Introduction
Hepatocellular carcinoma (HCC), the most common form
of liver cancer, is one of the most frequently diagnosed malignant
neoplasms worldwide (1,2). In recent decades, HCC has received a
significant amount of attention due to the rapid rise in
HCC-associated mortality (1,3).
Despite advances in diagnostic techniques and therapeutic
strategies over the last decade, the prognosis of patients with HCC
has remained poor (4,5). The majority of HCC patients are
diagnosed in the first instance with advanced-stage HCC; therefore,
the estimated 5-year overall survival rate is <20% (6). Previous studies have shown that the
prognosis of HCC is associated with several factors, including the
heterogeneity of HCC (7),
diagnostic challenges (8), limited
radiation therapy, and the development of resistance to HCC. Thus,
additional research into the molecular mechanisms driving the
malignant transformation of hepatocytes is urgently needed.
Aldehyde ketone reductase (AKR) family member B10
(AKR1B10) belongs to the AKR1B subgroup (9). AKR1B10 is a cytosolic NADPH-dependent
reductase, catabolizing various endogenous compounds by catalyzing
the corresponding redox reactions (2). According to recent studies, expression
of this enzyme is upregulated in several types of cancer, including
adrenocortical (10), breast
(11), colon (12), and hepatocellular carcinoma
(13,14). AKR1B10 has significant potential as
a relevant biomarker and therapeutic target for predicting tumor
progression. Based on a large-scale proteome quantification
analysis, HCC tissues showed dysregulated AKR1B10 expression
compared with normal liver (15,16).
However, the specific function of AKR1B10 in HCC pathology and its
molecular mechanisms remain to be determined.
The present study examined the diagnostic
performance of AKR1B10 in HCC tumor tissues. Furthermore, the role
and mechanism of AKR1B10 in the proliferation, migration, invasion,
and epithelial-mesenchymal transition (EMT) of HCC cells was
examined.
Materials and methods
Public data acquisition
The genomics data of GSE146719/GPL20795 was
downloaded from the Gene Expression Omnibus (GEO) database and
analyzed using the DESeq2 package (version 1.39.8; http://www.ncbi.nlm.nih.gov/geo/). Normalized
RNA-seq data (HTSeq-FPKM) and survival data of the Pan-Cancer Atlas
(PANCAN) and the liver hepatocellular carcinoma (LIHC) project of
The Cancer Genome Atlas (TCGA) were accessed from the GDC website
(https://portal.gdc.cancer.gov/) on March
1, 2023, and the data set is referred to as GDC-TCGA-LIHC in this
article. Normalized gene expression (normalized_read_count) and
donor information of the liver cancer project (named: LIRI-JP) of
the International Cancer Genome Consortium (ICGC) were downloaded
from the ICGC data portal (https://dcc.icgc.org/), and the data set is referred
to in this article as ICGC-LIRI. Immunohistochemical images of
AKR1B10 in tumor samples were analyzed and accessed from The Human
Protein Atlas database website (https://www.proteinatlas.org/). The gene effect and
expression of HCC cell lines were analyzed and downloaded from the
Broad Institute DepMap website (https://depmap.org/).
Public data processing
Normalized quantification of gene expression in
GDC-TCGA-LIHC was converted into transcripts per million (TPM) and
then logarithmically transformed [log2(TPM+1)]. In
ICGC-LIRI, the expression values were directly log2
(normalized_read_count) transformed since they were all >0
(17). No normal samples were
included in the prognosis analysis in GDC-TCGA-LIHC and ICGC-LIRI.
Missing values were considered in the GDC-TCGA dataset if the
clinical information of patients was unavailable or unknown.
Cells and agents
Human HCC cell lines Huh7 and Hep3B were obtained
from the Cell Bank of the Chinese Academy of Sciences and cultured
in DMEM (Thermo Fisher Scientific, Inc.), supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), and 1% penicillin/streptomycin
(HyClone, Cytiva; cat. no. SV30010) at 37°C with 5% CO2.
HCC cells were authenticated via short tandem repeat analysis.
Mycoplasma-free cells were used in all experiments.
Cells were seeded into six-well plates
(3–5×105 cells/well). Sh/oe-AKR1B10 in HCC cells was
performed using the lentivirus (DNA:16 µg/100 ml; Shanghai
GenePharma Co., Ltd.), when the cell confluency reached 60–70%. The
cells were treated with 100 µl/ml (1×109 TU/ml)
lentivirus for 8 h. Stably transduced cell lines were selected with
4 µg/ml puromycin (YEASEN, China). Full-length AKR1B10 sequences
were ligated into the lentivirus vectors pCMV-3×HA. The shRNA
sequences (5′-3′) were as follows: shAKR1B10,
CGCTCCTACTGACTCCTATTT, and Scramble, CCTAAGGTTAAGTCGCCCTCG. The
reagents and antibodies used are given in Table I.
 | Table I.Reagents and antibodies used in the
present study. |
Table I.
Reagents and antibodies used in the
present study.
| Reagent or
antibody | Source | Cat. no. |
|---|
| AKR1B10 | Abcam | ab192865 |
| EMT antibodies
kit | Cell Signaling
Technology, Inc. | 9782T |
| PI3K | Cell Signaling
Technology, Inc. | 84249T |
| p-PI3K | Cell Signaling
Technology, Inc. | 4228S |
| AKT | ProteinTech Group,
Inc. | 60203-2-lg |
| p-AKT | ProteinTech Group,
Inc. | 66444-1-lg |
| p-eIF4EBP1 | Abcam | ab259329 |
| p-RPS6 | Abcam | ab80158 |
| GAPDH | Abcam | ab181602 |
| GDC-0941 | Calbiochem (Merck
KGaA) | 957054-30-7 |
Cell proliferation and colony
formation assays
HCC cell proliferation was assessed using CCK-8
assays (Beyotime Institute of Biotechnology). Briefly,
3–5×103 stable cells transfected with sh/oeAKR1B10 or
corresponding scramble oligos were plated into 96-well plates and
incubated for 4 days.
The cells were seeded in 12-well plates
(1×104 cells/well) for 2 weeks to determine colony
formation. The methanol-fixed colonies were then stained with
crystal violet solution at room temperature.
Wound healing assay
Transfected Huh7 cells were seeded into six-well
plates (1×105 cells/well). With a 10 µl pipette tip,
wounds were made when the cell confluency reached 80–90%, and the
cells were cultured in serum-free DMEM at 37°C. The wounds were
observed under an inverted light microscope (×100). ImageJ v1.8.0
(National Institutes of Health) was used to evaluate the cell
migration rate as follows: Wound closure surface area/wound total
surface area ×100%.
Western blot and analysis
Using RIPA buffer and PMSF (Beyotime Institute of
Biotechnology) with phosphatase inhibitor (Beijing Solarbio Science
& Technology Co., Ltd.), total cell proteins were extracted and
analyzed by western blotting. Briefly, proteins (40 µg) were loaded
onto 10% SDS-PAGE for electrophoresis and transferred to
polyvinylidene difluoride membranes (MilliporeSigma). Membranes
were blocked with 5% BSA at room temperature for 1 h and then
incubated with primary antibodies overnight at 4°C and then
secondary antibodies at 37°C for 1 h, according to the
manufacturer's recommendations. Finally, the protein levels were
confirmed using enhanced chemiluminescence (Tanon 4800; Tanon
Science and Technology Co., Ltd.) and normalized with an anti-GAPDH
antibody. The images were analyzed using ImageJ (version 1.8.0.345,
National Institutes of Health).
Transwell migration and invasion
assays
Typically, 1×105 transfected Huh7/Hep3B
cells in serum-free media were seeded into the upper chambers with
(invasion) or without (migration) Matrigel coating (diluted in
DMEM; Corning, Inc.), and the bottom chambers were filled with 600
µl supplemented DMEM medium. After 36 h, the cells that had
migrated and invaded through to the bottom of the inserts were
fixed with methanol and stained with crystal violet for 30 min at
room temperature, respectively. In five random fields of view, the
numbers of cells that had migrated/invaded were viewed under an
inverted light microscope (magnification, ×200) and imaged and
quantified.
Flow cytometry analysis
For cell cycle analysis, cells were fixed with 70%
ethanol (4°C, >30 min), washed with PBS (5 min, 3 times), and
then stained with RNase/PI staining solution (room temperature, 30
min). Flow cytometry (Beckman Coulter, Inc.) was used to measure
the DNA content (>10,000 cells per sample), and ModFIT LT v3.1
software (Verity Software House, Inc.) was used for analysis.
Statistical analysis
GraphPad Prism version 8.0 (GraphPad Software, Inc.)
was used for statistical analysis. ImageJ was used for densitometry
analysis. Comparisons between groups were assessed using a
Student's t-test or an ANOVA followed by a post-hoc Tukey's test
for multi-group comparison. P<0.05 was considered to indicate a
statistically significant difference.
Results
AKR1B10 was screened out and was found
to be upregulated in HCC
To evaluate the biological implications of the
differential expression of genes in HCC vs. normal liver tissues,
expression analysis in the GSE146719 dataset was performed.
Moreover, 1,109 genes with q-value (p.adj) <0.05 were selected
for subsequent processing (Fig.
1A). Deep analysis of these genes was performed using a volcano
plot and mean differential plot maps (Fig. 1B). The top 10 genes from the
above-analyzed data were selected for repeated analysis.
Subsequently, the AKR1B10 was screened out. AKR1B10 status in HCC
by exploring its expression in this disease. Higher AKR1B10
expression in pan-cancerous tissues suggested that this oncogene
may be upregulated during the development of HCC. As shown in
Fig. 1E and F, AKR1B10 expression
in HCC tissues was significantly higher than in normal liver
tissues. Combined with the public IHC datasets from The Human
Protein Atlas database, the expression and localization of AKR1B10
were detected. AKR1B10 was primarily expressed in the cytoplasm
(Fig. 1G), consistent with its
function as an enzyme catalyzing redox reactions. HCC patients were
divided into two groups based on the median value of AKR1B10
expression in GDC-TCGA-PANCAN and GDC-TCGA-LIHC datasets, and
patients labeled as low-AKR1B10 had significantly longer survival
than the remaining high-AKR1B10 (Fig.
1H and I; P<0.0001 and P=0.0021, respectively). Similarly,
patients classified with the low-AKR1B10 group had a significantly
better prognosis in the ICGC-LIRI dataset (Fig. 1J; P=0.0071). Thus, high expression
of AKR1B10 in HCC patients was associated with poorer outcomes, and
AKR1B10 was shown to be a useful biomarker for the disease
(13,16).
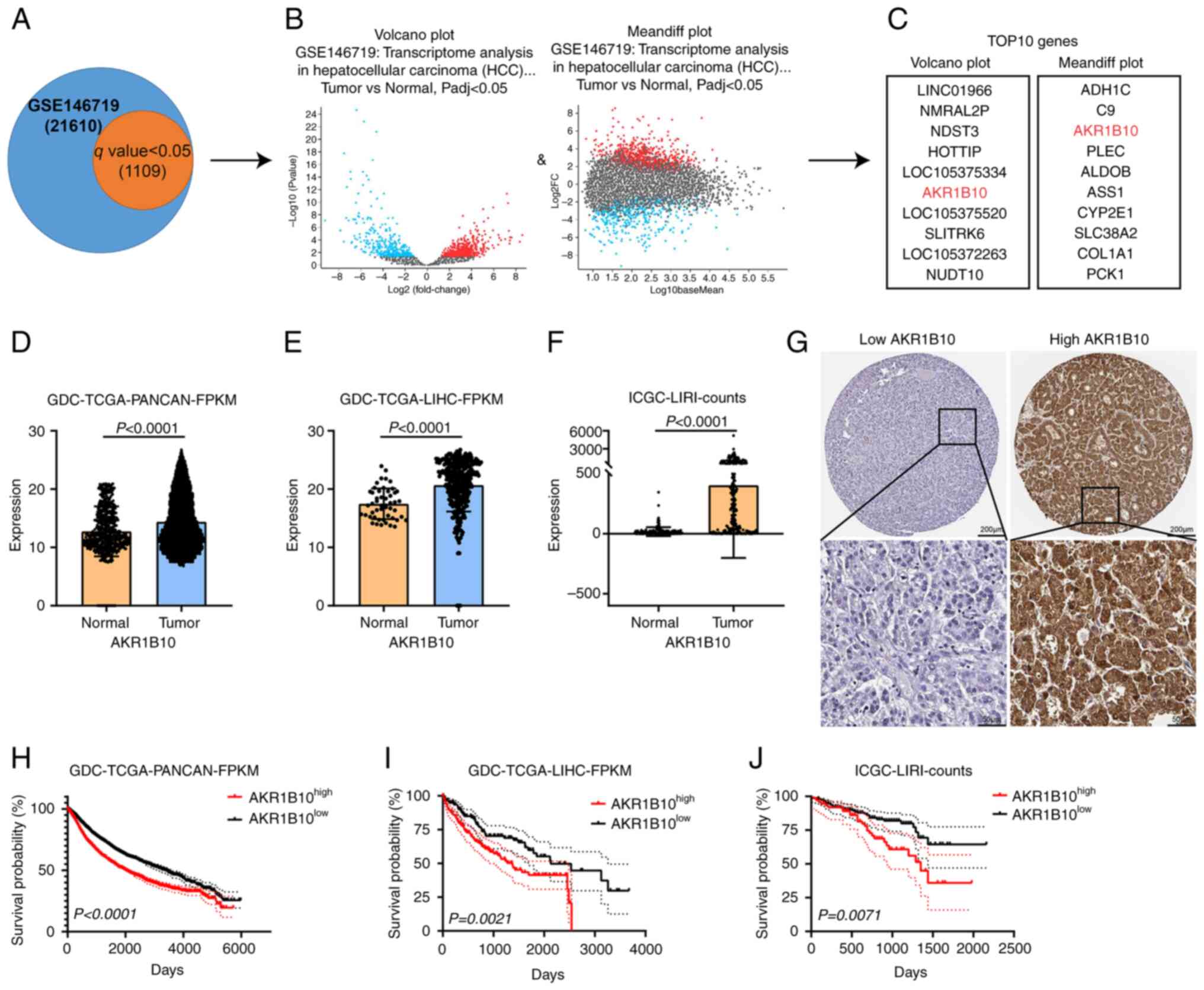 | Figure 1.AKR1B10 expression was screened out
and found to be upregulated in hepatocellular carcinoma. (A-C) Flow
chart of AKR1B10 filtering from the GSE146719 dataset (D-F)
Amplification of AKR1B10 was common in the GDC-TCGA and ICGC
provisional cohort. (G) Representative images of
immunohistochemical staining for low/high expression of AKR1B10 in
public tissue microarray datasets from The Human Protein Atlas
database (upper image: magnification, ×100, scale bar, 200 µm;
lower image: magnification, ×400; scale bar, 50 µm). (H-J)
Prognostic significance of AKR1B10 in (H) GDC-TCGA-PANCAN, (I)
GDC-TCGA-LIHC, and (J) ICGC-LIRI cohorts. AKR1B10, aldo-keto
reductase family 1 member B10; GDC-TCGA, Genomic Data Commons-The
Cancer Genome Atlas; ICGC, International Cancer Genome Consortium;
PANCAN, Pan-Cancer Atlas; LIHC, liver hepatocellular carcinoma;
LIRI, donor information of the liver cancer project; FPKM,
Fragments Per Kilobase Million. |
DepMap analysis of AKR1B10
Due to the differential expression of AKR1B10 and
its prognostic value in HCC, its potential functions in this
disease were examined. Using the DepMap dataset, a gene effect
CRISPR (DepMap Public 22Q4 + Score, Chronos) analysis of this gene
was performed. The results showed a significant gene effect of
AKR1B10 in most HCC cell lines (Fig.
2A), especially in Huh 7 cell line (Fig. 2B). Next, the expression of AKR1B10
in HCC cell lines was analyzed using the DepMap dataset. The
results showed that AKR1B10 was expressed at differentially high
levels in all HCC cell lines, including Huh7 (Fig. 2C and D). The above results indicated
that AKR1B10 was highly expressed in HCC tissues and showed a
significant gene effect in HCC and Huh7 cells, and thus these cells
were selected as an ideal HCC cell line for further experimental
validation.
AKR1B10 promotes the proliferation,
migration, and invasion of HCC cells
After transfection of sh-AKR1B10 into Huh7 cells,
AKR1B10 expression was significantly decreased (Fig. 3A). AKR1B10 knockdown significantly
reduced Huh7 cell viability and proliferation in CCK-8 and colony
formation assays (Fig. 3B and C).
Further studies illustrated that AKR1B10 promoted the migration and
invasion of Huh7 cells in wound healing and Transwell assays
(Fig. 3D-F). In addition, the above
effects of AKR1B10 overexpression on Hep3B, a relatively
low-expressing AKR1B10 HCC cell line, were assessed. The results
showed that cellular activity was significantly promoted in Hep3B
cells (Fig. 3G-I). The above
results suggested that AKR1B10 played a prominent role in the
proliferation, migration, and invasion of HCC.
 | Figure 3.AKR1B10 promoted the proliferation,
migration, and invasion in HCC cells. (A) AKR1B10 expression was
knocked down in Huh7 cells and confirmed by western blotting. (B-F)
AKR1B10 knockdown inhibits the proliferation, migration, and
invasion of Huh7 cells. (G) AKR1B10 expression was overexpressed in
Hep3B cells and confirmed by western blotting. (H-I) AKR1B10
overexpression promoted the proliferation, migration, and invasion
of Hep3B cells. Cell proliferation was evaluated using a CCK-8
assay (B and H) and colony formation assays (C). Cell migration and
invasion were investigated using Transwell assays (D, E, and I).
**P<0.01 and ****P<0.0001. AKR1B10, aldo-keto reductase
family 1 member B10; sh, short hairpin; oe, overexpression; NC,
negative control; OD, optical density. |
GDC-TCGA and DepMap analysis of
co-expressed genes of AKR1B10, CCND1, and EMT-related genes
Since AKR1B10 was upregulated in HCC tissues and
exhibited a notable influence on HCC cell behavior; therefore, its
potential mechanisms in promoting this disease were assessed. Next,
the association between AKR1B10 expression and the cell
proliferation-related gene CCND1 (18,19)
and EMT-related hub genes in GDC-TCGA tumor samples. The
EMT-related hub genes were selected as E-cadherin, N-cadherin,
Vimentin, and TWIST1. The results showed that all of the above
genes were significantly associated with the expression of AKR1B10
(P<0.0001; Fig. 4A-E). Next,
another dataset, DepMap, was selected for a similar correlation
analysis. The expression of E-cadherin (P=0.0217), N-cadherin
(P=0.0245), and Vimentin (P=0.0073) genes in DepMap showed varying
degrees of significant correlations with AKR1B10, apart from TWIST1
(P=0.7343; P-value cut-off=0.05; Fig.
4F-J).
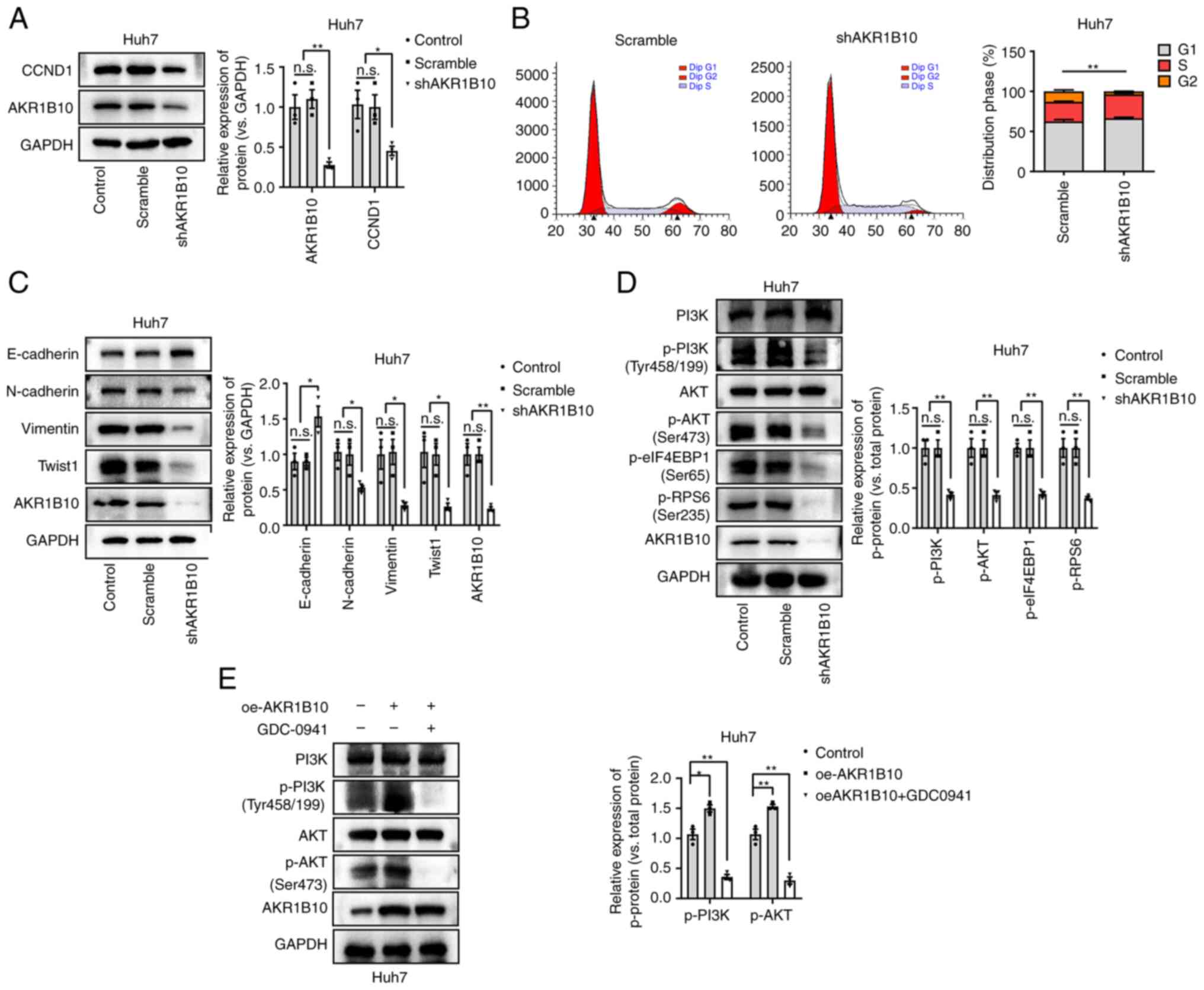
AKR1B10 promotes the expression of
CCND1 and EMT-related genes and activates the PI3K/AKT pathway
To further validate the above functions of AKR1B10,
we measured the effects of AKR1B10 on the gene expression of CCND1
and EMT-related genes in HCC cells. AKR1B10 knockdown suppressed
CCND1 expression and abrogated the cell cycle distribution in HCC
cells (Fig. 5A and B). In addition,
the expression of E-cadherin was increased, and other EMT
biomarkers, including N-cadherin, Vimentin, and TWIST-1, were
downregulated after AKR1B10 knockdown (Fig. 5C). Based on these results, it is
possible that AKR1B10 plays a significant role in the proliferation
and EMT of HCC, affecting its proliferation, migration, and
invasion.
As reported recently, AKR1B10 promotes tumor cell
proliferation and migration through PI3K/AKT signaling (11). Furthermore, the degree of
phosphorylation of proteins involved in the PI3K/AKT pathway was
assessed, and it was found that phosphorylation was significantly
reduced by AKR1B10 knockdown (Fig.
5D). Meanwhile, although cells transfected with AKR1B10 were
subsequently treated with GDC-0941 (100 nM; PI3K/AKT pathway
antagonist; MedChemExpress) for 24 h, western blotting showed that
the protein content of AKR1B10 failed to be significantly altered
(Fig. 5E). Based on these
experiments, it was suggested that the PI3K/AKT signaling was
involved in the pathological progression of HCC.
Discussion
HCC is one of the most common cancers in the world
and the third leading cause of cancer-associated death (20). Due to delayed diagnosis and frequent
cancer metastasis, the majority of HCC patients are diagnosed with
advanced-stage cancer at the initial diagnosis (21). With the recent developments in
biological therapies, the clinical treatment of HCC has seen
progress; however, the survival benefits of these drug treatments
for HCC patients still have several limitations. In particular,
with immune checkpoint inhibitors as monotherapy for HCC patients,
the outcomes to date have been disappointing (22). Furthermore, HCC is a heterogeneous
disease with limited chemoradiotherapy treatment (7). The 5-year survival rate for patients
with early-stage HCC has been reported to exceed 50% (2). Despite this, HCC has a poor 5-year
overall survival rate (6). The
limited value of existing biomarkers for diagnosing and treating
HCC requires urgent identification of better biomarkers. Therefore,
there is a crucial need to understand the molecular mechanisms
underlying HCC development to identify better biomarkers and
develop novel therapeutics.
As a member of the AKR1B family (9), the role of AKR1B10 in lung cancer
(23,24), Renal Cell Carcinoma (25), ovarian cancer (26) and bladder cancer (27) carcinogenesis has been studied
extensively. AKR1B10 is a cytosolic NADPH-dependent reductase,
catabolizing various endogenous compounds by catalyzing the
corresponding redox reactions (2).
Research has shown that AKR1B10 contributes to cancer progression
(11). Recent studies found that
the overexpression of the enzyme in several types of cancer,
including adrenocortical (10),
breast (11), colon (12), and HCC (13,14).
Subsequently, this study investigated its role in HCC. Through a
comprehensive analysis of data from GDC-TCGA-PANCAN, GDC-TCGA-LIHC,
and ICGC-LIRI databases, it was found that AKR1B10 expression was
significantly upregulated pan-cancer and in HCC, and high
expression was associated with shorter survival times (Fig. 2D-J), and confirmed that AKR1B10 was
a potential biomarker for prognostic prediction of HCC. Studies
have shown that cancer cells proliferate and migrate when AKR1B10
is knocked down (28,29). Additionally, studies have shown that
AKR1B10 expression is elevated in breast cancer and adrenocortical
carcinoma tissues, where it is involved in proliferation,
migration, and invasion (10,11).
It should be noted that AKR1B10 may exhibit differing functions in
different types of cells. Using the DepMap dataset, the gene effect
CRISPR analysis, and the expression analysis of AKR1B10 in HCC cell
lines, it was found that Huh7 cells were a potential excellent
candidate for HCC cells. It was found in the present study that by
knocking down AKR1B10 expression, Huh7 cells were less likely to
proliferate, migrate, invade, and undergo EMT.
Different cellular processes rely on PI3K/AKT
signaling, including cell proliferation, apoptosis, and migration
(30,31). The PI3K/AKT pathway is reported to
be regulated in the sustained activation of EMT in several types of
cancer (32,33). PI3K/AKT-mediated EMT has been shown
to promote HCC progression (34).
Recently, there have been reports that AKR1B10 affects the activity
of the PI3K pathway by regulating the substrate synthesis of PI3K
in cancer cells (35,36). Previous studies demonstrated that
AKR1B10-overexpressing breast cancer cells activated PI3K/AKT
(11), thus, the association
between AKR1B10 and PI3K/AKT signaling was assessed. Studies have
reported various ways by which AKR1B10 promotes HCC proliferation,
migration, and invasion (21,37).
Additionally, AKRB10 may play a significant role in metabolic
adaptation (2). While AKRB10 has
been studied in HCC, the results were inconsistent. For example,
AKR1B10 may play different roles depending on the HCC stage
(9). Together, AKR1B10 may
influence the progression of HCC through multiple signaling
pathways associated with cell proliferation and migration. Finally,
in the present study, it was found that AKR1B10 promoted HCC
progression in vitro by upregulating the activity of the
PI3K/AKT signaling pathway. Nevertheless, future studies are
required using in vivo tumor models to verify the validity
of the findings of the clinical tissue samples.
In conclusion, the results of the present study
showed that AKR1B10 activated the PI3K/AKT pathway in Huh7 cells,
and this resulted in increased proliferation, invasion, migration,
and EMT.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
KT, ZL and HZ performed the experiments. YD
collected the samples. ZL and HZ analyzed the data. HY conceived
the study experiments and wrote the manuscript. KT and HY confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ladd AD, Duarte S, Sahin I and Zarrinpar
A: Mechanisms of drug resistance in HCC. Hepatology. Jan
3–2023.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Z, Pei Y, Li W, Zhang J and Liu J:
Clinical value of AKR1B10 in hepatocellular carcinoma: A systematic
review and meta-analysis. PLoS One. 17:e02795912022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu M, Chen Z, Zhou Q, Zhang B, Huang J,
Jin L, Zhou B, Liu S, Yan J, Li X, et al: PARG inhibition limits
HCC progression and potentiates the efficacy of immune checkpoint
therapy. J Hepatol. 77:140–151. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Erstad DJ and Tanabe KK: Hepatocellular
carcinoma: Early-stage management challenges. J Hepatocell
Carcinoma. 4:81–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang MY, Hung TW, Wang CJ and Tseng TH:
Inhibitory Effect of Nelumbo nucifera Leaf extract on
2-acetylaminofluorene-induced hepatocarcinogenesis through
enhancing antioxidative potential and alleviating inflammation in
rats. Antioxidants (Basel). 8:3292019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang H, Tsui YM and Ng IO: Fueling HCC
dynamics: Interplay between tumor microenvironment and tumor
initiating cells. Cell Mol Gastroenterol Hepatol. 15:1105–1116.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cicinnati VR, Sotiropoulos GC and
Beckebaum S: Established and emerging therapies for hepatocellular
carcinoma. Minerva Med. 101:405–418. 2010.PubMed/NCBI
|
|
9
|
DiStefano JK and Davis B: Diagnostic and
prognostic potential of AKR1B10 in human hepatocellular carcinoma.
Cancers (Basel). 11:4862019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen D, Huang R, Ren F, Wang H, Wang C and
Zhang Y: FNDC5 and AKR1B10 inhibit the proliferation and metastasis
of adrenocortical carcinoma cells by regulating AMPK/mTOR pathway.
Exp Ther Med. 25:1362023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu J, Li J, Zhang Y, He R, Liu X, Gong K,
Duan L, Luo W, Hu Z, Wang G, et al: AKR1B10 promotes breast cancer
cell proliferation and migration via the PI3K/AKT/NF-κB signaling
pathway. Cell Biosci. 11:1632021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu C, Shi L, Li W, Huang Z, Wang S, Xu P,
Li T, Li Z, Luo F, Li W, et al: AKR1B10 accelerates the production
of proinflammatory cytokines via the NF-κB signaling pathway in
colon cancer. J Mol Histol. 53:781–791. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye X, Li C, Zu X, Lin M, Liu Q, Liu J, Xu
G, Chen Z, Xu Y, Liu L, et al: A large-scale multicenter study
validates aldo-keto reductase family 1 member B10 as a prevalent
serum marker for detection of hepatocellular carcinoma. Hepatology.
69:2489–2501. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie C, Ye X, Zeng L, Zeng X and Cao D:
Serum AKR1B10 as an indicator of unfavorable survival of
hepatocellular carcinoma. J Gastroenterol. 58:1030–1042. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu B, Wang F, Song C, Sun Z, Cheng K, Tan
Y, Wang H and Zou H: Large-scale proteome quantification of
hepatocellular carcinoma tissues by a three-dimensional liquid
chromatography strategy integrated with sample preparation. J
Proteome Res. 13:3645–3654. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim SY, Shen Q, Son K, Kim HS, Yang HD, Na
MJ, Shin E, Yu S, Kang K, You JS, et al: SMARCA4 oncogenic
potential via IRAK1 enhancer to activate Gankyrin and AKR1B10 in
liver cancer. Oncogene. 40:4652–4662. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang J, Zhang X, Li J and Song Z:
Systematic analysis of the ABC transporter family in hepatocellular
carcinoma reveals the importance of ABCB6 in regulating
ferroptosis. Life Sci. 257:1181312020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Z, Xu T, Xie T, Yang L, Wang G, Gao
Y, Xi G and Zhang X: CDC42EP3 promotes glioma progression via
regulation of CCND1. Cell Death Dis. 13:2902022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leontieva OV, Demidenko ZN and
Blagosklonny MV: MEK drives cyclin D1 hyperelevation during
geroconversion. Cell Death Differ. 20:1241–1249. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang T, Guan G, Zhang J, Zheng H, Li D,
Wang W, Lu F and Chen X: E2F1-mediated AUF1 upregulation promotes
HCC development and enhances drug resistance via stabilization of
AKR1B10. Cancer Sci. 113:1154–1167. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Romero D: Combination set to transform HCC
therapy. Nat Rev Clin Oncol. 17:3892020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim B, Lee HJ, Choi HY, Shin Y, Nam S, Seo
G, Son DS, Jo J, Kim J, Lee J, et al: Clinical validity of the lung
cancer biomarkers identified by bioinformatics analysis of public
expression data. Oncotarget. 67:7431–7438. 2007.
|
|
24
|
Zhu D, Nie Y, Zhao Y, Chen X, Yang Z and
Yang Y: RNF152 suppresses fatty acid oxidation and metastasis of
lung adenocarcinoma by inhibiting IRAK1-Mediated AKR1B10
expression. Am J Pathol. 193:1603–1617. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng L, Zhang X, Pan X, Huang Z, Zhang M,
Xian J, Wei Y, Nie L, Zhang M, Gong J, et al: AKR1B10 is a new
sensitive and specific marker for fumarate hydratase-deficient
renal cell carcinoma. Mod Pathol. 36:1003032023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hojnik M, Šuster NK, Smrkolj Š, Sisinger
D, Grazio SF, Verdenik I and Rižner TL: AKR1B1 as a prognostic
biomarker of high-grade serous ovarian cancer. Cancers (Basel).
14:8092022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang Z, Yan Y, Zhu Z, Liu J, He X,
Dalangood S, Li M, Tan M, Cai J, Tang P, et al: CBX7 suppresses
urinary bladder cancer progression via modulating AKR1B10-ERK
signaling. Cell Death Dis. 12:5372021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao X, Wu J, Yu S, Zhou Y and Zhou C:
AKR1B10 inhibits the proliferation and migration of gastric cancer
via regulating epithelial-mesenchymal transition. Aging (Albany
NY). 13:22298–22314. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yao Y, Wang X, Zhou D, Li H, Qian H, Zhang
J, Jiang L, Wang B, Lin Q and Zhu X: Loss of AKR1B10 promotes
colorectal cancer cells proliferation and migration via regulating
FGF1-dependent pathway. Aging (Albany NY). 12:13059–13075. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He Y, Sun MM, Zhang GG, Yang J, Chen KS,
Xu WW and Li B: Targeting PI3K/Akt signal transduction for cancer
therapy. Signal Transduct Target Ther. 6:4252021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Nie J, Ma X, Wei Y, Peng Y and Wei
X: Targeting PI3K in cancer: Mechanisms and advances in clinical
trials. Mol Cancer. 18:262019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang C, Yang Z, Xu E, Shen X, Wang X, Li
Z, Yu H, Chen K, Hu Q, Xia X, et al: Apolipoprotein C-II induces
EMT to promote gastric cancer peritoneal metastasis via
PI3K/AKT/mTOR pathway. Clin Transl Med. 11:e5222021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Li Y, Wang L, Chen B, Zhu M, Ma C,
Mu C, Tao A, Li S, Luo L, et al: Cinnamaldehyde suppressed
EGF-Induced EMT process and inhibits ovarian cancer progression
through PI3K/AKT pathway. Front Pharmacol. 13:7796082022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dian MJ, Li J, Zhang XL, Li ZJ, Zhou Y,
Zhou W, Zhong QL, Pang WQ, Lin XL, Liu T, et al: MST4 negatively
regulates the EMT, invasion and metastasis of HCC cells by
inactivating PI3K/AKT/Snail1 axis. J Cancer. 12:4463–4477. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang C, Cao Z, Ma J, Shen Y, Bu Y,
Khoshaba R, Shi G, Huang D, Liao DF, Ji H, et al: AKR1B10 activates
diacylglycerol (DAG) second messenger in breast cancer cells. Mol
Carcinog. 57:1300–1310. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Manning BD and Toker A: AKT/PKB signaling:
Navigating the network. Cell. 169:381–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu T, Ke Y, Tang H, Liao C, Li J and Wang
L: Fidarestat induces glycolysis of NK cells through decreasing
AKR1B10 expression to inhibit hepatocellular carcinoma. Mol Ther
Oncolytics. 23:420–431. 2021. View Article : Google Scholar : PubMed/NCBI
|