Introduction
Primary liver cancer (PLC) is one of the most common
types of malignant tumor worldwide. PLC incidence and mortality
rates rank sixth and third among malignant tumors globally,
respectively (1). There were
906,000 new cases and 830,000 deaths globally from PLC in 2020,
which highlights the worldwide healthcare burden caused by this
condition (2). The most common
histological type of PLC is hepatocellular carcinoma (HCC). The
typical clinical manifestations of HCC include pain in the liver
region, liver enlargement, ascites, etc. However, HCC has no
obvious symptoms in its early stages and is often diagnosed in the
mid to late stages of disease (3).
Typically, at the time of diagnosis, liver function and immune
system have been damaged, so the prognosis is poor (4,5).
Therefore, the search for potential targets to predict the
prognosis of patients with liver cancer is important to improve the
prognosis of these patients. Small nucleolar RNA (snoRNA) is a type
of small molecule RNA primarily expressed in the nucleolus with a
length of 60–300 nucleotides (6).
The role of snoRNA in the post-transcriptional modification and
maturation of ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs)
as well as other cellular RNAs has previously been reported
(7). Due to the need for new
ribosomes for cell growth, it is reasonable that tumor cells would
exploit the mechanisms involved in ribosome biogenesis to support
their fast growth (8). Recent
studies reported that, in addition to participating in the normal
biological functions of cells, snoRNA also serves an important role
in the occurrence and progression of multiple cancers, including
breast cancer (9), colorectal
cancer (10), and pancreatic ductal
adenocarcinoma (11). Previous
studies have reported that abnormal methylation of snoRNA H/ACA Box
51 (SNORA51) gene is associated with an increased risk of
lymphatic-hematopoietic cancer, including acute myelogenous
leukemia (12,13). However, further research is required
to elucidate the role of SNORA51 in HCC. Therefore, the present
study aimed to investigate expression of SNORA51 in HCC and its
potential clinical significance.
Materials and methods
Patients and specimens
Liver cancer and paired adjacent liver (≥1 cm from
the edge of the tumor) specimens were obtained from 136 patients
(78 male and 58 female) with HCC (age, 35–84 years) who underwent
hepatectomy at The First Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China) from January 1, 2016 to December 31,
2018. The inclusion criteria were as follows: Diagnosed with HCC by
clinical evidence, image finding, and histologic examination; No
serious heart, lung, kidney and coagulation dysfunction; With
written informed consent for the study. The exclusion criteria were
as follows: Metastatic tumors of the liver; radiotherapy,
chemotherapy or other treatment before surgery; Declined to
participate in this study.
Using the median expression of SNORA51 as the
cut-off point, 136 patients were divided into high (n=68) and low
expression (n=68) groups. Other clinical features measured included
age, sex, cirrhosis, portal vein tumor thrombus, vascular invasion,
tumor diameter, TNM stage, hepatitis B surface antigen (HBsAg) and
α-fetoprotein (AFP) levels and overall patient survival. The final
follow-up was December 31, 2021. The present study was approved by
the Xi'an Jiaotong University Health Science Center Ethics
Committee (Xi'an, China; approval no. XJTU-RE-2021644) and all
patients provided written, informed consent for participation in
the study.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from tissue using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and
the purity and concentration of RNA were determined using a
spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was
obtained using the HiScript® II Reverse Transcriptase
SuperMix (Vazyme Biotech Co., Ltd.) according to the manufacturer's
instructions. The expression levels of SNORA51 in tissue samples
were evaluated using FastStart Universal SYBR Green Master (Rox;
Roche Diagnostics) according to the manufacturer's instructions.
The thermocycling conditions were as follows: Initial denaturation
at 95°C for 1 min, followed by 40 cycles of 90°C for 20 sec, 55°C
for 30 sec and 68°C for 50 sec using the Bio-Rad CFX96 RT-qPCR
system (Bio-Rad Laboratories, Inc.). Amplification specificity was
assessed by melting curve analysis. U6 was used as an internal
control for standardization. All assays were performed three times
and data were analyzed using the comparative 2−ΔΔCq
method (14). The primer sequences
were as follows: SNORA51 forward (F), 5′-GCCTCCTGGTGCTTACCACA-3′
and reverse (R), 5′-GGGCCTGAGCTGAGGTGTAT-3′ and U6 F,
5′-CTCGCTTCGGCAGCACA-3′ and R, 5′-AACGCTTCACGAATTTGCGT-3′.
Database analysis
Data from patients with HCC from The Cancer Genome
Atlas Program (TCGA) (15) database
were analyzed using starBase 3.0 (starbase.sysu.edu.cn/) and The
University of Alabama at Birmingham Cancer Data Analysis Portal
(UALCAN; ualcan.path.uab.edu/index.html). A total of 362 patients
with HCC were divided into high and low SNORA51 expression groups
(both n=181) using the median of SNORA51 expression as the cut-off
point. Spearman's correlation coefficient between SNORA51
expression and survival time of patients with HCC was analyzed
using starBase. The methylation status of the SNORA51 promoter
region in HCC (n=377) and adjacent tissues (n=50) (also from TCGA)
was analyzed using UALCAN. The inclusion criteria were as follows:
i) Name of the disease listed as HCC and ii) available data on
expression levels of SNORA51.
Statistical analysis
SPSS software (version 16.0; SPSS, Inc.) was used
for statistical analysis. Data are presented as the mean ± standard
deviation (SD). A two-tail paired student's t test was used to
compare the means of the two groups of patients and χ2
test was applied to analyze categorical data. Cox regression model
was used for univariate and multivariate analysis. Overall survival
curves were plotted using the Kaplan-Meier method and log-rank test
was utilized for examining the differences in survival rates
between groups. All experiments were repeated three times
independently. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of SNORA51 are
significantly increased in HCC tissues
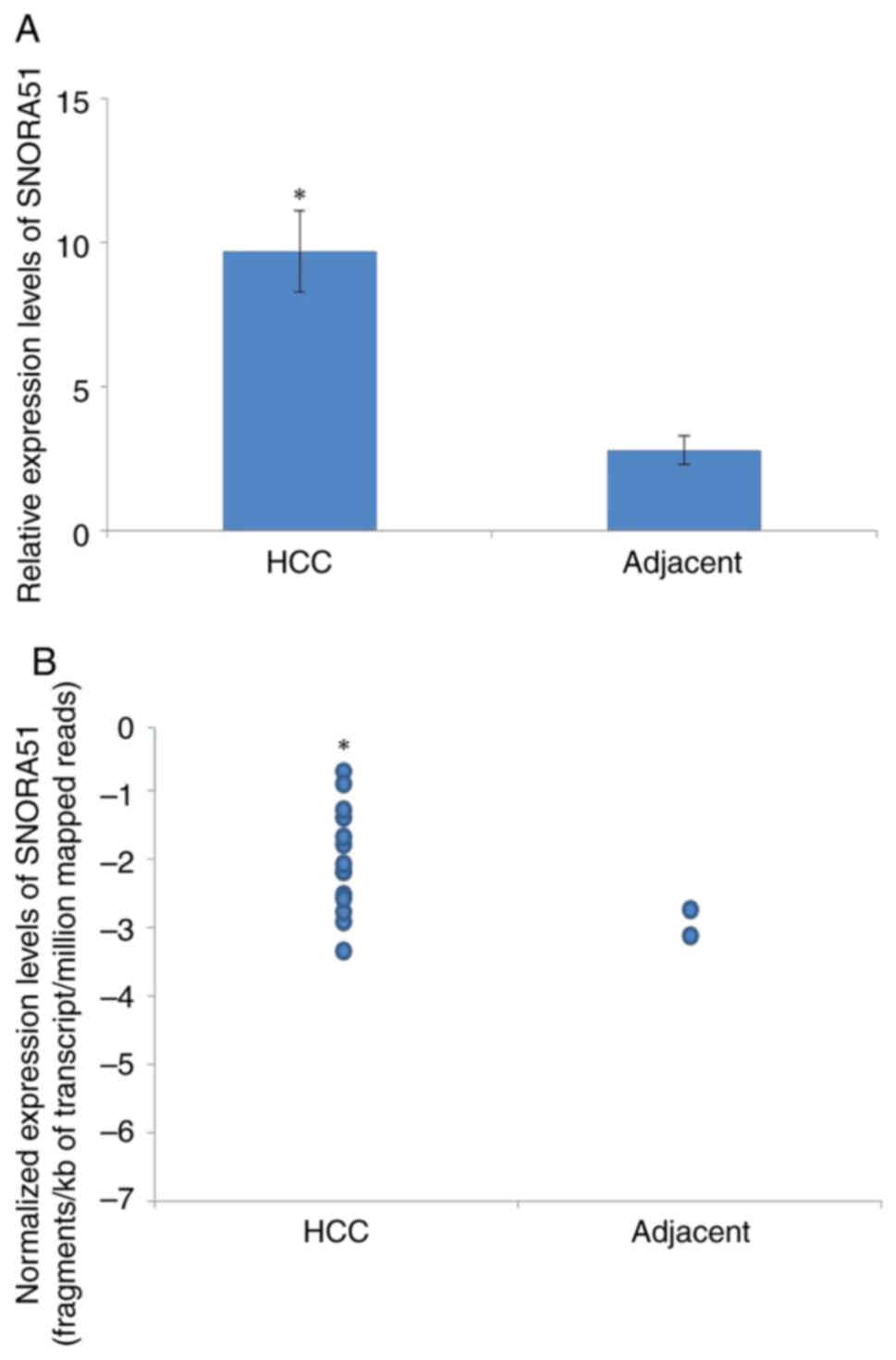
The results of the RT-qPCR demonstrated that the
expression levels of SNORA51 in HCC were significantly higher
compared with those in adjacent tissue (Fig. 1A). Bioinformatics analysis of data
from patients with HCC using starBase demonstrated an increased
expression of SNORA51 in cancer compared with adjacent tissues
(Fig. 1B).
High expression of SNORA51 was
associated with portal vein tumor thrombus, vascular invasion and
tumor stage
The association between SNORA51 expression levels
and clinicopathological characteristics of patients with HCC was
analyzed. These results demonstrated that high expression of
SNORA51 was significantly associated with portal vein tumor
thrombus, vascular invasion and tumor stage, but not with age, sex,
tumor diameter, HBsAg and AFP levels or cirrhosis (Table I).
 | Table I.Association between SNORA51 expression
and clinicopathological features of patients with hepatocellular
carcinoma. |
Table I.
Association between SNORA51 expression
and clinicopathological features of patients with hepatocellular
carcinoma.
|
|
| SNORA51
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
feature | Total number of
patients | High | Low |
χ2-value | P-value |
|---|
| Age, years |
|
|
| 0.472 | 0.606 |
|
>60 | 72 | 38 | 34 |
|
|
| ≤60 | 64 | 30 | 34 |
|
|
| Sex |
|
|
| 1.924 | 0.225 |
| Male | 78 | 43 | 35 |
|
|
|
Female | 58 | 25 | 33 |
|
|
| Cirrhosis |
|
|
| 1.666 | 0.268 |
|
Present | 43 | 18 | 25 |
|
|
|
Absent | 93 | 50 | 43 |
|
|
| Portal vein tumor
thrombus |
|
|
| 9.598 | 0.003 |
|
Present | 45 | 31 | 14 |
|
|
|
Absent | 91 | 37 | 54 |
|
|
| Vascular
invasion |
|
|
| 15.308 | <0.001 |
|
Present | 86 | 54 | 32 |
|
|
|
Absent | 50 | 14 | 36 |
|
|
| Tumor diameter,
cm |
|
|
| 0.491 | 0.599 |
| ≤3 | 54 | 29 | 25 |
|
|
|
>3 | 82 | 39 | 43 |
|
|
| TNM stage |
|
|
| 15.562 | <0.001 |
|
I–II | 67 | 22 | 45 |
|
|
|
III–IV | 69 | 46 | 23 |
|
|
| Hepatitis B surface
antigen |
|
|
| 1.074 | 0.388 |
|
Positive | 76 | 35 | 41 |
|
|
|
Negative | 60 | 33 | 27 |
|
|
| α-fetoprotein level
(ng/ml) |
|
|
| 0.319 | 0.779 |
|
≤20 | 14 | 6 | 8 |
|
|
|
>20 | 122 | 62 | 60 |
|
|
High expression of SNORA51 is
associated with a worse prognosis of HCC patients
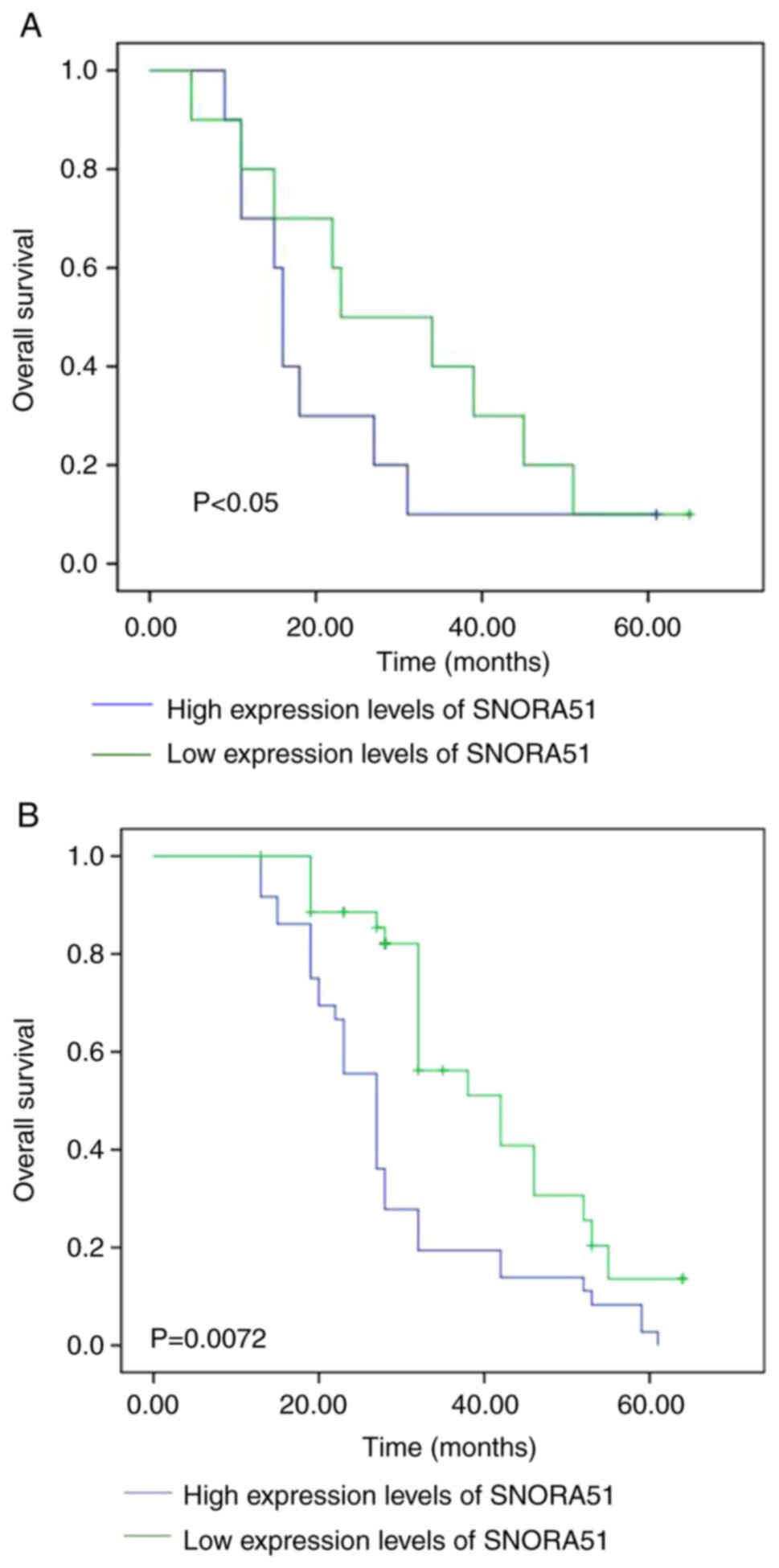
The relationship between SNORA51 expression and
overall survival of patients with HCC was assessed using
Kaplan-Meier curves. The median survival time of patients with HCC
in the high SNORA51 expression group [16 months (95% CI,
12.475–18.135 months)] was significantly shorter compared with that
in the low SNORA51 expression group [36 months (95% CI,
26.196–45.348 months)], which suggested that high expression of
SNORA51 was negatively associated with prognosis of patients with
HCC (Fig. 2A). Analysis of samples
from TCGA using starBase demonstrated that high expression levels
of SNORA51 were associated with poorer prognosis of patients with
HCC compared with those with low expression of SNORA51 (Fig. 2B).
Analysis of factors affecting the
prognosis of patients with HCC
Univariate and multivariate Cox regression analysis
demonstrated that high SNORA51 expression levels, portal vein tumor
thrombus, tumor diameter and TNM stage were independent risk
factors in predicting overall survival of patients with HCC
(Table II).
 | Table II.Uni- and multivariate Cox regression
analysis of prognostic factors in patients with hepatocellular
carcinoma. |
Table II.
Uni- and multivariate Cox regression
analysis of prognostic factors in patients with hepatocellular
carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≤60 vs. >60
years) | 0.852
(0.583–1.224) | 0.154 |
|
|
| Sex (female vs.
male) | 0.947
(0.687–2.564) | 0.322 |
|
|
| Cirrhosis (present
vs. absent) | 1.142
(0.864–2.953) | 0.188 |
|
|
| Portal vein tumor
thrombus (present vs. absent) | 2.417
(1.346–4.023) | 0.007 | 2.002
(1.159–3.873) | 0.013 |
| Vascular invasion
(present vs. absent) | 1.352
(0.808–2.193) | 0.224 |
|
|
| Tumor diameter
(>3 vs. ≤3 cm) | 2.295
(1.611–3.618) | <0.001 | 1.956
(1.447–3.126) | 0.009 |
| TNM stage (III–IV
vs. I–II) | 2.515
(1.366–4.465) | <0.001 | 2.214
(1.482–3.996) | 0.024 |
| Hepatitis B surface
antigen (negative vs. positive) | 0.986
(0.698–1.422) | 0.312 |
|
|
| α-fetoprotein (≤20
vs. >20 ng/ml) | 1.755
(0.842–4.422) | 0.256 |
|
|
| Small nucleolar RNA
H/ACA Box 51 expression (high vs. low) | 3.087
(1.690–5.214) | 0.001 | 2.856
(1.354–4.463) | 0.003 |
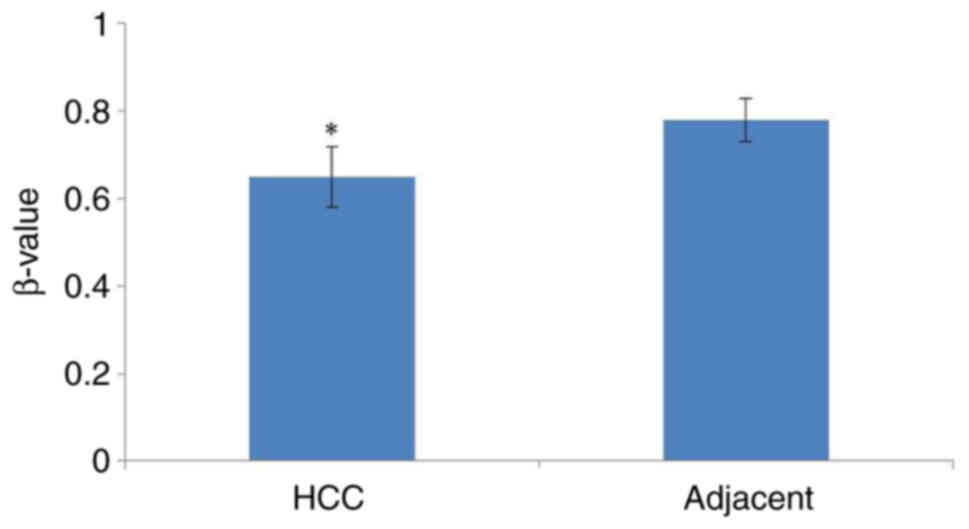
Methylation of SNORA51 promoter is
reduced in HCC tissue
Analysis using UALCAN demonstrated that the
methylation of the SNORA51 promoter region in HCC tissue was
significantly reduced compared with that in adjacent tissue
(Fig. 3).
Discussion
HCC is the most common type of malignant tumor of
the liver, accounting for >90% of PLC cases (16). The occurrence and development of HCC
is a complex process involving multiple factors and genes and the
mechanisms have not yet been fully elucidated. The early detection,
diagnosis and treatment of patients with HCC are vital to increase
the survival rate (17). Currently,
the clinical treatment plan for patients with HCC and prognostic
evaluation largely rely on tumor staging and there is a lack of
specific molecular targets associated with treatment and prognosis
of HCC (18).
snoRNAs are non-coding RNAs present in the nucleoli
of eukaryotic cells. They are primarily divided into C/D box
snoRNAs and H/ACA box snoRNAs. Both types of snoRNA combine with
ribonucleoproteins to form stable and functional snoRNP complexes
that participate in post-transcriptional maturation process of rRNA
and other types of small RNA (19).
Due to the unitary function of snoRNAs, their association with
tumors is unclear. However, snoRNAs participate in the occurrence
and development of tumors by regulating cell proliferation,
differentiation and apoptosis (20,21).
Yi et al (22) reported that
SNORA42 increases viability of prostate cancer cells and promotes
cancer cell migration and epithelial-mesenchymal transformation and
high expression of SNORA42 is significantly associated with poor
prognosis of patients with prostate cancer. Okugawa et al
(23) reported that overexpression
of SNORA42 enhances proliferation, migration and invasion of colon
cancer cells and high expression of SNORA42 is an independent risk
factor affecting the survival of patients with colorectal cancer.
In addition, snoRNA U50 is downregulated in breast cancer and
overexpression of U50 inhibits colony formation of breast cancer
cell lines (24). The specific
expression of snoRNAs in tumor tissue suggests that these molecules
may be a biomarker for tumor diagnosis and a potential therapeutic
target. The expression of snoRNAs in tumor tissue is also closely
related to the prognosis of patients, therefore, snoRNAs could be a
potential biomarker for tumor prognosis (25).
A previous study reported that SNORA51 may be
involved in progression of multiple myeloma (26). Bortezomib treatment and incubation
with vitamin D and K have inhibitory effects on expression of
SNORA51 in U266 myeloma cells and decrease tumor cell
proliferation. Liang et al (27) reported that SNORA51 is highly
expressed in breast cancer patients without non-sentinel lymph node
invasion. Another clinical study reported that SNORD51 and SNORD57,
in addition to small RNA fragments originating from their 3′ ends,
are detected in the plasma of patients with colorectal cancer,
which suggested that these snoRNAs may act as potential biomarkers
for the diagnosis of colorectal cancer (28). However, the role and mechanism of
SNORA51 in HCC is still unclear.
Systematic treatment regimens have made significant
progress in the treatment of advanced HCC. Nowadays, patients with
stage III–IV advanced HCC receive systemic treatment rather than
surgery (29); however, at time of
patient recruitment, a number of patients with advanced HCC still
received aggressive surgical treatment (30). In this study, liver cancer and
paired adjacent liver specimens obtained from 136 HCC patients
(including 69 III–IV advanced HCC) underwent hepatectomy were
compared. The present study demonstrated that expression of SNORA51
in HCC was significantly higher compared with that in adjacent
tissue; data from patients with HCC using starBase demonstrated the
same results. These results were consistent with the aforementioned
studies. The present study also demonstrated that expression of
SNORA51 was related to portal vein tumor thrombus, vascular
invasion and TNM staging, which indicated that SNORA51 may be
involved in tumor invasion and metastasis. The prognostic analysis
demonstrated that the median survival time of patients with HCC
with high SNORA51 expression was significantly shorter compared
with that of patients with low SNORA51 expression; data from
patients with HCC using starBase demonstrated the same results.
Uni- and multivariate Cox regression analysis demonstrated that
high SNORA51 expression was an independent risk factor affecting
the prognosis of patients with HCC, which indicated that SNORA51
could be a biomarker for the prognosis of HCC and potentially be a
future therapeutic target. UALCAN analysis demonstrated that
methylation of the SNORA51 promoter region in HCC was significantly
reduced compared with adjacent tissues, which indicated that
upregulation of SNORA51 in HCC may be related to decreased
methylation of its promoter region. This may serve as a future
research direction for further study of the mechanism of SNORA51 in
HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Research and
Development Program of Shaanxi Province (grant no. 2020SF-060).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY, MZ and ZM analyzed and interpreted the data. ZM
performed RT-qPCR and database analysis. LY wrote the manuscript.
SW designed the experiments and reviewed the manuscript. LY and SW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Xi'an Jiaotong
University Health Science Center Ethics Committee (Xi'an,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chakraborty E and Sarkar D: Emerging
therapies for hepatocellular carcinoma (HCC). Cancers (Basel).
14:27982022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Teo EK and Fock KM: Hepatocellular
carcinoma: An Asian perspective. Dig Dis. 19:263–268. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Orcutt ST and Anaya DA: Liver resection
and surgical strategies for management of primary liver cancer.
Cancer Control. 25:10732748177446212018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kufel J and Grzechnik P: Small nucleolar
RNAs tell a different tale. Trends Genet. 35:104–117. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Z, Yu H, Yao W, Zhu N, Miao R, Liu
Z, Song X, Xue C, Cai C, Cheng M, et al: RRP9 promotes gemcitabine
resistance in pancreatic cancer via activating AKT signaling
pathway. Cell Commun Signal. 20:1882022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pecoraro A, Pagano M, Russo G and Russo A:
Ribosome biogenesis and cancer: Overview on ribosomal proteins. Int
J Mol Sci. 22:54962021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gee HE, Buffa FM, Camps C, Ramachandran A,
Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al: The
small-nucleolar RNAs commonly used for microRNA normalisation
correlate with tumour pathology and prognosis. Br J Cancer.
104:1168–1177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang X, Li Y, Li L, Liu J, Wu M and Ye M:
SnoRNAs are involved in the progression of ulcerative colitis and
colorectal cancer. Dig Liver Dis. 49:545–551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui L, Nakano K, Obchoei S, Setoguchi K,
Matsumoto M, Yamamoto T, Obika S, Shimada K and Hiraoka N: Small
nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic
ductal adenocarcinoma, regulates expression of spectrin
repeat-containing nuclear envelope 2 to promote growth and
metastasis of xenograft tumors in mice. Gastroenterology.
153:292–306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Domingo-Relloso A, Huan T, Haack K,
Riffo-Campos AL, Levy D, Fallin MD, Terry MB, Zhang Y, Rhoades DA,
Herreros-Martinez M, et al: DNA methylation and cancer incidence:
Lymphatic-hematopoietic versus solid cancers in the Strong Heart
Study. Clin Epigenetics. 13:432021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng J, Zhang T, Guo W, Zhou C, Cui X,
Gao L, Cai C and Xu Y: Integrative analysis of Multi-Omics
identified the prognostic biomarkers in acute myelogenous leukemia.
Front Oncol. 10:5919372020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Jensen MA and Zenklusen JC: A
practical guide to the cancer genome atlas (TCGA). Methods Mol
Biol. 1418:111–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar AR, L A, Nair B, Mathew B, Sugunan S
and Nath LR: Decoding the Mechanism of drugs of heterocyclic nature
against hepatocellular carcinoma. Anticancer Agents Med Chem.
23:882–893. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singal AG, Zhang E, Narasimman M, Rich NE,
Waljee AK, Hoshida Y, Yang JD, Reig M, Cabibbo G, Nahon P, et al:
HCC surveillance improves early detection, curative treatment
receipt, and survival in patients with cirrhosis: A meta-analysis.
J Hepatol. 77:128–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ayuso C, Rimola J, Vilana R, Burrel M,
Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C,
Barrufet M, et al: Diagnosis and staging of hepatocellular
carcinoma (HCC): Current guidelines. Eur J Radiol. 101:72–81. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen CL, Chen CJ, Vallon O, Huang ZP, Zhou
H and Qu LH: Genomewide analysis of box C/D and box H/ACA snoRNAs
in Chlamydomonas reinhardtii reveals an extensive organization into
intronic gene clusters. Genetics. 179:21–30. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Romano G, Veneziano D, Acunzo M and Croce
CM: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang D, Zhou J, Gao J, Wu RY, Huang YL,
Jin QW, Chen JS, Tang WZ and Yan LH: Targeting snoRNAs as an
emerging method of therapeutic development for cancer. Am J Cancer
Res. 9:1504–1516. 2019.PubMed/NCBI
|
|
22
|
Yi C, Wan X, Zhang Y, Fu F, Zhao C, Qin R,
Wu H, Li Y and Huang Y: SNORA42 enhances prostate cancer cell
viability, migration and EMT and is correlated with prostate cancer
poor prognosis. Int J Biochem Cell Biol. 102:138–150. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okugawa Y, Toiyama Y, Toden S, Mitoma H,
Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR and Goel A:
Clinical significance of SNORA42 as an oncogene and a prognostic
biomarker in colorectal cancer. Gut. 66:107–117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou
W and Dong JT: Implication of snoRNA U50 in human breast cancer. J
Genet Genomics. 36:447–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wan R, Bai L, Cai C, Ya W, Jiang J, Hu C,
Chen Q, Zhao B and Li Y: Discovery of tumor immune
infiltration-related snoRNAs for predicting tumor immune
microenvironment status and prognosis in lung adenocarcinoma.
Comput Struct Biotechnol J. 19:6386–6399. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Łuczkowska K, Kulig P, Baumert B and
Machaliński B: The evidence that 25(OH)D3 and VK2 MK-7 vitamins
influence the proliferative potential and gene expression profiles
of multiple myeloma cells and the development of resistance to
bortezomib. Nutrients. 14:51902022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang F, Qu H, Lin Q, Yang Y, Ruan X,
Zhang B, Liu Y, Yu C, Zhang H, Fang X and Hao X: Molecular
biomarkers screened by next-generation RNA sequencing for
non-sentinel lymph node status prediction in breast cancer patients
with metastatic sentinel lymph nodes. World J Surg Oncol.
13:2582015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tosar JP, García-Silva MR and Cayota A:
Circulating SNORD57 rather than piR-54265 is a promising biomarker
for colorectal cancer: Common pitfalls in the study of somatic
piRNAs in cancer. RNA. 27:403–410. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang C, Zhang H, Zhang L, Zhu AX, Bernards
R, Qin W and Wang C: Evolving therapeutic landscape of advanced
hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol.
20:203–222. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Mo H, Jiang Y, Wang Y, Sun L, Yao
B, Chen T, Liu R, Li Q, Liu Q and Yin G: MicroRNA-519c-3p promotes
tumor growth and metastasis of hepatocellular carcinoma by
targeting BTG3. Biomed Pharmacother. 118:1092672019. View Article : Google Scholar : PubMed/NCBI
|