terms of both
diagnosing BC and differentiating BC from the normal controls
(Fig. 7).
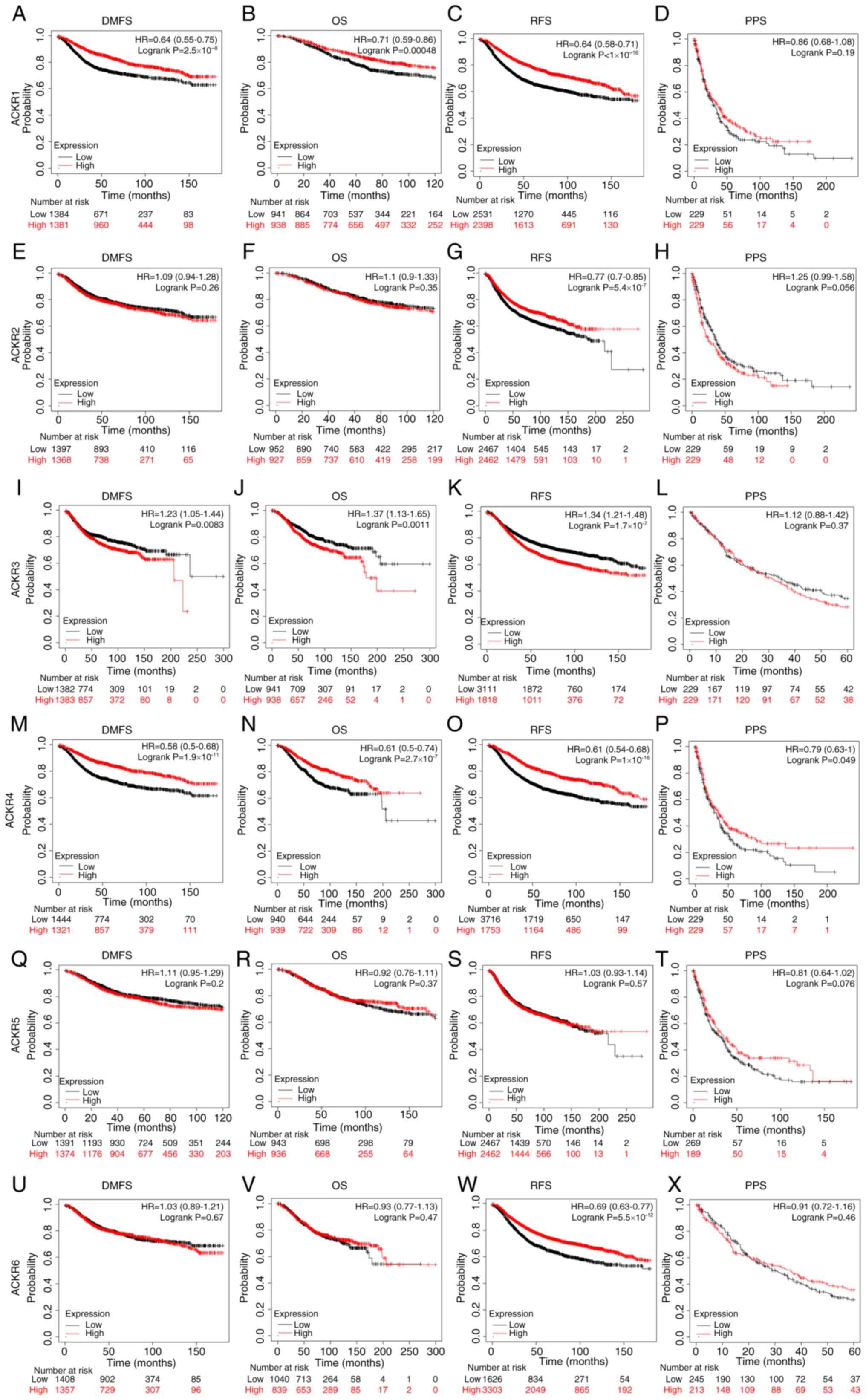 | Figure 6.The prognostic value of ACKRs
according to Kaplan-Meier curves of (A, E, I, M, Q and U) DMFS, (B,
F, J, N, R and V) OS, (C, G, K, O, S and W) RFS and (D, H, L, P, T
and X) PPS in patients with BRCA. The significance was computed
using the Cox-Mantel test to compare two cohorts. BRCA, breast
carcinoma; ACKR, atypical chemokine receptor; DMFS, distant
metastasis-free survival; OS, overall survival; RFS,
recurrence-free survival; PFS, progression-free survival. |
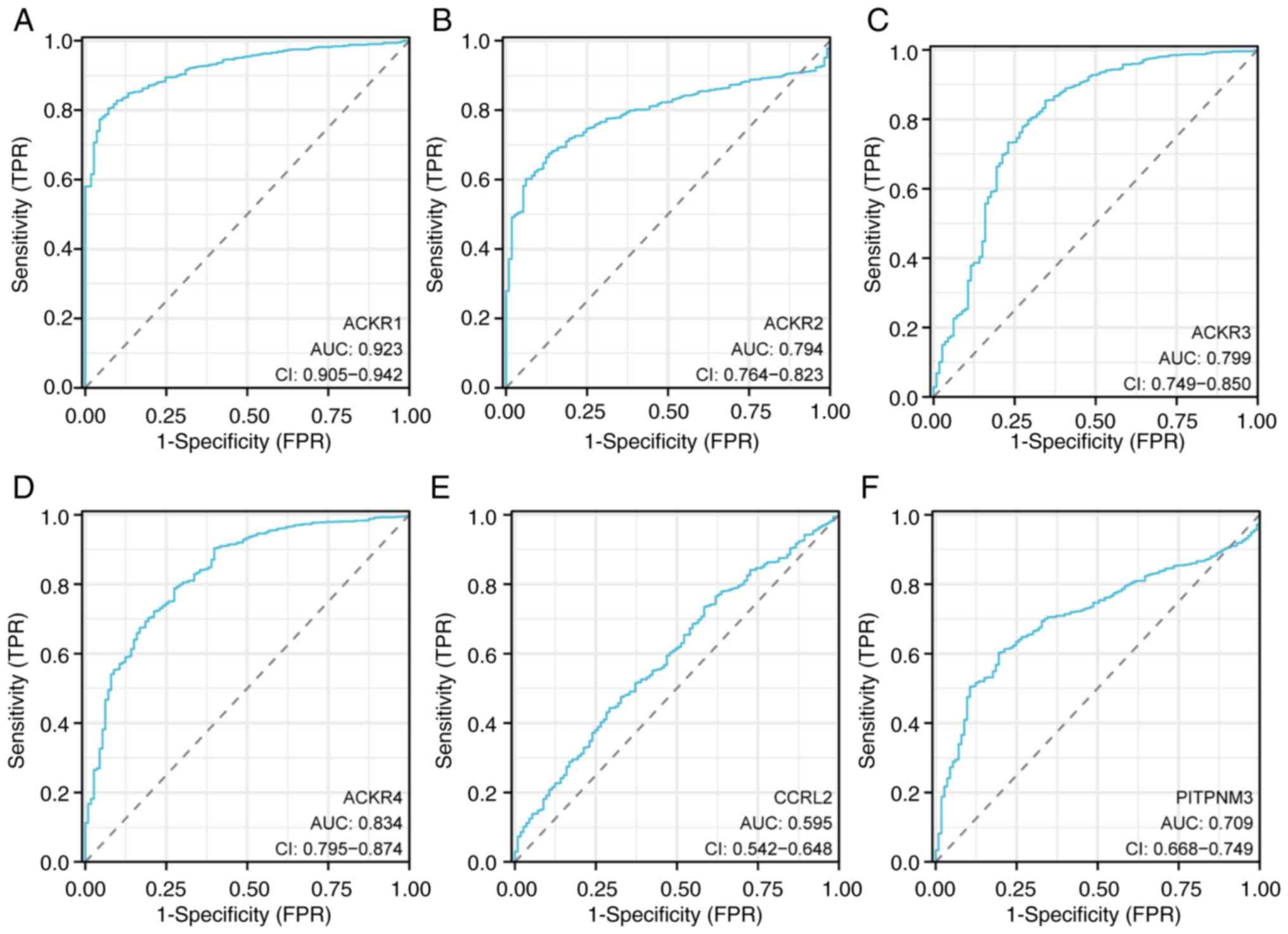 | Figure 7.ROC curves for breast cancer.
Sensitivity and specificity values for (A) ACKR1, (B) ACKR2, (C)
ACKR3, (D) ACKR4, (E) ACKR5 and (F) ACKR6 in BRCA are shown. BRCA,
breast carcinoma; ACKR, atypical chemokine receptor; ROC, receiver
operating characteristic; AUC, area under the curve; CI, confidence
interval; TPR, true-positive rate; FPR, false-positive rate. |
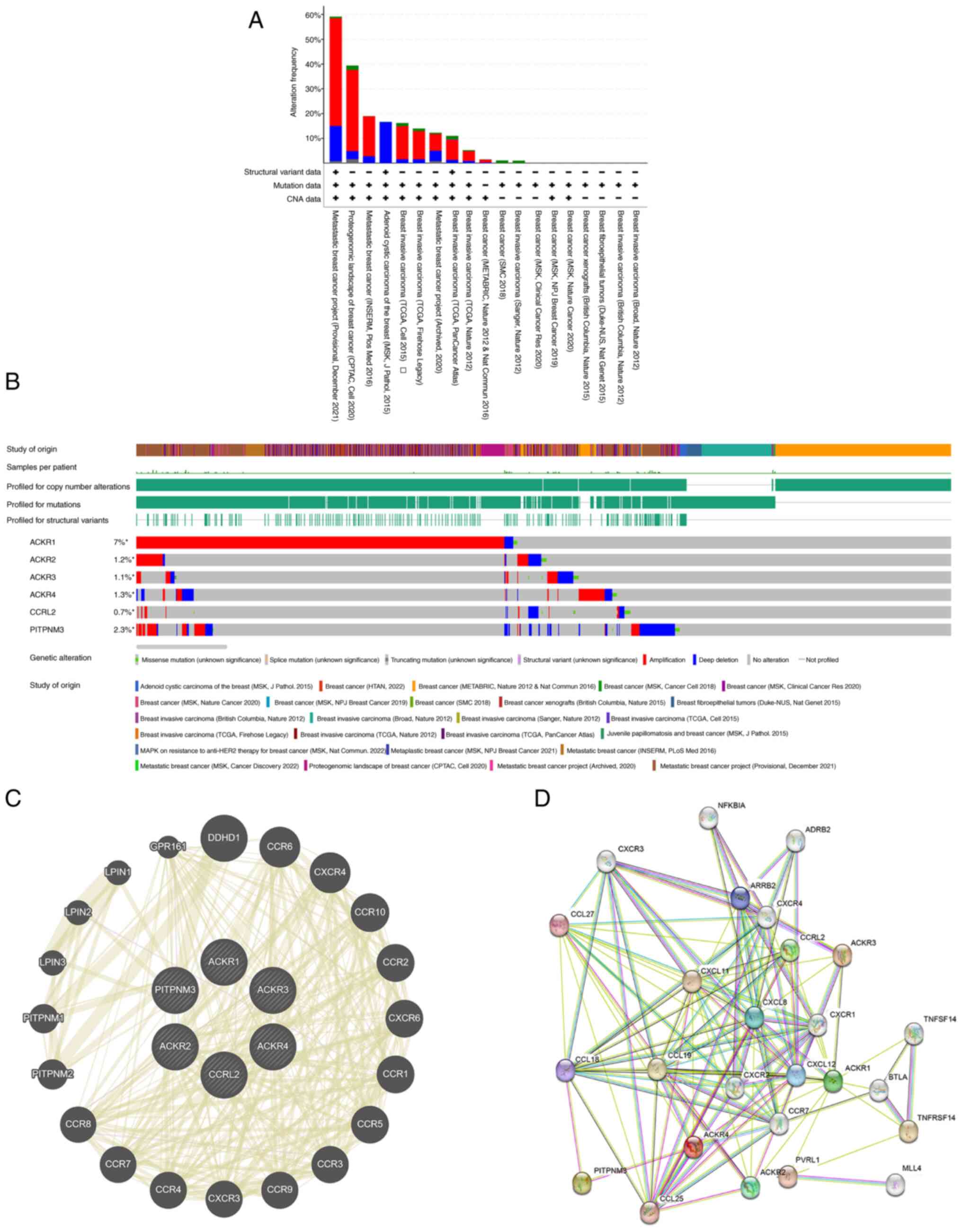
Changes in expression of ACKR genes,
and expression and interaction analysis of ACKR genes in patients
with BRCA
The genetic alterations affecting the expression of
ACKR family genes in BC were examined using the cBioPortal web
tool. The ACKR family members (ACKR1, ACKR2, ACKR3, ACKR4, ACKR5
and ACKR6) were changed in 7.0, 1.2, 1.1, 1.3, 0.7 and 2.3% of the
BC samples, respectively (Fig. 8A and
B). A PPI network analysis of PPIs was subsequently performed
on the differentially expressed ACKRs using the STRING website, and
possible interactions between the ACKRs were investigated. Multiple
nodes and edges were presented in the PPI network (Fig. 8C). The GeneMANIA database was used
to establish the gene interaction networks. Fig. 8D indicated that the network involved
links between 6 ACKRs and several other closely related
genes, including DDHD1, CCR8, CXCR4, CCR10, CCR2, CXCR6, CCR1,
CCR5, CCR3 and CCR9.
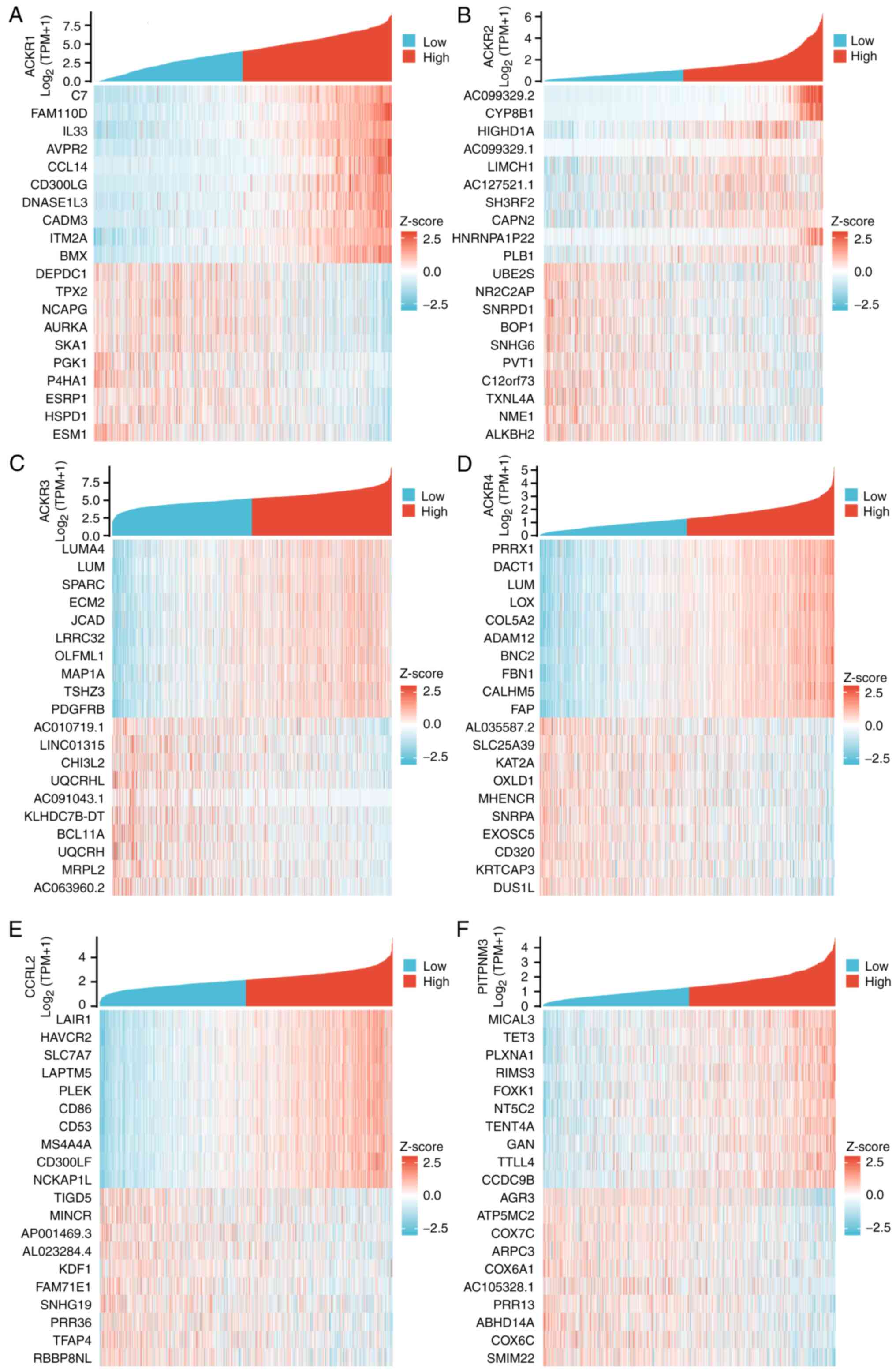
BC TCGA, gene ontology (GO) and Kyoto
Encyclopedia of Genomes (KEGG) analyses of ACKRs and their
co-expressed genes
The TCGA database software package DEseq2 R was used
to analyze differentially expressed genes in patients with BC with
either high or low expression of ACKR. The top 10 top 10 genes that
were positively or negatively correlated with ACKRs in BC were
identified (Fig. 9). GO and KEGG
enrichment analyses using the top 100 differentially expressed
genes that were mainly positively correlated with ACKRs were
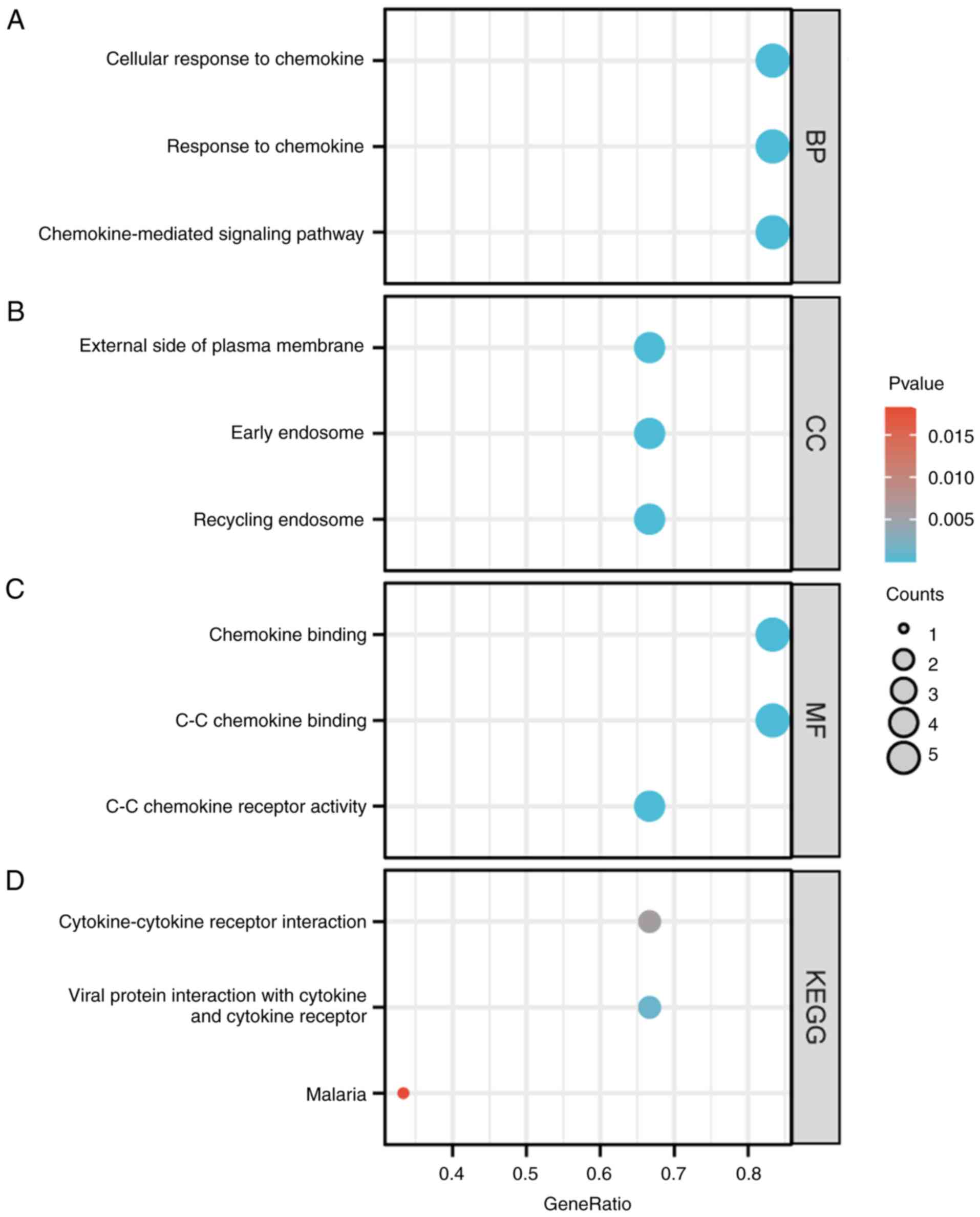
subsequently performed. Condensed GO analysis information for
biological processes (BP), cellular composition (CC) and molecular
functions (MF). From the perspective of BP, ‘chemokine-mediated
signaling pathways’, ‘cellular response to chemokine’ and ‘response
to chemokine’ were found to be significantly enriched for ACKRs and
their associated genes. KEGG analysis terms, were enriched in
‘cytokine-cytokine receptor interactions’, ‘viral protein
interaction with cytokine and cytokine receptor’ and ‘malaria’
(Fig. 10).
ACKR immune cell infiltration in
patients with BC
The concentration of immune cells has previously
been reported to be associated with the proliferation and
progression of cancer cells (28).
In the present study, the TIMER database was used to examine the
association between ACKR members and immune cell infiltration. The
expression of ACKR1 was demonstrated to be significantly associated
with the infiltration of CD4+ T cells, CD8+ T
cells and dendritic cells in patients with BC (Fig. 11A). ACKR2 was significantly
positively correlated with the infiltration of CD8+ T
cells, B cells, CD4+ T cells, macrophages and
neutrophils in patients with BC (Fig.
11B). The expression of ACKR3 was significantly positively
correlated with the infiltration of CD8+ T cells,
CD4+ T cells, macrophages and neutrophils in BC
(Fig. 11C). Furthermore, the
expression of ACKR4 in patients with BC was markedly positively
correlated with the infiltration rates of CD4+ T cells,
CD8+ T cells, macrophages and neutrophils (Fig. 11D). The expression of ACKR5 was
found to be significantly positively correlated with all six host
immune cell types (B cells, CD8+ T cells,
CD4+ T cells, macrophages, neutrophils and dendritic
cells) in patients with BC (Fig.
11E). Finally, the expression of ACKR6 in colorectal cancer was
found to be significantly positively correlated with the
infiltration of CD8+ T cells, CD4+ T cells,
neutrophils and dendritic cells (Fig.
11F).
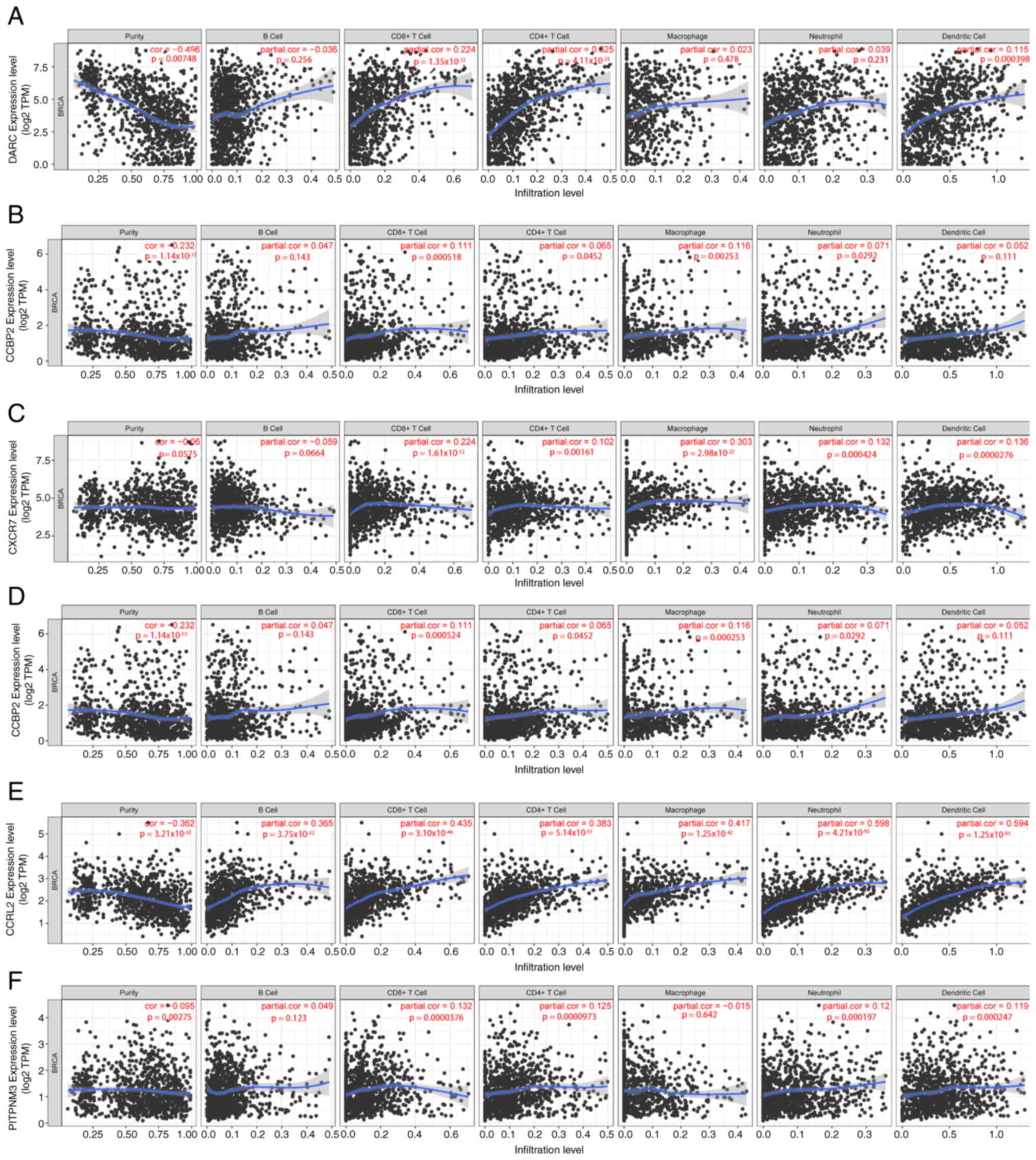 | Figure 11.Correlation of differentially
expressed ACKR with immune cell infiltration. The expression of (A)
ACKR1 was significantly positively correlated with CD4+
T cells, CD8+ T cells and dendritic cell infiltration;
(B) ACKR2 expression was significantly positively correlated with
infiltration of CD4+ T cells, CD8+ T cells,
macrophages and neutrophils; (C) ACKR3 expression was significantly
positively correlated with the infiltration of CD4+ T
cells, CD8+ T cells, macrophages, neutrophils and
dendritic cells; (D) ACKR4 expression was significantly positively
correlated with the infiltration of CD4+ T cells,
CD8+ T cells, macrophages and neutrophils; (E) ACKR5
expression was significantly positively correlated with the
infiltration of B cells, CD8+ T cells, CD4+ T
cells, macrophages, neutrophils and dendritic cells; and (F) ACKR6
expression was significantly positively correlated with the
infiltration of CD4+ T cells, CD8+ T cells,
neutrophils and dendritic cells. TPM, transcripts per million;
BRCA, breast carcinoma; ACKR, atypical chemokine receptor; TIMER,
Tumor Immune Estimation Resource. |
To further assess the effect of ACKRs on the tumor
microenvironment (TME), the correlation between ACKRs and immune
cell infiltration was evaluated using Spearman's correlation
analysis. This demonstrated that ACKRs were correlated both
positively and negatively with numerous types of immune cells; for
example, Tcm, T helper cells and Tem were consistently positive
correlated with all ACKRs (Fig.
12).
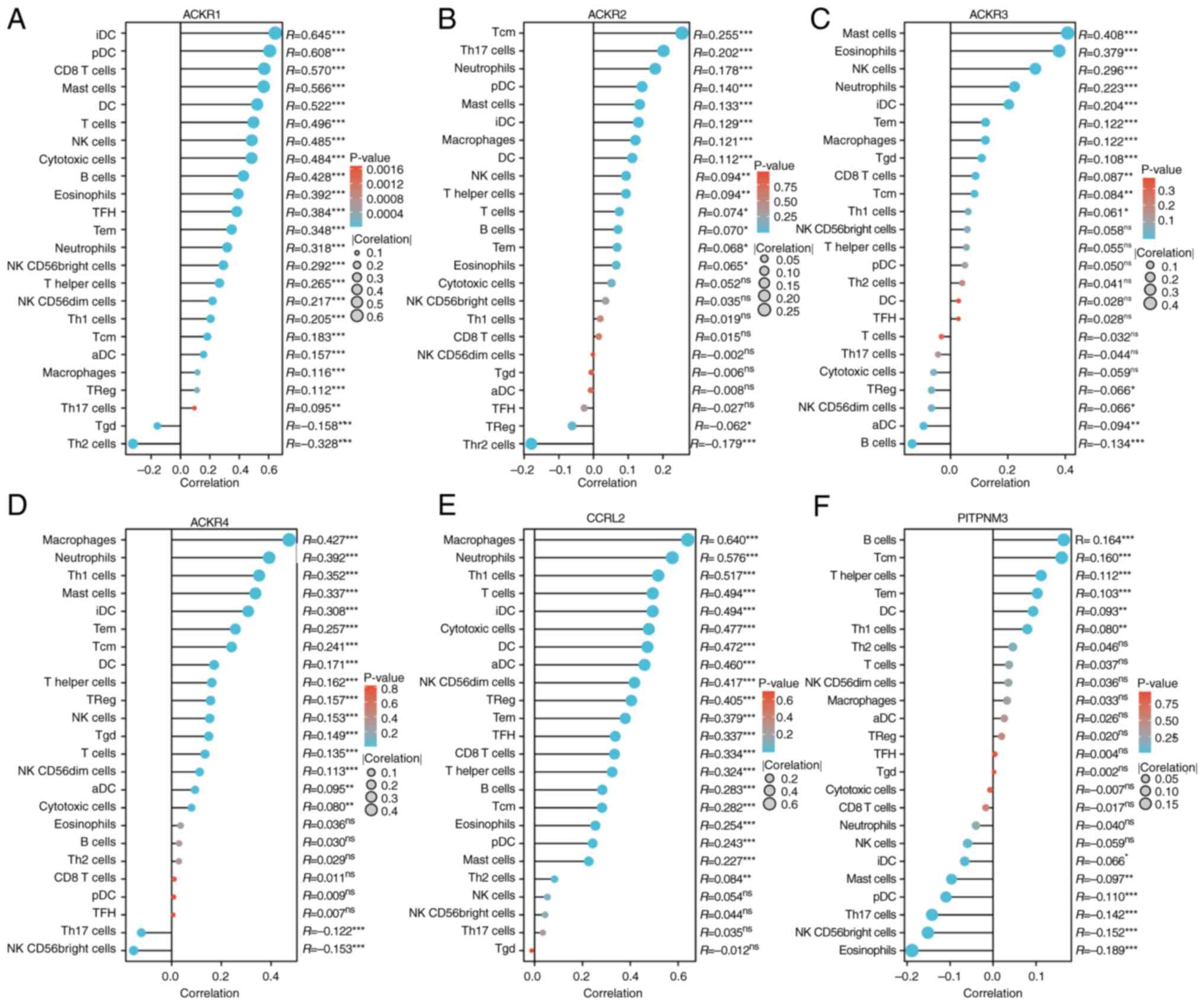 | Figure 12.Expression levels of (A) ACKR1, (B)
ACKR2, (C) ACKR3, (D) ACKR4, (E) ACKR5 and (F) ACKR6 were
significantly correlated with immune cell infiltration in BRCA
using the Lollipop chart. *P<0.05, **P<0.01 and
***P<0.001. BRCA, breast carcinoma; ACKR, atypical chemokine
receptor; Th, T helper; aDC, activated dendritic cell; Treg,
regulatory T cells; TFH, T follicular helper cell; Tgd, γδ T cell;
NK, natural killer; iDC, immature dendritic cell; pDC, plasmacytoid
dendritic cell. |
Discussion
BC is known to be a highly heterogeneous malignancy,
which presents challenges in terms of its diagnosis and treatment
due to the differences among individual cases. A previous study
suggested that the identification of novel therapeutic targets for
BC may be a viable strategy to address its heterogeneity (28). Therefore, it is critical to explore
the mechanisms which underlie BC initiation and progression, and to
identify disease-associated biomarkers in order to develop
effective strategies for the diagnosis, treatment and prognostic
assessment of BC.
Traditional chemokines and chemokine receptors
fulfill key roles in the regulation of immune cell migration and
the control of immune cell composition in the TME, and have been
reported as potential targets for cancer therapy (29). However, ACKRs, as a subfamily of
chemokine receptors, have received less attention as cancer
therapeutic targets due to their unclear biological function
(12). ACKRs were originally
considered to function solely as decoy or scavenger receptors as
they were able to bind chemokines without activating signaling
coupled to downstream G-proteins, which differentiates them from
classical chemokine receptors. In the present study, the
expression, mutation status, prognostic value and immune cell
infiltrates of ACKRs in BC were evaluated.
ACKRs have also been previously assessed in other
settings, in addition to their potential role in BC. ACKR1, for
example, has been reported to regulate hematopoiesis in nucleated
erythroid cells, and its deficiency has been reported to be
associated with reduced atherogenesis in ApoE-knockout mice
(30,31). These studies suggested that ACKRs
may serve important functions in addition to their role in cancer,
and further investigation is required to fully understand their
biological significance. Graham (32) speculated that ACKR1 could be a tumor
suppressor, which could serve as a predictive factor for the
treatment of cervical cancer. ACKR2 receptor has been reported to
have anti-oncogenic properties in several human tumor types
(32). Indeed, its expression was
previously shown to be correlated with improved outcomes in gastric
cancer and cervical squamous cell carcinoma, and to lead to an
inhibition of proliferation of lung cancer cells and tumorigenesis
(32). ACKR2 has also been reported
to delay tumor progression in Kaposi's sarcoma through the
inhibition of the inflammatory chemokines, chemokine ligand
(CCL)-2, CCL-5 and CCL-3, thereby reducing the infiltration of
macrophages and angiogenesis (33,34).
Although further studies are required to fully elucidate the
underlying mechanism of the action of ACKR2 in BC, as well as its
potential as a therapeutic target (16). ACKR4 is an important member of the
ACKR family, which functions as a scavenger receptor for
homoeostatic chemokines, such as CCL19, CCL21, CCL25 and CXCL13
(17). Although the mechanism by
which ACKRs function as scavengers has yet to be fully elucidated,
it is well established that ACKR4 spontaneously moves between the
plasma membrane and endosomes, with β-arrestins regulating its
trafficking at the steady state and promoting chemokine uptake
(17). It has been suggested that
ACKR4 may impair the migratory and invasive properties of
colorectal cancer cells, potentially through a reduction in the
protein levels of CCR7 (34). In
addition, PITPNM3 is the functional receptor for CCL18, which may
become phosphorylated and subsequently initiate a cascade of
events, including the activation of praline-rich tyrosine kinase
(Pyk2), a crucial component of the focal adhesion complex (FAC)
(19). ACKR3 (CXCR7) is an atypical
chemokine receptor with seven transmembrane domains, belonging to
the AG class of G-protein-coupled receptors (35). Notably, altered ACKR3 expression
levels have also been reported in numerous types of cancers,
including prostate, kidney, liver, cervix, brain, lung cancer and
BC (36,37).
It is noteworthy that, in the present study, we
found that ACKR family members were downregulated in breast cancer
tissues, which suggested that ACKRs may serve a role in tumor
development. It was also demonstrated that ACKRs may serve
important roles in the development of different pathological stages
of breast cancer based on the correlation between the differential
expression of the ACKR gene family and the pathological stages of
patients with breast cancer. These may serve as targets for guiding
the treatment of patients with different stages in future clinical
practice.
To evaluate the link between prognosis and the ACKR
gene families in patients with breast cancer, biological analysis
was performed, which demonstrated that higher expression of certain
ACKRs were correlated with better survival in patients with all
types of breast cancer. For example, the present study demonstrated
that elevated transcriptional levels of ACKR2 were significantly
associated with longer OS times in patients with BC, which
suggested a potential role for ACKR2 as a favorable prognostic
biomarker. However, the expression level of ACKR3 was found to be
significantly lower in BC tumor tissues compared with normal breast
tissues, although in the prognostic analysis, the OS rate of the
ACKR3 high-expression group was worse compared with that of the
low-expression group. In tumor tissues, there are not only tumor
cells, but also mesenchymal tissue and immune cells (38). In breast tumor tissues, ACKR3 was
positively correlated with the levels of immune cells, including
mast cells, eosinophils, neutrophils, natural killer cells,
macrophages and CD8+ T cells, and may have a potential
immunomodulatory role, which could be associated with poor
prognosis for patients with BC. In conclusion, the TME is an area
of research which has attracted an increasing level of interest, as
it has been reported to have an impact on tumor progression and
recurrence (38,39). The presence of immune cells in the
TME can either promote or suppress tumor growth, and therefore
these are an important determinant of clinical outcome and response
to immune therapy (29,40). The present study demonstrated that
the expression of ACKRs was highly correlated with the infiltration
of six types of immune cells in BC, which suggested that ACKRs may
serve as a marker of immune status, in addition to predicting the
disease prognosis of BC. This study may be useful in the field of
breast cancer treatment and may assist in the development of new
immunotherapies.
There were some limitations to the present study.
All the data analyzed in the study were retrieved from online
databases and cell lines, and further studies consisting of
clinical studies and additional cell experiments are required to
validate the findings and to further explore the potential
mechanisms, molecules interactions and clinical applications of
distinct ACKRs in BC.
Although further studies are needed to validate
these results, the present study has provided a rationale for the
discovery of novel targets and prognostic markers for BRCA
therapy.
Acknowledgements
Not applicable.
Funding
This study was funded by Xingtai City Key Research and
Development Plan (grant no. 2021ZC148) and the Scientific Research
Fund of Health Commission of Hebei Province (grant no.
20220224).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY designed the study and wrote the article. SZ and
PP performed the literature searches and data analysis. SZ and PP
confirm the authenticity of all the raw data. All the authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: The landscape of cancer genes and mutational processes
in breast cancer. Nature. 486:400–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saad ED, Squifflet P, Burzykowski T,
Quinaux E, Delaloge S, Mavroudis D, Perez E, Piccart-Gebhart M,
Schneider BP, Slamon D, et al: Disease-free survival as a surrogate
for overall survival in patients with HER2-positive, early breast
cancer in trials of adjuvant trastuzumab for up to 1 year: A
systematic review and meta-analysis. Lancet Oncol. 20:361–370.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nik-Zainal S, Alexandrov LB, Wedge DC, Van
Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J,
Stebbings LA, et al: Mutational processes molding the genomes of 21
breast cancers. Cell. 149:979–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lischka A, Doberstein N, Freitag-Wolf S,
Koçak A, Gemoll T, Heselmeyer-Haddad K, Ried T, Auer G and
Habermann JK: Genome instability profiles predict disease outcome
in a cohort of 4,003 patients with breast cancer. Clin Cancer Res.
26:4606–4615. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aushev VN, Lee E, Zhu J, Gopalakrishnan K,
Li Q, Teitelbaum SL, Wetmur J, Degli Esposti D, Hernandez-Vargas H,
Herceg Z, et al: Novel predictors of breast cancer survival derived
from miRNA activity analysis. Clin Cancer Res. 24:581–591. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shachar SS, Deal AM, Weinberg M, Williams
GR, Nyrop KA, Popuri K, Choi SK and Muss HB: Body composition as a
predictor of toxicity in patients receiving anthracycline and
taxane-based chemotherapy for early-stage breast cancer. Clin
Cancer Res. 23:3537–3543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duraker N, Hot S, Akan A and Nayır PÖ: A
comparison of the clinicopathological features, metastasis sites
and survival outcomes of invasive lobular, invasive ductal and
mixed invasive ductal and lobular breast carcinoma. Eur J Breast
Health. 16:22–31. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lobbezoo DJA, van Kampen RJ, Voogd AC,
Dercksen MW, van den Berkmortel F, Smilde TJ, van de Wouw AJ,
Peters FP, van Riel JM, Peters NA, et al: Prognosis of metastatic
breast cancer subtypes: The hormone receptor/HER2-positive subtype
is associated with the most favorable outcome. Breast Cancer Res
Treat. 141:507–514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang J: Lack of robust prognostic
biomarkers for immunotherapy in breast cancer-adverse events. JAMA
Oncol. 5:1639–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lokeshwar BL, Kallifatidis G and Hoy JJ:
Atypical chemokine receptors in tumor cell growth and metastasis.
Adv Cancer Res. 145:1–27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Griffith JW, Sokol CL and Luster AD:
Chemokines and chemokine receptors: Positioning cells for host
defense and immunity. Annu Rev Immunol. 32:659–702. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li XX, Lee JD, Kemper C and Woodruff TM:
The complement receptor C5aR2: A powerful modulator of innate and
adaptive immunity. J Immunol. 202:3339–3348. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu S, Tang J, Wang C, Liu J, Fu Y and Luo
Y: CXCR7 promotes melanoma tumorigenesis via Src kinase signaling.
Cell Death Dis. 10:1912019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sjöberg E, Meyrath M, Milde L, Herrera M,
Lövrot J, Hägerstrand D, Frings O, Bartish M, Rolny C, Sonnhammer
E, et al: A novel ACKR2-dependent role of fibroblast-derived CXCL14
in epithelial-to-mesenchymal transition and metastasis of breast
cancer. Clin Cancer Res. 25:3702–3717. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Whyte CE, Osman M, Kara EE, Abbott C,
Foeng J, McKenzie DR, Fenix KA, Harata-Lee Y, Foyle KL, Boyle ST,
et al: ACKR4 restrains antitumor immunity by regulating CCL21. J
Exp Med. 217:e201906342020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Del Prete A, Martínez-Muñoz L, Mazzon C,
Toffali L, Sozio F, Za L, Bosisio D, Gazzurelli L, Salvi V, Tiberio
L, et al: The atypical receptor CCRL2 is required for
CXCR2-dependent neutrophil recruitment and tissue damage. Blood.
130:1223–1234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin Z, Li W, Zhang H, Wu W, Peng Y, Zeng
Y, Wan Y, Wang J and Ouyang N: CCL18/PITPNM3 enhances migration,
invasion, and EMT through the NF-κB signaling pathway in
hepatocellular carcinoma. Tumour Biol. 37:3461–3468. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Le Mercier A, Bonnavion R, Yu W, Alnouri
MW, Ramas S, Zhang Y, Jäger Y, Roquid KA, Jeong HW, Sivaraj KK, et
al: GPR182 is an endothelium-specific atypical chemokine receptor
that maintains hematopoietic stem cell homeostasis. Proc Natl Acad
Sci USA. 118:e20215961182021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin W, Li Y, Song Y, Zhang J, Wu C, Chen
Y, Miao Y, Lin C, Lin Y, Yan D, et al: CCRL2 promotes antitumor
T-cell immunity via amplifying TLR4-mediated immunostimulatory
macrophage activation. Proc Natl Acad Sci USA. 118:e20241711182021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bachelerie F, Ben-Baruch A, Burkhardt AM,
Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH,
Locati M, Luster AD, et al: International union of basic and
clinical pharmacology. [corrected]. LXXXIX. Update on the extended
family of chemokine receptors and introducing a new nomenclature
for atypical chemokine receptors. Pharmacol Rev. 66:1–79. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao Y and Yu D: Tumor microenvironment as
a therapeutic target in cancer. Pharmacol Ther. 221:1077532021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wan W, Liu Q, Lionakis MS, Marino AP,
Anderson SA, Swamydas M and Murphy PM: Atypical chemokine receptor
1 deficiency reduces atherogenesis in ApoE-knockout mice.
Cardiovasc Res. 106:478–487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duchene J, Novitzky-Basso I, Thiriot A,
Casanova-Acebes M, Bianchini M, Etheridge SL, Hub E, Nitz K,
Artinger K, Eller K, et al: Atypical chemokine receptor 1 on
nucleated erythroid cells regulates hematopoiesis. Nat Immunol.
18:753–761. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Graham GJ: D6/ACKR2. Front Immunol.
6:2802015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hansell CAH, Fraser AR, Hayes AJ, Pingen
M, Burt CL, Lee KM, Medina-Ruiz L, Brownlie D, Macleod MKL,
Burgoyne P, et al: The atypical chemokine receptor Ackr2 constrains
NK cell migratory activity and promotes metastasis. J Immunol.
201:2510–2519. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Massara M, Bonavita O, Savino B, Caronni
N, Mollica Poeta V, Sironi M, Setten E, Recordati C, Crisafulli L,
Ficara F, et al: ACKR2 in hematopoietic precursors as a checkpoint
of neutrophil release and anti-metastatic activity. Nat Commun.
9:6762018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Behnam Azad B, Lisok A, Chatterjee S,
Poirier JT, Pullambhatla M, Luker GD, Pomper MG and Nimmagadda S:
Targeted imaging of the atypical chemokine receptor 3 (ACKR3/CXCR7)
in human cancer xenografts. J Nucl Med. 57:981–988. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Neves M, Fumagalli A, van den Bor J, Marin
P, Smit MJ and Mayor F: The role of ACKR3 in breast, lung, and
brain cancer. Mol Pharmacol. 96:819–825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Smit MJ, Schlecht-Louf G, Neves M, van den
Bor J, Penela P, Siderius M, Bachelerie F and Mayor F Jr: The
CXCL12/CXCR4/ACKR3 axis in the tumor microenvironment: Signaling,
crosstalk, and therapeutic targeting. Annu Rev Pharmacol Toxicol.
61:541–563. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ngambenjawong C, Gustafson HH and Pun SH:
Progress in tumor-associated macrophage (TAM)-targeted
therapeutics. Adv Drug Deliv Rev. 114:206–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu T and Dai Y: Tumor microenvironment and
therapeutic response. Cancer Lett. 387:61–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vitale I, Manic G, Coussens LM, Kroemer G
and Galluzzi L: Macrophages and metabolism in the tumor
microenvironment. Cell Metab. 30:36–50. 2019. View Article : Google Scholar : PubMed/NCBI
|