Introduction
Liver cancer is one of the tumors with the highest
mortality rate worldwide. According to the latest Cancer
Statistical yearbook, 830,000 individuals succumbed to liver cancer
worldwide in 2020, accounting for 8.3% of all cancer deaths, second
only to lung cancer (1). Likewise,
China is also characterized by a high incidence of liver cancer and
related mortality. Among all the new cases of liver cancer and
liver cancer-related deaths registered worldwide, ~50% of them
occurred in China, accounting for 409,000 and 391,000,
respectively, and ranking second only to lung cancer in China
(2). The high incidence and
mortality of liver cancer pose a serious disease burden on the
country and its population. Tumor resection, local ablation or
liver transplantation can be used for the early treatment of liver
cancer, while there is no effective treatment for advanced liver
cancer due to liver metastasis. Although chemotherapy or
multikinase inhibitors show some positive effects on patient
survival, the prognosis remains poor (3); therefore, it is urgent to deeply
analyze the pathogenesis and treatment of liver cancer (4).
Upstream transcription factor (USF) 1 belongs to the
basic helix-loop-helix leucine zipper family and serves as a
cellular transcription factor (TF). As a TF, USF1 has a
bidirectional regulatory function, being able to regulate gene
expression by activating or suppressing the promoter region of
target genes (5,6). For instance, a previous study by the
authors validated that USF1 binds to the core promoter of APOBEC3G
and increases its transcription level in hepatocytes (7). Furthermore, it has been demonstrated
that the binding of USF1 to the Chitinase 3-Like 1 (Chi3L1)
promoter region can enhance the transcriptional activity of Chi3L1.
However, paradoxically, USF1 reduces the expression of Chi3L1 in
both mouse lung tissue and human lung cancer cells (8). USF (including USF1 and USF2) is a
negative transcriptional suppressor of human telomerase reverse
transcriptase (hTERT) in oral cancer cells, which exerts its
inhibitory effect by directly binding to the E-box site of the
hTERT promoter. Loss of USF's inhibitory effect on hTERT expression
may induce the reactivation of telomerase and the occurrence of
oral cancer (9). The defective
expression of USF1 in gastric cancer could drive p53 degradation
during Helicobacter pylori infection and is associated with
gastric carcinogenesis (10). USF1
is involved in signaling pathways, including NF-κB and inflammatory
signaling (11,12). In addition to regulating the
expression of protein-coding genes, USF1 can also regulate long
non-coding (lnc)RNA and micro (mi)RNA that are involved in cancer
development and other diseases (13,14).
For example, USF1 can directly bind to the promoter region of
lncRNA GAS6-AS2 and overexpress GAS6-AS2, thereby promoting the
progression of osteosarcoma (13).
Single nucleotide polymorphism of rs2516839 in the 5′ untranslated
region of USF1 is significantly associated with an increased risk
of liver cancer (15). LncRNA TUG1
could recruit USF1 protein and enhance its transcriptional function
activity to increase ROMO1 expression and finally promote the
growth and metastasis of Huh7 cells (16). Meanwhile, USF1 promotes the
expression of lncRNA-FASRL by super-enhancer, and the latter could
promote de novo fatty acid biosynthesis to exacerbate
hepatocellular carcinoma (HCC) (17). USF1 expression is increased in
patients with liver cirrhosis, poorly differentiated tissues,
advanced stage and metastatic recurrence, suggesting its potential
as a novel marker for metastatic recurrence in patients with liver
cancer (18).
To investigate the clinical role of USF1 and
elucidate the mechanisms involving genome-wide USF1 binding sites
and downstream gene alterations in cancer, the present study
followed a comprehensive research approach. The study first
examined the correlation between USF1 expression and patient
prognosis within a tissue microarray cohort. This initial analysis
was aimed to determine whether a significant association exists
between USF1 expression levels and clinical outcomes in cancer.
Subsequently, chromatin immunoprecipitation followed by
high-throughput sequencing (ChIP-seq), a cutting-edge technique
enabling the high-resolution, genome-wide identification of
DNA-binding protein sites was employed to precisely pinpoint the
binding sites of the USF1 protein throughout the entire genome. To
further advance the present research, cellular models involving
USF1 overexpression (USF1-OE) were established. Finally,
RNA-sequencing (RNA-seq) was used to obtain transcriptomic data.
This approach allowed a comprehensive investigation of the impact
of USF1 on gene expression and function at the genomic level.
Materials and methods
Cloning and plasmid construction
The pcDNA3.1-USF1 (cat. no. NM_007122) plasmid was
purchased from Youbio Biotech (Changsha, China).
Cell culture and transfections
Huh7 cells, an adult HCC cell line (cat. no.
CL-0120; Procell Life Science & Technology Co., Ltd.), were
cultured at 37°C with 5% CO2 in DMEM (cat. no. PM150210;
Procell Life Science & Technology Co., Ltd.) with 10% FBS (cat.
no. 10091148; Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml
streptomycin and 100 U/ml penicillin (cat. no. SV30010, HyClone;
Cytiva). For USF1-OE, 500 ng empty plasmid or USF1 overexpression
plasmid were transfected into Huh7 cells at 37°C using
Lipofectamine™ 2000 (cat. no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.) for 48 h, according to the
manufacturer's protocol. The plasmid backbone used was pcDNA
3.1-C-FLAG, purchased from Youbio. The transfected cells were
harvested for reverse transcription-quantitative (RT-q) PCR and
western blotting analyses 1 week after transfection.
Western blotting
Huh7 cells were lysed in ice-cold RIPA buffer (cat.
no. PR20001; Wuhan Sanying Biotechnology) supplemented with a
protease inhibitor cocktail (cat. no. 4693116001; Sigma-Aldrich;
Merck KGaA) and incubated on ice for 30 min. Samples were kept for
10 min in boiling water with protein loading buffer (cat. no.
P1040; Beijing Solarbio Science & Technology Co., Ltd.). A
total of 25 µg protein (as determined by the BCA method) was loaded
per lane of a 10% gel and the proteins were separated by SDS-PAGE,
then transferred onto 0.45-mm PVDF membranes (cat. no. ISEQ00010;
MilliporeSigma). The PVDF membranes were then blocked using 5% skim
milk for 1 h at room temperature and incubated overnight at 4°C
with primary antibody against FLAG tag (anti-FLAG; 1:2,000;
antibody produced in rabbit; cat. no. F7425; Sigma-Aldrich; Merck
KGaA) and GAPDH (1:1,000; antibody produced in rabbit; cat. no.
A19056; ABclonal Biotech Co., Ltd.), followed by an incubation with
horseradish peroxidase-conjugated secondary antibody (anti-rabbit;
1:5,000, cat. no. SA00001-2; Wuhan Sanying Biotechnology) or
anti-mouse, 1:5,000; cat. no. AS003; ABclonal Biotech Co., Ltd.)
for 45 min at room temperature. Subsequently, the protein bands on
the membranes were visualized through chemiluminescence using ECL
reagent (cat. no. P0018FM; Beyotime Institute of
Biotechnology).
RNA-seq and data analysis
RNA-seq assays were performed by Wuhan Ruixing
Biotechnology Co., Ltd. (http://www.rxbio.cc). The collected Huh7 cells were
subjected to total RNA extraction using TRIzol® (cat.
no. 15596-018, Ambion; Thermo Fisher Scientific, Inc.). The total
RNA was further purified with two phenol-chloroform treatments and
then treated with RQ1 DNase (cat. no. M6101; Promega Corporation)
to remove DNA. The quality and quantity of the purified RNA were
determined by measuring the ratio of the absorbances measured at
260 and 280 nm (A260/A280=1.9–2.1) using an Ultrafine
spectrophotometer (N50 touch; Implen GmbH). The integrity of RNA
was further verified using 1.5% agarose gel electrophoresis
followed by staining with gel red (cat. no. GR501-01; Vazyme
Biotech Co., Ltd.) for visualization. For each sample, 1 µg of the
total RNA was used for RNA-seq library preparation using the VAHTS
Stranded mRNA-seq Library Prep kit (cat. no. NR605-02, Vazyme
Biotech Co., Ltd.). Polyadenylated mRNAs were purified and
fragmented, and then converted into double-strand cDNA. After end
repair and A tailing, the DNAs were ligated to VAHTS RNA Adapters.
Purified ligation products corresponding to 200–500 bps were
digested with heat-labile uracil-DNA glycosylase and the
single-strand cDNA was amplified, purified, quantified and stored
at −80°C before sequencing. For high-throughput sequencing, the
libraries were prepared following the manufacturer's instructions
and applied to the Illumina Nova6000 system (Illumina, Inc.) for
150 nt paired-end sequencing.
For data analysis, raw reads containing >2-N
bases were first discarded, and then adaptors and low-quality bases
were trimmed from raw sequencing reads using the FASTX Toolkit
(Version 0.0.13; hannonlab.cshl.edu/fastx_toolkit/). The short
reads <16 nt were also dropped. After that, clean reads were
aligned to the human GRCh38 genome using HISAT2 (version 2.2.1)
(19) allowing no more than four
mismatches. Uniquely mapped reads were used for gene reads number
counting and fragments per kilobase of transcript per million
fragments mapped (FPKM) (20). The
R Bioconductor package ‘edgeR’ (21) was utilized to screen out the
differentially expressed genes (DEGs). P<0.01 and fold change
>2 or <0.5 were set as the cut-off criteria for identifying
DEGs.
ChIP-seq and data analysis
ChIP assay was performed by Wuhan Ruixing
Biotechnology Co., Ltd. (http://www.rxbio.cc). A total of ~1×107
cells were crosslinked in 1% formaldehyde for 10 min and the
reaction was quenched with 0.125 M glycine for 5 min at room
temperature. The cross-linked cells were lysed in Lysis buffer (1X
PBS, 0.1% SDS, 0.5% NP-40 and 0.5% sodium deoxycholate) and
sonicated to generate DNA fragments of 200–1,000 bp in length. For
immunoprecipitation, protein-DNA complex was immunoprecipitated by
incubating with ChIP-grade Protein A/G Magnetic Beads (50 µl for IP
and 10 µl for IgG; cat. no. 26162; Invitrogen; Thermo Fisher
Scientific, Inc.) conjugated with anti-FLAG antibody (cat. no.
F7425; Sigma-Aldrich; Merck KGaA), or IgG (cat. no. B900610; Wuhan
Sanying Biotechnology) at 4°C for 2 h. The beads were washed with
LiCl Immune Complex wash buffer (0.5 M LiCl) five times, and TE
buffer [10 mM Tris (pH 8.0), 1 mM EDTA (pH 8.0)] for one time. The
beads were resuspended with 100 µl Elution Buffer (100 mM
NaHCO3 and 1% SDS) and reverse cross-linked by overnight
incubation at 65°C. After sequential RNase A and proteinase K
treatments, DNA fragments were purified through phenol extraction
and ethanol precipitation. The libraries were performed by using
VAHTS Universal DNA Library Prep Kit for Illumina V3 (cat. no.
ND607; Vazyme Biotech Co., Ltd.), according to the manufacturer's
instructions, and PCR products corresponding to 200–500 bps were
enriched, quantified and finally sequenced on Novaseq 6000
sequencer (Illumina, Inc.) with PE150 model. Reads were aligned to
the human GRCh38 genome using bowtie2 (22). Only uniquely mapped reads were used
for the following analysis. To identify the binding sites of USF1,
Model-based Analysis for ChIP-seq (MACS version 1.4; http://github.com/macs3-project/MACS/blob/macs_v1) was
employed (23). The input samples
without immunoprecipitation were used as controls. DeepTools
(Version 3.4.3) (24) was used for
the assignment of genomic features, such as relative location to
the transcription start site (TSS) to the peaks and visualization
of binding profiles. Hypergeometric Optimization of Motif
Enrichment (HOMER) software (http://homer.ucsd.edu/homer/motif/) (25) was used to search the enriched
binding motifs in peaks.
RT-qPCR and ChIP-qPCR
cDNA synthesis was performed using a reverse
transcription kit (cat. no. R323-01; Vazyme Biotech Co., Ltd.) in
the thermocycler T100 (Bio-Rad Laboratories, Inc.) with the
following thermocycling conditions: 42°C for 5 min, 37°C for 15
min, 85°C for 5 sec. ChIP assay was performed according to the
aforementioned method. The qPCR was performed on an ABI QuantStudio
5 (Thermo Fisher Scientific, Inc.) with Hieff™ qPCR
SYBR® Green Master Mix (Low Rox Plus) (cat. no. Q431-02;
Shanghai Yeasen Biotechnology Co., Ltd.) and the following
thermocycler conditions: Denaturing at 95°C for 10 min, 40 cycles
of denaturing at 95°C for 15 sec and annealing and extension at
60°C for 1 min. Each sample was analyzed in three technical
replicates. The concentration of each transcript was then
normalized to the internal reference gene GAPDH and mRNA levels
were quantified using the 2−ΔΔCq method (26). Comparisons were performed with the
paired Student's t-test by using GraphPad Prism software (version
8.0; Dotmatics). The primer sequences for PCR experiments were
provided in Table SI. Comparisons
were performed with the paired Student's t-test and two-way ANOVA
by using GraphPad Prism software (Version 8.0; Dotmatics). The data
analyzed in the present study are available under Gene Expression
Omnibus database series accession no. GSE232263.
Statistical analysis
Expression and prognosis analyses of genes from
patients with liver hepatocellular carcinoma (LIHC) were performed
using Gene Expression Profiling Interactive Analysis 2 (GEPIA2)
online server (27) and
Kaplan-Meier (K-M) Plotter webpage, respectively (28). Principal component analysis (PCA)
was performed with the R package ‘factoextra’ (https://cloud.r-project.org/package=factoextra). The
‘pheatmap’ and ‘ggplot2’ packages (https://cran.r-project.org/web/packages/) in R were
used to generate figures.
Results
USF1 is highly expressed in patients
with liver cancer and is associated with prognosis
To explore the clinical influence of USF1 in liver
cancer, its expression pattern and prognostic effect were
investigated by analyzing data retrieved from The Cancer Genome
Atlas (TCGA) database using GEPIA2 and K-M Plotter software,
respectively. A significant increase in USF1 expression level was
observed in LIHC compared with that in normal samples (Fig. 1A). Expression pattern sorted by
tumor stages showed that the USF1 expression level was high in
stage I–III while it decreased in stage IV (Fig. 1C). Meanwhile, patients with higher
expression of USF1 showed worse prognosis than patients with a
lower level of USF1 (Fig. 1B).
These results indicate that USF1 influences the clinical features
of patients with liver cancer and the underlying mechanisms should
be further investigated.
Construction of Huh7 cell line
overexpressing USF1 and following RNA-seq analysis
To further explore the molecular mechanisms of USF1
in liver cancer cells, Huh7 cells were transiently transfected with
USF1-OE plasmid to construct USF1-OE Huh7 cells. The overexpression
efficiency was confirmed using RT-qPCR and western blotting. The
RT-qPCR results showed that the mRNA levels of USF1 in USF1-OE Huh7
cells were >15 times higher than that in normal Huh7 cells
(Fig. 2A) and the western blotting
results revealed that the protein levels of USF1 were also
increased (Fig. 2B). RNA-seq
experiments were then performed to identify the genes
differentially expressed by USF1-OE in Huh7 cells. After aligning
quality-filtered reads onto the human genome, the expression levels
of all detected genes were obtained. Compared with the control
cells, the FPKM value of USF1 in USF1-OE cells significantly
increased from 18.80 to 678.38 (Fig.
2C), indicating that the experiment was successful. PCA of all
expressed genes demonstrated the clear separation between USF1-OE
and control cells (Fig. 2D). These
results demonstrated that the USF1-OE cell line was successfully
constructed, and it was found that USF1-OE globally regulates the
transcriptome profile of Huh7 cells.
Analyses of DEGs in USF1-OE Huh7
cells
To explore genes that USF1 may regulate, RNA-seq was
used to characterize the changes in gene expression in Huh7 cells
after USF1 OE. The results showed that there were 350 DEGs after
USF1-OE, including 171 upregulated and 179 downregulated genes
(Fig. 3A). From the numbers of
upregulated and downregulated genes, there were no significant
differences in USF1 promoting or restraining gene expression.
However, regarding the types of genes, there were significant
differences. The most upregulated genes were protein-coding genes,
and TFs, while the most downregulated genes were long intergenic
non-coding (linc)RNAs and other types (Fig. 3B), indicating that USF1 could
activate the expression of protein-coding genes. The changes in the
expression level of the top 10 upregulated and downregulated genes
were then shown. USF1 displayed the most significant upregulation,
whereas the lincRNA NEAT1 exhibited the most pronounced
downregulation. Notably, the extent of change in NEAT1 expression
was comparatively less than that observed for USF1 (Fig. 3C).
Subsequently, the enriched functions of upregulated
and downregulated DEGs which were found following USF1-OE were
analyzed. USF1-OE led to the upregulation of several inflammation
and immune-associated pathways, including inflammatory response,
immune response, signal transduction and negative regulation of
cell proliferation (Fig. 3D).
Surprisingly, most of the downregulated genes were only enriched in
one biological process pathway (small molecule metabolic process;
Fig. 3E), which may be attributed
to the less protein-coding genes among downregulated DEGs. Kyoto
Encyclopedia of Genes and Genomes (KEGG) analysis showed that
upregulated genes were also mainly related to signaling pathways,
including the TNF signaling pathway, AGE-RAGE signaling pathway and
Toll-like receptor signaling pathway (Fig. 3F). The downregulated genes were
enriched in ABC transporters, Chagas disease, T cell receptor
signaling pathway and other pathways (Fig. 3G). These results demonstrated that
USF1-OE mainly regulates the expression of
immune/inflammation-associated genes, as well as cell
proliferation-associated genes.
Analyses of the potential gene targets
of USF1
To identify the potential gene targets of USF1,
ChIP-seq was used to characterize all DNA directly bound by USF1
and two replicate libraries were constructed to improve accuracy.
The results demonstrated that most of the binding sites were in the
TSS, intergenic and intron regions (Fig. 4A). The binding profile around the
TSS region demonstrated that the binding peaks of USF1 were highly
enriched in the TSS region (Fig.
4B). A total of 10,891 genes were involved in binding with
USF1, indicating that the USF1 binding to target genes is extensive
(Fig. 4C). The subsequent analysis
focused on the 2,718 genes which were detected in both sets of
experiments (Fig. 4B). There was a
variety of gene types in these 2,718 genes, including protein
coding genes, lincRNA, microRNA, small nucleolar RNA and others,
indicating that the USF1 binding targeted an extensive range of
genes (Fig. 4D). Gene Ontology (GO)
analysis showed that these genes were mainly related to the
biological processes associated with the occurrence and development
of cancer, such as apoptotic process, cell death and mitotic cell
cycle (Fig. 4E). KEGG analysis
showed that these genes were mainly related to signaling pathway
(Sphingolipid and FoxO signaling pathways), cancer and protein
processing (Fig. 4E). Subsequently,
HOMER identified the potential motifs in the target sequences,
revealing that the motif G(C)GTCACGTGA(G), G(A)TCACG(A)TGGT and
G(A)T(A)CACGTG were the top three most frequent sequences
overexpressed among the USF1-binding sites (Fig. 4F). The identified motifs were
canonical cognate E-box regulatory elements of USF proteins
(29).
USF1 binds to the promoter of a few
genes and regulates their expression in Huh7 cells
To identify the genes which were bound and regulated
by USF1, the data of RNA-seq and ChIP-seq were combined for further
analysis. Only 16 genes could be bound by USF1 in 350 DGEs,
including 10 protein-coding genes, three antisense genes, two
lincRNA and one sense intronic gene (Fig. 5A; Table
SII). The expression levels of these overlapped genes were
investigated and it was found that there were only two
downregulated genes, while all the remaining genes were upregulated
(Fig. 5B). The results revealed
that USF1 significantly binds to the promoter region of NEAT1 by
exhibiting the reads density of ChIP-seq data (Fig. 5C). Meanwhile, the expression level
of NEAT1 was significantly decreased by USF1 (Fig. 5D). NEAT1 is a canonical lincRNA and
a novel target for diagnosis and therapy in human tumors (30). The present study further explored
the expression level and prognostic influence of NEAT1 in liver
cancer. Based on the TCGA LIHC RNA-seq data, the tumor samples
exhibited a lower expression level of NEAT1 compared with that in
normal samples (Fig. 5E).
Meanwhile, the overall survival analysis by the K-M method
demonstrated that patients with higher NEAT1 expression levels
showed improved prognostic results compared with those patients
with lower NEAT1 expression level (Fig.
5F). These results were consistent with those of USF1
demonstrated in Fig. 1 after
considering that USF1 negatively regulated NEAT1 expression. In
summary, the aforementioned results indicated that USF1 regulates
the expression of several genes by directly binding to their
promoter region and this regulation may be associated with its
biological functions in liver cancer.
USF1 modulates gene expression by
regulating TFs in Huh7 cells
Very few overlapped genes between USF1-bound genes
and DEGs were detected, which were not significant after
calculation (P=0.97, hypergeometric test). This result indicated
that USF1 may indirectly regulate gene expression in Huh7 cells. To
confirm this hypothesis, TFs were extracted from DEGs. Several
genes among the overlapped genes were TFs, including GBX2, FOSB,
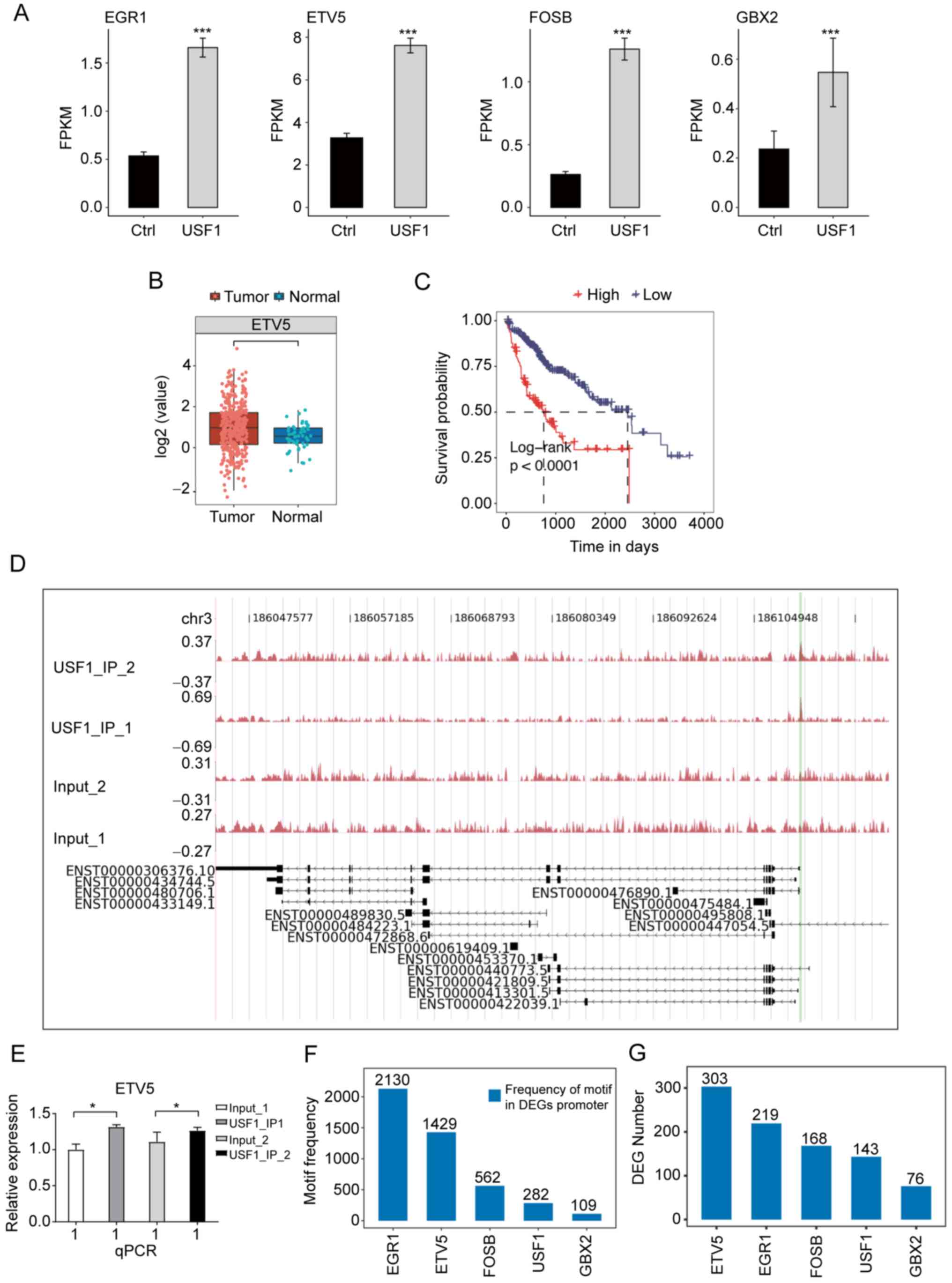
ETV5 and EGR1. Among these TFs, ETV5 had the highest expression
level and a significant expression increase after USF1-OE (Fig. 6A), indicating that ETV5 may have
important roles in Huh7 cells and HCC. The expression level and
prognostic effect of ETV5 in patients with LIHC were investigated
and it was found that the EVT5 expression was significantly
increased in tumor samples (Fig.
6B), while patients with higher ETV5 expression showed worse
prognosis results (Fig. 6C).
ChIP-qPCR experiments validated that USF1 significantly binds to
the promoter region of ETV5 (Fig. 6D
and E). To further explore the underlying mechanisms of the
differentially expressed TFs (DETFs), the occurrence of the motif
sites of these four TFs within the promoter region of DEGs in
USF1-OE Huh7 cells were analyzed to identify which TF had the
highest frequency of motif sites and the number of DEGs. This
analysis was also performed for USF1. The results demonstrated that
EGR1 and ETV5 have the highest frequency of their motifs, which was
markedly higher than that of USF1 (Fig.
6F). Although EGR1 had more motif sites than ETV5, the
expression level of EGR1 in LIHC tumor samples was lower than that
in normal samples (Fig. S1A), and
patients with higher EGR1 expression level showed improved
prognosis compared with that in patients with lower EGR1 expression
level (Fig. S1B), indicating that
USF1 may not promote HCC progression by mediating EGR1 expression.
Furthermore, DEG number analysis of these identified motifs
demonstrated that ETV5 occupies more DEGs than other TFs and USF1
(Fig. 6G), suggesting that ETV5 may
regulate the expression of these DEGs and USF1 may indirectly
affect gene expression by ETV5 in Huh7 cells.
Discussion
USF1 is a canonical TF that affects the expression
of numerous genes by binding to their promoter region or
super-enhancers to control the transcription process. Previous
studies demonstrated the critical roles of USF1 in HCC exacerbation
and identified several downstream targets of USF1 (16,17),
while the global binding profile on DNA and the
downstream-regulated genes of USF1 have not been deeply
investigated in liver cancer. In the present study, whole
transcriptome sequencing and ChIP-seq experiments were performed to
systematically explore how USF1 affects gene expression and the
biological functions of USF1 in HCC Huh7 cells. USF1 globally
enhanced the expression level of immune and inflammatory-associated
genes, while the downregulated genes by USF1-OE were mainly
non-coding. By integrating the ChIP-seq data, it was found that
USF1 may regulate the expression of a few genes by binding to their
promoters, while a larger number of DEGs were not bound by USF1 and
may be regulated by other DETFs, suggesting a novel regulatory
mechanism of USF1 in liver cancer cells.
Consistent with a previous study (17), USF1 was upregulated and associated
with a worse prognosis in patients with LIHC. To explore the
underlying mechanisms, USF1 was overexpressed in Huh7 cells and
DEGs were identified. Interestingly, DEGs upregulated by USF1-OE
were significantly enriched in inflammatory and immune response
pathways. It was also demonstrated that USF1 promotes TNFAIP3/A20
expression, and thus inhibits inflammatory NF-κB pathway activity;
this regulatory axis is a potential anti-inflammatory strategy for
the treatment of several diseases (31). The USF1/A20 regulatory axis was also
validated in mitigating vascular inflammation by NF-κB inactivation
(32). In HCC, the inflammation can
be orchestrated by the tumor itself by secreting factors that
recruit inflammatory cells to the tumor favoring the buildup of a
microenvironment, and inflammation promotes HCC development by
promoting a series of cancer-promoting biological processes
(33). At present, immunotherapies
have shown their advantages in HCC treatment and several other
cancer immunotherapies are also in early-stage clinical trials for
the treatment of advanced HCC (34). Among the upregulated DEGs, several
chemokine and interleukin genes were detected, including CXCL1,
CXCL3, CXCL10, CXCL11, IL1A, IL1B and IL6, several of which were
associated with the progression of HCC. A previous study
demonstrated that in HCC cells, CXCL1 is secreted in response to
metabolic syndrome signals and may promote the progression of HCC
through apoptosis recovery or the metastasis pathway (35). IL6 cooperates with IL6R and triggers
the activation of the JAK-STAT3 signaling pathway, which could
participate in the processes of anti-apoptosis, angiogenesis,
proliferation, invasion, metastasis and drug resistance of cancer
cells (36). In HCC cells, it was
found that CXCL3 and CD133 form a positive feedback loop to
maintain the CD133+ cancer stem cell populations via
Erk1/2 activation, indicating the potential of CXCL13 as a
therapeutic target for HCC (37).
It was hypothesized that USF1 could promote HCC progression by
upregulating the expression of these genes and modulating the
immune microenvironment of tumor. Further studies are necessary to
deeply explore how USF1 regulates the proportion and infiltration
dysregulation, as well as gene expression alteration of immune
cells in HCC.
The global interacting DNA targets of USF1 were then
identified using ChIP-seq data and 2,492 target genes were
detected. The canonical binding motifs of USF1 were identified,
consistently with the previous result in the HepG2 cell line, a
hepatoblastoma cell line, using the ChIP-chip method (5), indicating the high confidence of the
ChIP-seq result. For the overlapped peak genes in the two
replicates, it was found that they were enriched in several types
of cancer and cancer-associated pathways, suggesting that USF1 may
modulate HCC progression by affecting the transcription of
cancer-associated genes. The most enriched GO Biological Process
subontology pathway was the viral process; meanwhile, the
pathogenesis of HCC is tightly associated with hepatitis B and C
viral infections, especially for hepatitis B virus infection
(38). The viral process-associated
genes bound by USF1 indicated that USF1 may modulate the
replication and functions of the hepatitis B or C virus to
accelerate the progression of HCC. Multiple transcription
regulation and RNA splicing pathways were also enriched, implying
that USF1 binds to the promoter regions of TFs and splicing
regulators. A previous study demonstrated that USF1 epigenetically
modulates TF-HoxB4 transcription to control the lineage
differentiation of embryonic stem cells (39). These downstream regulators of USF1
could also regulate their downstream targets, thus forming a
multiple-level regulatory network of USF1; this regulatory network
was reflected in the RNA-seq data of USF1. In addition to these
pathways, autophagy, lysosome and apoptosis KEGG pathways were
enriched by USF1-bound genes. USF1 was reported to suppress
autophagy-related gene expression via positive regulation of mTOR
transcription in HepG2 cells (40).
In multiple cancer cell lines, including HepG2, USF1 could
cooperate with RAD51 to regulate transcription of genes associated
with the autophagy pathway, such as ATG3 and ATG5, by binding to
their promoter regions (41), which
was consistent with the present results. In summary, the present
results suggested that USF1 could broadly bind to the promoters of
various cancer-associated genes in multiple cancer cell lines and
USF1 probably plays important roles in the progression of multiple
liver cancer types, including HCC and hepatoblastoma. As for the
spatial and functional relationship between USFs and H3ac at
protein-coding gene promoters (5),
it was hypothesized that the inhibitors of histone deacetylase may
modulate the functions of USF1 and thus these could be drug
candidates in HCC by targeting USF1.
Lastly, an interaction analysis between DEGs and
bound genes by USF1 was performed. However, only a few genes passed
the criteria of both DEGs and USF1-bound genes, suggesting that
USF1 may also indirectly regulate gene expression. Among the
directly bound DEGs by USF1, several TFs were detected and focused
on ETV5, which acts as an oncoprotein and is implicated in numerous
cancers (42). ETV5 gene fusions
with TMPRSS2 and SLC45A3 were detected in patients with prostate
cancer, and ETV5 overexpression promoted the invasion of RWPE
cells, indicating the biomarker potential of ETV5 in prostate
cancer (43). In hepatocytes, ETV5
regulates hepatic fatty acid metabolism by binding to the PPAR
response element region of downstream genes (44), indicating the critical role of ETV5
in liver diseases. In the present study, a dramatic increase in
ETV5 expression by USF1-OE was detected. Meanwhile, the binding
motif of ETV5 was presented in more DEGs than that of USF1 and
other USF1-bound TFs, suggesting that ETV5 may directly regulate
the expression of DEGs obtained through USF1-OE. These results
could indicate that ETV5 is a target of USF1 and participates in
the USF1-induced progression of HCC. The USF1-ETV5 regulatory axis
may be treated as a target for HCC therapy in future.
There were certain limitations to the present study.
Experiments performed on other HCC cell lines would strengthen the
significance of the present results and conclusion. The experiments
using in vivo animal models or more clinical cases could
also provide more robust results for the important functions of
USF1 in HCC. At the same time, the exact functions and molecular
mechanisms of USF1 downstream targets, including NEAT1 and ETV5,
could be identified using additional experiments in HCC cell lines,
although there are several studies summarizing their functions in
cancers. Thus, deeper studies are necessary to solve the
aforementioned questions in future.
In conclusion, the present deeply investigated the
downstream targets of USF1 and their potential functions associated
with HCC in Huh7 cells. The present results demonstrated that USF1
binds to the promoter region of thousands of genes and
significantly affects the expression of part genes. The downstream
genes, including lncRNA-NEAT1 and TF-ETV5, may have essential
regulatory functions in the USF1-regulated network and be involved
in the progression of HCC. Further studies are also needed to
deeply explore the underlying molecular mechanism and biological
functions of USF1 and identify the potential value of USF1 for
clinical treatment of HCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Henan Young and
Middle-aged Health and Medical Science and Technology Innovation
Outstanding Young Talent Development Program (grant no.
YXKC2020042) and the Beijing iGandan Foundation (grant no.
iGandanF-1082023-RGG003).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets generated and/or
analyzed during the current study are available in the Gene
Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232263).
Authors' contributions
YLZ and FG conceived and designed the study,
conducted experiments, analyzed the data and wrote the manuscript.
PPR, CZ and LM participated in the study design, performed data
analysis and provided critical revisions to the manuscript. XW and
RW contributed to the experimental work, data interpretation and
manuscript revisions. YK and KL provided technical support,
performed data analysis and contributed to the manuscript
preparation. All authors have read and approved the final version
of the manuscript. YLZ and FG confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakagawa H, Fujita M and Fujimoto A:
Genome sequencing analysis of liver cancer for precision medicine.
Semin Cancer Biol. 55:120–127. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rada-Iglesias A, Ameur A, Kapranov P,
Enroth S, Komorowski J, Gingeras TR and Wadelius C: Whole-genome
maps of USF1 and USF2 binding and histone H3 acetylation reveal new
aspects of promoter structure and candidate genes for common human
disorders. Genome Res. 18:380–392. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu TC, Wang Z, Feng X, Chuang P, Fang W,
Chen Y, Neves S, Maayan A, Xiong H, Liu Y, et al: Retinoic acid
utilizes CREB and USF1 in a transcriptional feed-forward loop in
order to stimulate MKP1 expression in human immunodeficiency
virus-infected podocytes. Mol Cell Biol. 28:5785–5794. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng Y, Li H, Zhang X, Shang J and Kang Y:
Basal transcription of APOBEC3G is regulated by USF1 gene in
hepatocyte. Biochem Biophys Res Commun. 470:54–60. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim KC, Yun J, Son DJ, Kim JY, Jung JK,
Choi JS, Kim YR, Song JK, Kim SY, Kang SK, et al: Suppression of
metastasis through inhibition of chitinase 3-like 1 expression by
miR-125a-3p-mediated up-regulation of USF1. Theranostics.
8:4409–4428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang JTC, Yang HT, Wang TCV and Cheng AJ:
Upstream stimulatory factor (USF) as a transcriptional suppressor
of human telomerase reverse transcriptase (hTERT) in oral cancer
cells. Mol Carcinog. 44:183–192. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Costa L, Corre S, Michel V, Le Luel K,
Fernandes J, Ziveri J, Jouvion G, Danckaert A, Mouchet N, Da Silva
Barreira D, et al: USF1 defect drives p53 degradation during
Helicobacter pylori infection and accelerates gastric
carcinogenesis. Gut. 69:1582–1591. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song X, Zhu M, Li H, Liu B, Yan Z, Wang W,
Li H, Sun J and Li S: USF1 promotes the development of knee
osteoarthritis by activating the NF-κB signaling pathway. Exp Ther
Med. 16:3518–3524. 2018.PubMed/NCBI
|
|
12
|
Zhang L, Handel MV, Schartner JM, Hagar A,
Allen G, Curet M and Badie B: Regulation of IL-10 expression by
upstream stimulating factor (USF-1) in glioma-associated microglia.
J Neuroimmunol. 184:188–197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei G, Zhang T, Li Z, Yu N, Xue X, Zhou D,
Chen Y, Zhang L, Yao X and Ji G: USF1-mediated upregulation of
lncRNA GAS6-AS2 facilitates osteosarcoma progression through
miR-934/BCAT1 axis. Aging (Albany NY). 12:6172–6190. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun Q, Li J, Li F, Li H, Bei S, Zhang X
and Feng L: LncRNA LOXL1-AS1 facilitates the tumorigenesis and
stemness of gastric carcinoma via regulation of miR-708-5p/USF1
pathway. Cell Prolif. 52:e126872019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X and Wang T, Liu B, Wu Z, Yu S and
Wang T: Significant association between upstream transcription
factor 1 rs2516839 polymorphism and hepatocellular carcinoma risk:
A case-control study. Tumour Biol. 36:2551–2558. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu S, Qiu J, He W, Geng C, He G, Liu C,
Cai D, Liu X, Tian B and Pan H: TUG1 long non-coding RNA enlists
the USF1 transcription factor to overexpress ROMO1 leading to
hepatocellular carcinoma growth and metastasis. MedComm (2020).
1:386–399. 2020.PubMed/NCBI
|
|
17
|
Peng JY, Cai DK, Zeng RL, Zhang CY, Li GC,
Chen SF, Yuan XQ and Peng L: Upregulation of superenhancer-driven
LncRNA FASRL by USF1 promotes de novo fatty acid biosynthesis to
exacerbate hepatocellular carcinoma. Adv Sci (Weinh).
10:e22047112022.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen B, Chen XP, Wu MS, Cui W and Zhong M:
Expressions of heparanase and upstream stimulatory factor in
hepatocellular carcinoma. Eur J Med Res. 19:452014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ramírez F, Ryan DP, Grüning B, Bhardwaj V,
Kilpert F, Richter AS, Heyne S, Dündar F and Manke T: deepTools2: A
next generation web server for deep-sequencing data analysis.
Nucleic Acids Res. 44((W1)): W160–W165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heinz S, Benner C, Spann N, Bertolino E,
Lin YC, Laslo P, Cheng JX, Murre C, Singh H and Glass CK: Simple
combinations of lineage-determining transcription factors prime
cis-regulatory elements required for macrophage and B cell
identities. Mol Cell. 38:576–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagy Á, Munkácsy G and Győrffy B:
Pancancer survival analysis of cancer hallmark genes. Sci Rep.
11:60472021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Corre S and Galibert MD: Upstream
stimulating factors: Highly versatile stress-responsive
transcription factors. Pigment Cell Res. 18:337–348. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dong P, Xiong Y, Yue J, Hanley SJB,
Kobayashi N, Todo Y and Watari H: Long non-coding RNA NEAT1: A
novel target for diagnosis and therapy in human tumors. Front
Genet. 9:4712018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tiruppathi C, Soni D, Wang DM, Xue J,
Singh V, Thippegowda PB, Cheppudira BP, Mishra RK, Debroy A, Qian
Z, et al: The transcription factor DREAM represses the
deubiquitinase A20 and mediates inflammation. Nat Immunol.
15:239–247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cho MJ, Lee DG, Lee JW, Hwang B, Yoon SJ,
Lee SJ, Park YJ, Park SH, Lee HG, Kim YH, et al: Endothelial PTP4A1
mitigates vascular inflammation via USF1/A20 axis-mediated NF-κB
inactivation. Cardiovasc Res. 119:1265–1278. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu LX, Ling Y and Wang HY: Role of
nonresolving inflammation in hepatocellular carcinoma development
and progression. NPJ Precis Oncol. 2:62018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu JKH, Irvine AF, Jones RL and Samson A:
Immunotherapies for hepatocellular carcinoma. Cancer Med.
11:571–591. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dahlquist KJV, Voth LC, Fee AJ and
Stoeckman AK: An autocrine role for CXCL1 in progression of
hepatocellular carcinoma. Anticancer Res. 40:6075–6081. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu J, Lin H, Wu G, Zhu M and Li M:
IL-6/STAT3 is a promising therapeutic target for hepatocellular
carcinoma. Front Oncol. 11:7609712021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Zhang L, Li H, Ge C, Zhao F, Tian
H, Chen T, Jiang G, Xie H, Cui Y, et al: CXCL3 contributes to
CD133(+) CSCs maintenance and forms a positive feedback regulation
loop with CD133 in HCC via Erk1/2 phosphorylation. Sci Rep.
6:274262016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ringelhan M, McKeating JA and Protzer U:
Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol
Sci. 372:201602742017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng C, Li Y, Liang S, Cui K, Salz T, Yang
H, Tang Z, Gallagher PG, Qiu Y, Roeder R, et al: USF1 and hSET1A
mediated epigenetic modifications regulate lineage differentiation
and HoxB4 transcription. PLoS Genet. 9:e10035242013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo J, Fang W, Chen X, Lin Y, Hu G, Wei J,
Zhang X, Yang C and Li J: Upstream stimulating factor 1 suppresses
autophagy and hepatic lipid droplet catabolism by activating mTOR.
FEBS Lett. 592:2725–2738. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kang K, Choi Y, Moon H, You C, Seo M, Kwon
G, Yun J, Beck B and Kang K: Epigenomic analysis of RAD51 ChIP-seq
data reveals cis-regulatory elements associated with autophagy in
cancer cell lines. Cancers (Basel). 13:25472021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oh S, Shin S and Janknecht R: ETV1, 4 and
5: An oncogenic subfamily of ETS transcription factors. Biochim
Biophys Acta. 1826:1–12. 2012.PubMed/NCBI
|
|
43
|
Helgeson BE, Tomlins SA, Shah N, Laxman B,
Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, et
al: Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions
in prostate cancer. Cancer Res. 68:73–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mao Z, Feng M, Li Z, Zhou M, Xu L, Pan K,
Wang S, Su W and Zhang W: ETV5 regulates hepatic fatty acid
metabolism through PPAR signaling pathway. Diabetes. 70:214–226.
2021. View Article : Google Scholar : PubMed/NCBI
|