Introduction
Acute myeloid leukemia (AML) is a heterogenous
clonal hematopoietic malignancy characterized by arrest of myeloid
cell maturation and disorder of differentiation, resulting in
abnormal accumulation of immature malignant cells in the bone
marrow (BM) and disruption of the normal hematopoietic process
(1). Consequently, patients with
AML exhibit a range of clinical symptoms such as fatigue, weight
loss and frequent infections. The incidence of AML is about 3.4 to
5.0 cases per 100,000 individuals and the 5-year survival is poor
(32.0–33.1%) (2). The clinical and
genomic diversity make therapy challenging, and the development of
novel biomarkers and treatment strategies is in urgent need. Over
the past decades, the risk assessment and treatment selection for
AML have relied on morphology, immunophenotype, cytogenetics and
molecular features (1). Molecular
testing is crucial because molecular changes often precede
morphological abnormalities. However, clinical molecular testing
mainly focuses on gene mutations and fusion genes (3). Recent studies have highlighted the
critical roles of transcriptional dysregulation in AML
leukemogenesis (4–6), and genome sequencing might serve as an
alternative (7) or complement
(8) to traditional testing in the
diagnosis and prognosis of the disease.
Cytohesins, including cytohesins 1–4 (CYTH1-4), are
a subfamily of guanine nucleotide exchange factors. Cytohesins
activate ADP-ribosylation factor family GTPases which are involved
in several essential biological functions, such as cytoskeletal
organization (9), cell migration
(10,11) and cell signaling (12). CYTH1-4 share a similar structural
organization with an N-terminal coiled-coil motif, a central Sec7
domain and a C-terminal pleckstrin homology domain (13). Data from several sources have
demonstrated the effects of cytohesins in carcinogenesis and cancer
progression. A study by Lee et al (14) showed that CYTH2 was upregulated in
malignant melanoma and contributed to tumor growth. CYTH2 was also
reported to be upregulated in colorectal cancer (15) and hepatocellular carcinoma (16), and it was associated with poor
prognosis (15,16). This could be because CYTH2 enhanced
the epidermal growth factor pathway (17). A study by Fu et al (18) demonstrated that CYTH3 was
upregulated in hepatocellular carcinoma, and it was associated with
tumor progression. Inhibiting cytohesins could inhibit the
proliferation of gefitinib-resistant lung cancer cells, as reported
by Bill et al (19).
Moreover, Zhang et al (20)
comprehensively analyzed public datasets and revealed that high
CYTH4 expression was associated with worse survival in ovarian
cancer.
Although numerous studies have reported the clinical
and pathological implications of cytohesins in cancer, their roles
in leukemia remain largely unexplored. A recent study reported that
CYTH1 promotes leukemogenesis, and targeting CYTH1 overcomes
resistance to venetoclax (21).
Therefore, the present study aimed to investigate the expression of
CYTH4 in AML and explore its potential clinical implications.
Another aim was to identify genes associated with CYTH4 in AML to
provide a promising prognostic biomarker for AML.
Material and methods
Gene expression analysis of CYTH4
In the current study, gene expression analysis of
CYTH4 was carried out using various public datasets and online
platforms. The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/ENSG00000100055-CYTH4/tissue,
accessed on 14 October 2022) (22)
was used to analyze the expression of CYTH4 in different healthy
human tissues and cancer cell lines. The Cancer Cell Line
Encyclopedia (CCLE) database (depmap.org/portal/interactive/,
accessed on 15 October 2022) (23)
was used to compare the expression of CYTH4 among various cancer
types.
The Cancer Genome Atlas (TCGA) database (TCGA-LAML,
https://portal.gdc.cancer.gov/, accessed
on 26 October 2022) is a large public database containing both
genome and clinical information spanning 33 cancer types (24). The Tumor Immune Estimation Resource
version 2.0 (TIMER2.0) web resource (http://timer.cistrome.org/, accessed on 20 October
2022) (25) was used in the present
study to compare the expression of CYTH4 between tumor and adjacent
normal tissues from TCGA. The University of Alabama at Birmingham
CANcer (UALCAN) data analysis portal (http://ualcan.path.uab.edu, accessed on 24 October
2022) (26) was used to visualize
the expression data among different AML French-American-British
(FAB) subtypes in TCGA database. The Gene Expression Omnibus (GEO)
dataset GSE30029 (27) was adopted
to compare the expression of CYTH4 between AML and healthy BM
CD34+ cells.
Survival analysis
TCGA database was used to investigate the survival
significance of CYTH4 (24). A
total of 151 AML samples from TCGA-LAML dataset with intact RNA
sequencing data and survival status were included in the current
study (24). Receiver Operating
Characteristic (ROC) analysis was conducted, and the Youden index
was calculated as the sum of sensitivity and specificity minus one.
The expression level that achieves the maximum of the Youden index
is referred to as the cut-off value to divide patients into the
low- and the high-CYTH4 groups. Overall survival (OS) and
event-free survival (EFS) were calculated using the Kaplan-Meier
method and comparisons were carried out using the log-rank test.
Univariate and multivariate survival analyses were performed using
the Cox regression model and described with the hazard ratio (HR)
and 95% confidence interval (CI). A stepwise forward procedure was
used in the multivariate analysis. The datasets GSE10358 (28) and GSE14468 (29) from the GEO database were also used
in the survival analysis.
Differential gene expression analysis
and gene association analysis
DESeq2 package (Version 1.42.0,
github.com/thelovelab/DESeq2) (30)
in R software was used to screen differentially expressed genes
between the low- and the high-CYTH4 groups in AML. Significantly
differentially expressed genes were defined using an adjusted
P<0.05 and |fold change (FC)|>2. Gene association analysis
was carried out using LinkedOmics (http://www.linkedomics.org/login.php, accessed on 1
November 2022) (31), a publicly
available portal for analyzing multi-omics data based on TCGA
dataset. Pearson's correlation coefficient was calculated to search
for CYTH4-associated genes. Significantly-associated genes were
determined based on the criteria of False Discovery Rate
(FDR)<0.05 and |r|>0.5.
Gene Ontology (GO), Gene Set
Enrichment Analysis (GSEA), Search Tool for the Retrieval of
Interacting Genes/Proteins (STRING) and CIBERSORT analyses
GO enrichment analysis and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analysis were carried out using the
Database for Annotation, Visualization and Integrated Discovery
online tool (Version v2023q3, https://david.ncifcrf.gov/, accessed on 20 October
2023) (32). GSEA was performed
using the Molecular Signatures Database (Version 2023.1, https://www.gsea-msigdb.org/gsea/index.jsp, accessed
on 21 October 2023) (33). Genes
interacting with CYTH4 were also investigated using STRING (Version
11.5, https://string-db.org/, accessed on 19
November 2022). The CIBERSORTx tool (https://cibersortx.stanford.edu/, accessed on 22
October 2023) (34) was used to
compare the difference in immune cell infiltration between the low-
and high-CYTH4 groups.
Cell culture
MV4-11, HL-60, THP-1, U-937, Kasumi-1, K-562, RS4;11
and 293T cells were purchased from American Type Culture
Collection. HEL, Reh and MOLT-4 cells were purchased from the
Chinese National Collection of Authenticated Cell Cultures. NOMO-1,
MOLM-13, NB4, BALL-1, NALM-6, and SUP-B15 cells were purchased from
Procell Life Science & Technology Co., Ltd. MV4-11, HL-60,
THP-1, U-937, Kasumi-1, K-562, RS4;11, HEL, Reh, MOLT-4, NOMO-1,
MOLM-13, NB4, BALL-1, NALM-6, and SUP-B15 leukemia cell lines were
maintained in RPMI-1640 medium (VivaCell BIOSCIENCES), supplemented
with 10% fetal bovine serum (FBS, TransGen Biotech Co., Ltd) and 1%
penicillin-streptomycin (VivaCell BIOSCIENCES). 293T cells were
maintained in DMEM medium (VivaCell BIOSCIENCES) supplemented with
10% FBS and 1% penicillin-streptomycin. Cells were cultured in a
humidified incubator (Esco Lifesciences) with 5% CO2 at
a temperature of 37°C. All the cell lines were tested and
authenticated by using short tandem repeat matching analysis. No
mycoplasma contamination was detected.
cDNA and short-hairpin (sh)RNA
construction, lentivirus preparation and infection
Human CYTH4 was amplified from cDNA and cloned into
the pLV3-EF1α-MCS-puro lentiviral construct (Wuhan MiaoLing Biotech
Science Co., Ltd.). shRNA-targeting CYTH4 and non-targeting control
were constructed using synthesized shRNA-encoded DNA oligos and
cloned into the pLKO.1-puro vector (Addgene, Inc.). The designed
target sequences were as follows: Scramble (TGAGGAAATTGCGGCTTATTT),
shCYTH4 #1 (TRCN0000242587, CCGCCAAGGGTATCCAGTATT), shCYTH4 #2
(TRCN0000242586, TTGCACGGTTCCTGTATAAAG). The lentivirus was
produced in 293T cells by transfecting the designed plasmid
together with the packing vectors pLP/VSVG and psPAX2 (Addgene,
Inc.). Cells were subsequently infected with lentiviral particles
via two rounds of ‘spinoculation’ with 8 µg/ml polybrene.
RNA isolation, complementary (c)DNA
preparation and quantitative (q)PCR
The detection of mRNA expression level was carried
out on day 2 after lentiviral infection to assess the
overexpression and knockdown of CYTH4. Total RNA was isolated from
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions.
Isolated RNA was converted into cDNA using the
TransScript® All-in-One First-Strand cDNA Synthesis
SuperMix for qPCR kit (One-Step gDNA Removal; cat. no. AT341;
TransGen Biotech Co., Ltd.). The reaction was carried out by
incubating the mixture at 42°C for 15 min, followed by inactivation
at 85°C for 5 sec. The expression of CYTH4 was detected by qPCR
using the KAPA SYBR® Fast Universal kit (cat. no.
KK4601; Sigma-Aldrich; Merch KGaA) on the ABI Prism 7500 sequence
detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The process included 3 parts: initial denaturation at 95°C
for 2 min, cycling stage (35 cycles) with denaturation at 95°C for
15 sec and annealing plus extension at 60°C for 1 min, and melt
curve stage with 95°C for 15 sec, 60°C for 1 min, 95°C for 30 sec
and 60°C for 15 sec. Expression of CYTH4 was determined by the
comparative Cq method (35) using
GAPDH for normalization. The following CYTH4 primer sequences were
used: Forward, ATTGGGCGCAAGAAGTTCAAC; Reverse,
TTTATACAGGAACCGTGCAATGT. The following GAPDH primer sequences were
used: Forward, CTCTGCTCCTCCTGTTCGAC; Reverse,
GCCCAATACGACCAAATCC.
Western blotting
Western blotting was performed on day 2 after
lentiviral infection. Cells were lysed using RIPA buffer
supplemented with 1 mM phenylmethane sulfonyl fluoride (both
Beijing Solarbio LIFE SCIENCES). Total protein concentration was
measured using a BCA assay kit (Solarbio LIFE SCIENCES). Equal
amounts of protein (~30 µg) were separated by 12% SDS-PAGE and
transferred onto polyvinylidene fluoride membranes. The membrane
was blocked with 5% non-fat milk at room temperature for 1 h, then
incubated with primary antibody CYTH4 (cat. no. H00027128-B01P;
Novus Biologicals, LLC; Bio-Techne) at 4°C overnight for about 12
h. The incubation of HRP-linked secondary antibody (cat. no. 7076;
Cell Signaling Technology, Inc.) was carried out at room
temperature for 1 h. After detecting CYTH4, the membrane was washed
with stripping buffer (Solarbio LIFE SCIENCES) at room temperature
for 30 min and blocked again. It was then incubated with primary
β-Actin (HRP conjugate; cat. no. 5125; Cell Signaling Technology,
Inc.) antibody for 2 h at room temperature. The primary antibody
was diluted at 1:1,000 and the secondary antibody 1:3,000. TBST
buffer with 0.05% Tween 20 (Solarbio LIFE SCIENCES) was used for
washing. The immobilon western chemiluminescent HRP substrate (cat.
no. WBKLS0100; MilliporeSigma) was added to the membrane, and blot
signals were detected using a ChemiDoc XRS+ System (Bio-Rad
Laboratories, Inc.).
Cell proliferation, cell cycle,
apoptosis and in vitro colony formation assay
Cell proliferation was assessed using the Cell
Counting Kit-8 (CCK8; MedChemExpress) assay according to the
manufacturer's instructions. At 72 h after infection, 5,000 viable
cells counted by Trypan blue staining were seeded into 96-well
plates. After incubating the media with CCK-8 reagent for 3 h, the
absorbance was measured at 450 nm using a Multiskan FC Microplate
Photometer (Thermo Fisher Scientific, Inc.). The CCK-8 assay was
performed at the same time for 5 consecutive days. At 72 h after
infection, cells were harvested and fixed with 75% ice-cold ethanol
at 4°C for about 12 h. Cell cycle analysis was conducted using a
Fluorescence-activated cell sorting (FACS) flow cytometer (Beckman
Coulter, Inc.) after staining the samples with propidium iodide
(PI) for 30 min. At 96 h after infection, apoptosis was detected
using Annexin V-fluorescein isothiocyanate (FITC)/PI apoptosis
detection kit (Dojindo Molecular Technologies, Inc.) according to
the manufacturer's instructions. Briefly, cells were stained with
Annexin V-FITC and PI at room temperature for 15 min and analyzed
with a FACS flow cytometer (Beckman Coulter, Inc.). In FACS
analysis, 10,000 cells were gated for cell cycle and apoptosis
detection. Regarding the colony formation assay, cells were harvest
at 72 h after infection with scramble or shCYTH4 lentivirus.
Variable cells were seeded in methylcellulose medium (MethoCult™
H4434, Stemcell Technologies, Inc.) at a density of 1,000 cells/ml.
Colonies (≥50 cells) were counted manually after 10 days.
Statistical analysis
SPSS (version 22.0; IBM Corp.) and GraphPad Prism
(version 7; Dotmatics) were used for statistical analyses. Clinical
features between the two groups were compared using the
χ2 test for categorical variables and the Fisher's exact
test for the expected frequency of an event (<5 in any cell of
2×2 tables). Continuous variables were compared using the
non-parametric Mann-Whitney U test. One-way ANOVA was used to
compare scramble cells and CYTH4-knockdown cells, with scramble
cells serving as the control. Dunnett's multiple comparison test
was used for testing. Two-way ANOVA followed by Sídák's multiple
comparisons test were used to compare the cell viability between
different groups on each day. Data are presented as mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
CYTH4 was upregulated in AML cell
lines
The present study focused on CYTH4 because its
expression was much higher than other cytohesins in AML (Fig. S1).
The HPA dataset was first explored to examine the RNA tissue
specificity of CYTH4 in healthy humans. Results showed that the
expression of CYTH4 was enhanced in the BM and lymphoid tissues,
while being expressed at low levels in other tissues (Fig. 1A).
The level of CYTH4 expression was then analyzed in
cell lines based on the latest next-generation sequencing data from
the CCLE dataset (Table SI). Results showed that CYTH4 was
expressed at high levels in lymphoma and leukemia cell lines,
followed by thyroid cancer (Fig.
1B). The analysis of the HPA dataset also revealed that CYTH4
was expressed at high levels in myeloid cancer cells compared with
other cancer cell lines such as brain, liver and kidney cancer cell
lines (Fig. S2). In addition, the data from the CCLE leukemia cell
lines were used to compare the expression level of CYTH4 in
different types of leukemia, and it was found that CYTH4 was
expressed at high levels in AML compared with acute lymphoblastic
leukemia (ALL) and chronic myeloid leukemia (CML; Fig. 1C). The AML cell lines NOMO-1,
MV4-11, HL-60, THP-1, MOLM-13, Kasumi-1, HEL, NB4 and U-937, the
ALL cell lines RS4;11, BALL-1, Reh, NALM-6, MOLT-4 and SUP-B15, and
the CML cell line K-562 were used in the present study to examine
the expression of CYTH4. RNA was extracted from these cell lines
and qPCR analysis was performed. Results showed that CYTH4 was
expressed at high levels in NOMO-1, MV4-11, HL-60, THP-1 and
MOLM-13 AML cell lines (Fig.
1D).
CYTH4 is upregulated in patients with
AML
To investigate CYTH4 expression in human cancers,
TCGA-LAML dataset was analyzed. Fig.
2A displays an overview of the different expression of CYTH4
between tumors and adjacent normal tissues across TCGA dataset,
suggesting that CYTH4 expression was higher in patients with AML
compared with that in patients with other tumors. These results for
the AML samples matched those obtained from the cell lines
(Fig. 1B). Analysis of the GSE30029
dataset showed CYTH4 expression was significantly upregulated in
AML BM CD34+ cells compared with that in normal BM
CD34+ cells (Fig. 2B).
The French-American-British (FAB) classification system divides AML
into 8 subtypes, designated Myeloid 0–7 (M0-M7), based on the
morphology and the appearance of the leukemia cells. We compared
the CYTH4 expression among different AML subtypes and patients with
M3-AML had the lowest expression of CYTH4 (Fig. 2C).
 | Figure 2.Expression of CYTH4 in patients with
AML. (A) Expression of CYTH4 in tumor and adjacent normal tissues
across all TCGA tumors, analyzed by TIMER 2.0. (B) Comparison of
CYTH4 expression between AML BM CD34+ cells and normal
BM CD34+ cells, analyzed using the GSE30029 dataset. (C)
Expression of CYTH4 in different AML FAB subtypes, analyzed using
UALCAN. *P<0.05, **P<0.01 and ***P<0.001. TCGA, The Cancer
Genome Atlas; ACC, adrenocortical cancer; BLCA, bladder cancer;
BRCA, breast cancer; CESC, cervical cancer; CHOL, bile duct cancer;
COAD, colon cancer; DLBC, large B-cell lymphoma; ESCA, esophageal
cancer; GBM, glioblastoma; HNSC, head and neck cancer; KICH, kidney
chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney
papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, lower
grade glioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC,
lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian
cancer; PAAD, pancreatic cancer; PCPG, pheochromocytoma &
paraganglioma; PRAD, prostate cancer; READ, rectal cancer; SARC,
sarcoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular
cancer; THCA, thyroid cancer; THYM, thymoma; UCEC, endometrioid
cancer; UCS, uterine carcinosarcoma; UVM, ocular melanomas; BM,
bone marrow; FAB, French-American-British classification system;
CYTH4, cytohesin-4; TIMER 2.0, Tumor Immune Estimation Resource
version 2.0; UALCAN, University of Alabama at Birmingham CANcer
data analysis Portal. |
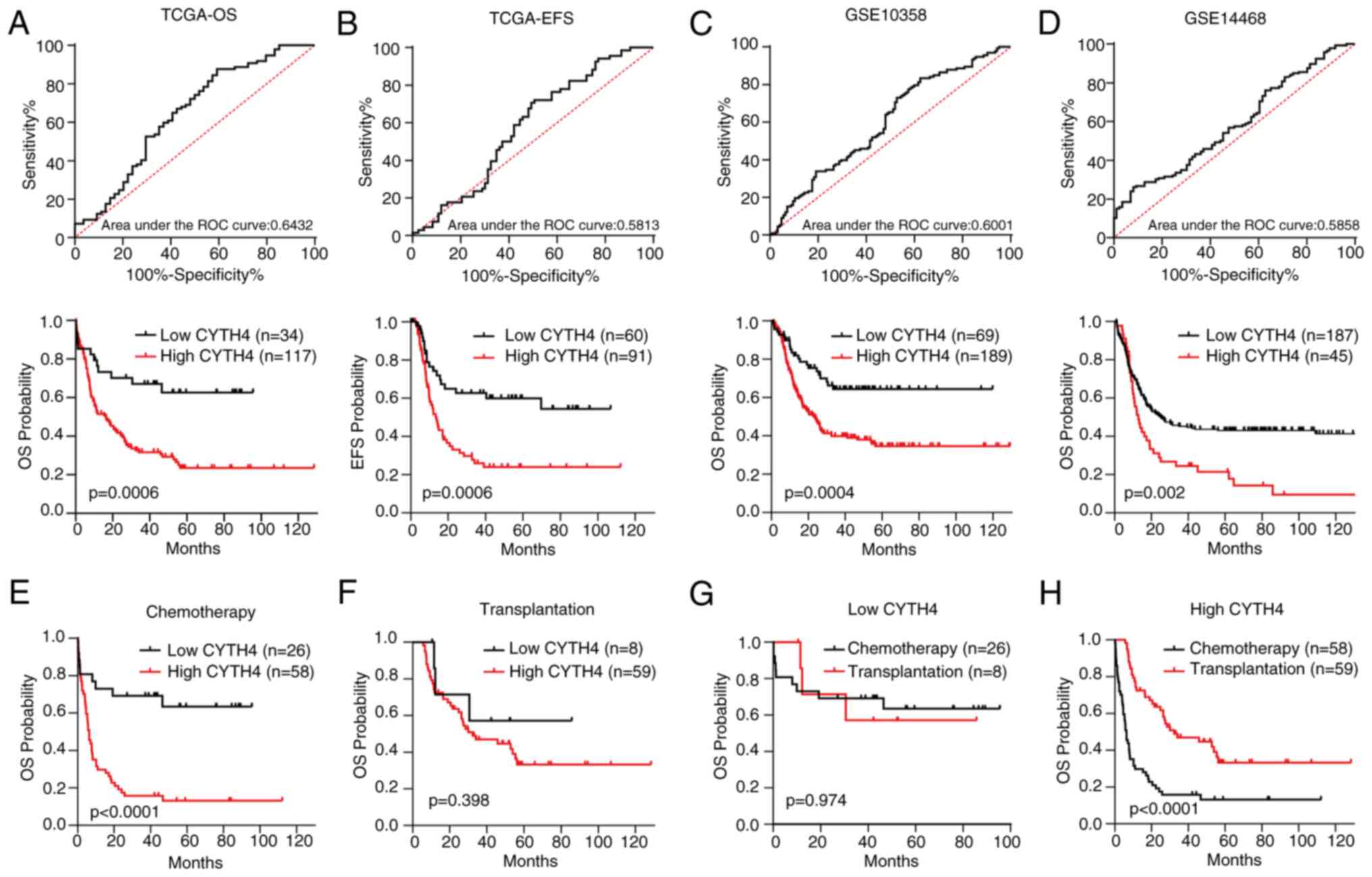
High expression of CYTH4 is associated
with poor survival in AML
To investigate the significance of CYTH4 expression
in AML prognosis, survival was compared between the high- and
low-CYTH4 expression groups in the different datasets. ROC analysis
was performed to determine the cut-off value between the low- and
high-CYTH4 expression groups (Fig.
3A). As shown in Fig. 3A and B,
high CYTH4 expression was significantly associated with unfavorable
OS (high vs. low; HR=2.19; 95% CI, 1.40–3.44; P=0.0006) and EFS
(high vs. low; HR=2.32; 95% CI, 1.43–3.75; P=0.0006). This
conclusion was validated in the GSE10358 dataset (Fig. 3C) and in the GSE14468 dataset
(Fig. 3D). Next, survival was
compared between the low- and high-CYTH4 expression groups by
treatment using TCGA dataset. The results showed that high CYTH4
expression was associated with poor OS (Fig. 3E; high vs. low; HR=3.12; 95% CI,
1.82–5.34; P<0.0001) in patients treated with chemotherapy
alone. However, in cases of patients who received both chemotherapy
and transplantation, no significant difference was found (Fig. 3F; P=0.398). Survival was then
compared between patients treated with chemotherapy alone and
patients treated with chemotherapy plus transplantation grouped by
CYTH4 expression level. Transplantation did not show a significant
difference in the OS of the low-CYTH4 group (Fig. 3G; P=0.974), but significantly
improved OS in the high-CYTH4 expression group compared with
chemotherapy alone (Fig. 3H;
transplantation vs. chemotherapy; HR=0.29; 95% CI, 0.18–0.47;
P<0.0001). These findings suggest that transplantation may
attenuate the adverse effect of high CYTH4 expression on patient
survival in AML.
Clinical features of the low- and the
high-CYTH4 groups
Based on the cut-off value in Fig. 3A, the characteristics of patients in
the low- and the high-CYTH4 group were analyzed (Table SII). Both
clinical features and gene mutations were listed in Table I. It was observed that the white
blood cell count (WBC) varied significantly between the two groups,
with patients in the high-CYTH4 group exhibiting higher WBC than
those in the low-CYTH4 group (median WBC, 25.9 vs. 9.7; P=0.004).
Moreover, in the M4-AML subtype, there were more patients with high
CYTH4 expression than patients with low expression (P=0.001), while
all patients with M3-AML were in the low-CYTH4 expression group
(P<0.0001). This discovery is in line with the previous result
that patients with M3-AML had the lowest CYTH4 expression (Fig. 2C). The low-CYTH4 group had higher
percentages of cases with PML-RARA and RUNX1-RUNX1T1 karyotypes
(P<0.0001). Regarding risk status, low expression of CYTH4 was
significantly associated with a good-risk status (low vs. high,
58.8 vs. 9.4; P<0.0001). Besides, patients in the high-CYTH4
group tended to be older than those in the low-CYTH4 group (57 vs.
51 years; P=0.071). Patients in the high-CYTH4 group had a higher
percentage of RUNX1 mutation (12% vs. 0; P=0.034). The percentage
of patients with more than one mutation did not differ
significantly between the low- and high-CYTH4 groups. No
significant difference was found between the low- and high-CYTH4
group concerning sex, and BM and peripheral blood (PB) blasts.
 | Table I.Characteristics of patients with AML
(n=151) in TCGA dataset grouped by CYTH4 expression; low-CYTH4
(n=34) and high-CYTH4 (n=117). |
Table I.
Characteristics of patients with AML
(n=151) in TCGA dataset grouped by CYTH4 expression; low-CYTH4
(n=34) and high-CYTH4 (n=117).
| Clinicopathological
characteristics | Low-CYTH4 | High-CYTH4 | P-value |
|---|
| Sex |
|
| 0.567 |
|
Male | 17 | 65 |
|
|
Female | 17 | 52 |
|
| Median age, years
(range) | 51 (25–76) | 57 (21–88) | 0.071 |
| Median BM blasts, %
(range) | 79 (33–100) | 71 (30–99) | 0.212 |
| Median WBC,
×109 cells/l (range) | 9.7 (0.4–90.4) | 25.9
(0.7–223.8) | 0.004 |
| Median PB blasts, %
(range) | 36 (0–97) | 39 (0–96) | 0.471 |
| FAB
classifications, n (%) |
|
|
|
| M0 | 1 (2.9) | 14 (12.0) | 0.192 |
| M1 | 7 (20.6) | 29 (24.8) | 0.613 |
| M2 | 10 (29.4) | 27 (23.1) | 0.450 |
| M3 | 14 (41.2) | 1 (0.9) | <0.0001 |
| M4 | 0 (0.0) | 29 (24.8) | 0.001 |
| M5 | 0 (0.0) | 15 (12.8) | 0.028 |
| M6 | 0 (0.0) | 2 (1.7) | 1.000 |
| M7 | 1 (2.9) | 0 (0.0) | 0.225 |
| NA | 1 (2.9) | 0 (0.0) | 0.225 |
| Fusion gene, n
(%) |
|
|
|
| Normal
karyotype | 7 (20.6) | 55 (47.0) | 0.006 |
|
BCR-ABL1 | 2 (5.9) | 1 (0.9) | 0.064 |
|
MYH11-CBFB | 0 (0.0) | 10 (8.5) | 0.170 |
|
PML-RARA | 14 (41.2) | 1 (0.9) | <0.0001 |
| MLL
translocation | 0 (0.0) | 7 (6.0) | 0.319 |
|
RUNX1-RUNX1T1 | 7 (20.6) | 0 (0.0) | <0.0001 |
| Complex
karyotype | 1 (2.9) | 17 (14.5) | 0.125 |
|
Others | 3 (8.8) | 26 (22.2) | 0.141 |
| Molecular risk
level, n (%) |
|
|
|
|
Good | 20 (58.8) | 11 (9.4) | <0.0001 |
|
Intermediate | 9 (26.5) | 72 (61.5) | <0.001 |
|
Poor | 5 (14.7) | 31 (26.5) | 0.156 |
| NA | 0 (0.0) | 3 (2.6) | 1.000 |
| Gene mutation, n
(%) |
|
|
|
|
TET2 | 1 (2.9) | 11 (9.4) | 0.220 |
|
DNMT3A | 4 (11.8) | 32 (27.4) | 0.060 |
|
IDH1/IDH2 | 6 (17.6) | 23 (19.7) | 0.793 |
|
CEBPA | 2 (5.9) | 11 (9.4) | 0.520 |
|
RUNX1 | 0 (0.0) | 14 (12.0) | 0.034 |
|
NPM1 | 6 (17.6) | 32 (27.4) | 0.251 |
|
TP53 | 3 (8.8) | 8 (6.8) | 0.695 |
|
WT1 | 3 (8.8) | 7 (6.0) | 0.558 |
|
FLT3 | 9 (26.5) | 34 (29.1) | 0.768 |
| >1
mutation | 31 (91.2) | 102 (87.2) | 0.765 |
To further explore the prognostic effect of CYTH4 in
AML, univariate and multivariate survival analyses were performed
using the Cox regression model (Table
II). In the univariate analysis, high CYTH4, older age, high
WBC, poor cytogenetics risk, FLT3, DNMT3A and TP53 mutations, and
non-transplantation were associated with poor OS. High CYTH4, WBC,
PB blasts, poor cytogenetics risk and DNMT3A mutation were
identified as inferior prognostic factors for EFS. When age, WBC,
cytogenetics risk, FLT3, DNMT3A and TP53 mutations, and
transplantation were combined in the multivariate Cox regression
analysis, results confirmed that high expression of CYTH4 was
independently associated with inferior OS (HR=2.49; 95% CI,
1.28–4.83; P=0.007) and EFS (HR=2.56; 95% CI, 1.48–4.42;
P=0.001).
 | Table II.Univariate and multivariate analyses
of OS and EFS in patients with AML in TCGA dataset (n=151). |
Table II.
Univariate and multivariate analyses
of OS and EFS in patients with AML in TCGA dataset (n=151).
|
| OS | EFS |
|---|
|
|
|
|
|---|
|
Characteristics | Univariate, HR (95%
CI) | P-value | Multivariate, HR
(95% CI) | P-value | Univariate, HR (95%
CI) | P-value | Multivariate, HR
(95% CI) | P-value |
|---|
| CYTH4 high vs.
low | 2.79
(1.52–5.12) | 0.001 | 2.49
(1.28–4.83) | 0.007 | 2.47
(1.45–4.21) | 0.001 | 2.56
(1.48–4.42) | 0.001 |
| Sex | 0.98
(0.66–1.46) | 0.924 |
|
| 1.07
(0.66–1.72) | 0.796 |
|
|
| Age | 2.07
(1.33–3.20) | 0.001 | 1.02
(1.00–1.04) | 0.028 | 1.39
(0.85–2.26) | 0.187 |
|
|
| WBC | 1.00
(1.00–1.01) | 0.020 |
|
| 1.01
(1.00–1.01) | 0.003 |
|
|
| BM blast | 1.00
(0.99–1.01) | 0.977 |
|
| 1.00
(0.98–1.01) | 0.529 |
|
|
| PB blast | 1.00
(0.99–1.00) | 0.485 |
|
| 1.01
(1.00–1.02) | 0.007 | 1.01
(1.00–1.02) | 0.006 |
| Cytogenetics
risk |
|
|
|
| 1.11
(1.01–1.23) | 0.033 |
|
|
| Inter vs. good | 3.12
(1.59–6.14) | 0.001 | 2.71
(1.29–5.68) | 0.008 | 2.94
(1.48–5.84) | 0.002 |
|
|
| Poor vs. good | 4.42
(2.13–9.16) | <0.001 | 4.43
(1.81–10.83) | 0.001 | 1.76
(0.71–4.33) | 0.222 |
|
|
| Gene mutations |
|
|
|
|
|
|
|
|
|
FLT3 | 1.54
(1.01–2.38) | 0.045 | 2.30
(1.45–3.65) | <0.001 | 1.59
(0.95–2.67) | 0.078 |
|
|
|
DNMT3A | 1.74
(1.11–2.71) | 0.015 |
|
| 1.76
(1.03–3.01) | 0.039 | 1.71
(1.00–2.92) | 0.050 |
|
NPM1 | 0.87
(0.56–1.37) | 0.554 |
|
| 0.71
(0.43–1.20) | 0.200 |
|
|
|
TP53 | 5.09
(2.64–9.85) | <0.001 | 3.80
(1.69–8.57) | 0.001 | 3.18
(0.98–10.36) | 0.054 | 7.55
(2.16–26.41) | 0.002 |
|
Transplantation | 0.53
(0.36–0.81) | 0.003 | 0.38
(0.23–0.63) | <0.001 | 1.55
(0.95–2.53) | 0.082 |
|
|
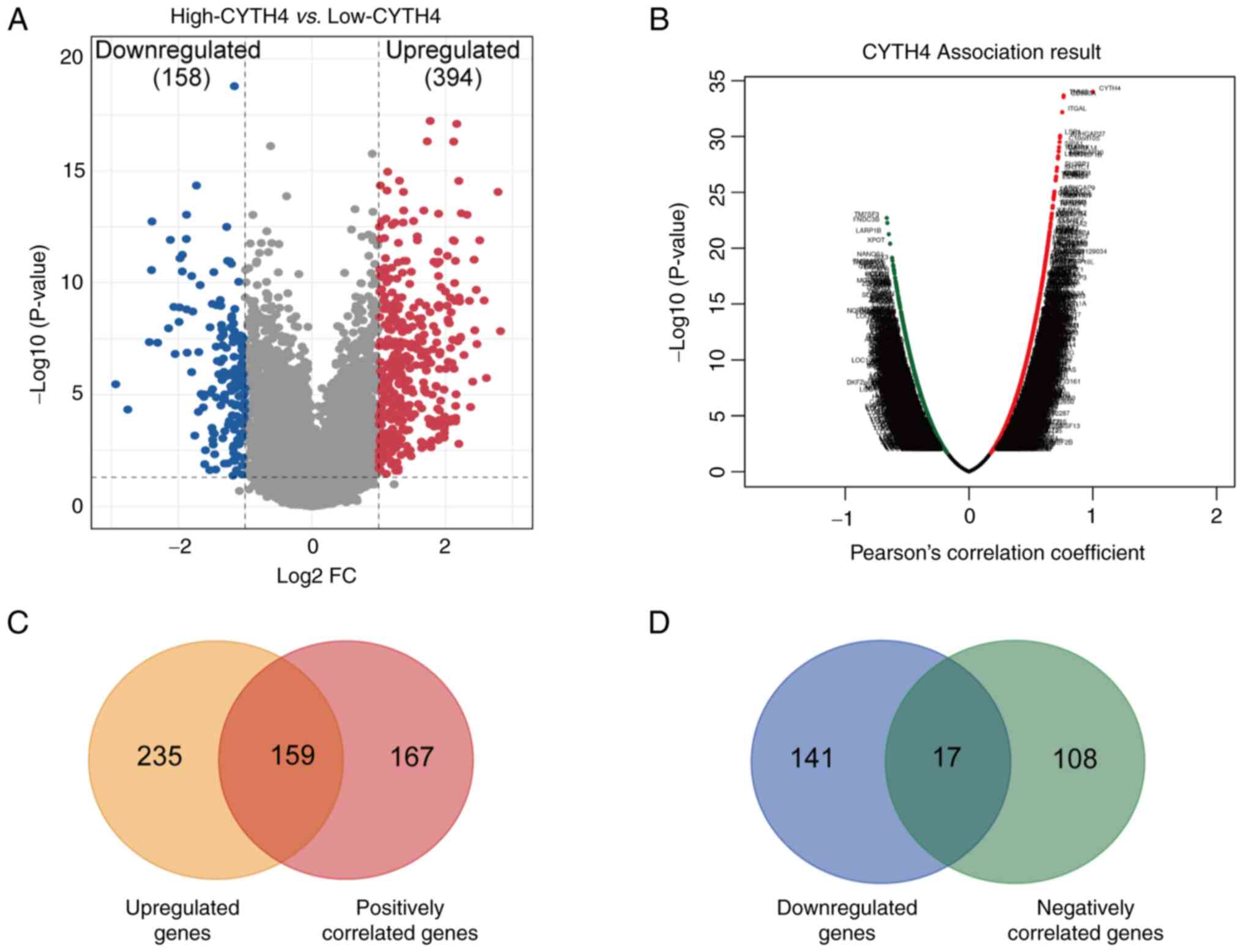
CYTH4-associated gene analysis
To better understand the role of CYTH4 in AML, the
transcriptomes were compared between the high- and the low-CYTH4
groups in TCGA dataset. A total of 552 genes showed significantly
different expression (adjusted P<0.05; |FC|>2) including 394
and 158 genes significantly upregulated and downregulated in the
high-CYTH4 group, respectively (Fig.
4A; Table SIII). LinkedOmics tools were then used to perform
correlation analysis, and a total of 451 significantly co-expressed
genes with a cut-off value of FDR<0.05 and |r|>0.5 were
identified (Fig. 4B; Table SIV). Of
these co-expressed genes, 326 and 125 genes were positively and
negatively correlated with CYTH4 expression, respectively (Table
SIV). By integrating the results of these two analyses, 159 genes
were identified that were upregulated in the high-CYTH4 group and
positively correlated with CYTH4 expression (Fig. 4C). By contrast, only 17 genes were
found to be both downregulated in the high-CYTH4 group and
negatively correlated with CYTH4 expression (Fig. 4D). The overlapping genes were
further analyzed for their biological functions.
Functional enrichment analysis of
overlapping genes
Next. the possible biological function of CYTH4 was
explored. GO and KEGG pathway enrichment analyses were performed
(Fig. 5A and B). These overlapping
genes were significantly associated with ‘innate immune response’,
‘inflammatory response’, ‘signal transduction’, ‘apoptotic
process’, ‘phagosome’ and ‘allograft rejection’. GSEA and the
enrichment plot showed that these overlapping CYTH4-associated
genes were significantly enriched in the gene set related to the
immune response (Fig. 5C).
Additionally, STRING analysis was used to investigate the genes
that interacted with CYTH4 (Fig.
5D). Then, the fractions of 22 distinct immune cell types were
estimated using the CIBERSORTx algorithm. Results showed that high
CYTH4 expression was significantly correlated with CD4+
memory T cells resting (P<0.01), monocytes (P<0.0001) and
mast cells resting (P<0.0001; Fig.
5E).
In vitro validation of the function of
CYTH4
To further investigate the function of CYTH4 in AML,
in vitro validation was carried out. CYTH4 knockdown was
conducted in the AML cell lines MOLM-13, NOMO-1 and THP-1. These
cell lines were chosen because they exhibit relatively high
expression of CYHT4 (Fig. 1D).
Lentivirus-expressing shRNA significantly reduced the mRNA and
protein expression levels of CYTH4 (Fig. 6A; Fig. S3). Cell proliferation
analysis showed that CYTH4 knockdown significantly suppressed the
cell growth of AML cells (Fig. 6B).
Cell cycle assays revealed a significant G0/G1 phase arrest in all
three AML cell lines (Fig. 6C). The
results of the colony-forming assays demonstrated that the
silencing of CYTH4 significantly impaired the clonogenic potential
of the three leukemia cell lines (Fig.
6D). Increased apoptosis was also recorded in the leukemia cell
lines following transfection with CYTH4 shRNA (Fig. 6E). Furthermore, CYTH4 was
overexpressed by lentivirus in the U-937 (acute monocytic leukemia)
and Kasumi-1 (acute myeloblastic leukemia with maturation) AML cell
lines (Fig. S4A and B), in which the CYTH4 expression was low
(Fig. 1D). Results showed that the
overexpression of CYTH4 enhanced the cell growth of the U-937 and
Kasumi-1 cell lines (Fig. S4C). Taken together, these results
indicated that CYTH4 plays an oncogenic role in AML cells.
Discussion
AML is a clonal hematological malignancy
characterized by abnormally rapid proliferation, maturation arrest
and differentiation block of myeloid precursors (36). Patients diagnosed with AML usually
have poor outcomes and high mortality rates (2). At present, the diagnosis of AML mainly
relies on the analysis of BM morphology, immunophenotype,
cytogenetics and molecular features, which also form the basis for
risk stratification (1,8). The complexity and heterogeneity of AML
shed light on the importance of precision medicine and the
detection of robust biomarkers. Recent studies highlight the
feasibility of gene expression assay in AML management, with an
improvement in risk stratification efficiency and prognostic
capacity (7,8). In the present study, OS and EFS were
used to evaluate the prognosis of patient survival. It was shown
that high expression of CYTH4 was associated with poor survival in
AML, and it might be used as a prognostic biomarker. Other clinical
outcomes such as the chemotherapy response, graft-versus-host
disease (GVHD), and relapse rate were not discussed in the present
study. Whether these outcomes are influenced by CYTH4 requires
further investigation.
Cytohesins have been reported to play a pivotal role
in various cancers, including but not limited to hepatocellular
carcinoma, colorectal, lung and ovarian cancer (10,14,16–19,37).
The present study focused on CYTH4 due to its higher expression in
AML compared with that of other cytohesins. The tissue distribution
of CYTH4 showed that CYTH4 expression was predominantly enhanced in
the BM and lymphoid tissues. The distribution partly contributes to
the high expression of CYTH4 in leukemia and lymphoma. Furthermore,
CYTH4 expression in AML BM is higher than that in healthy people.
The high expression and BM specificity provided prerequisites for
CYTH4 to be a possible biomarker in AML. Furthermore, the limited
tissue specificity made it reasonable to hypothesize that it might
be used as a therapeutic target. The study by Bill et al
(19) found that inhibition of
cytohesins improved the treatment of gefitinib-resistant lung
cancer. A recent study reported that the cytohesin inhibitor
SecinH3 showed anti-leukemic effects both in vitro and in
vivo (21). Furthermore,
reduced expression of CYTH4 was observed in patients with PML-RARA
and RUNX1-RUNX1T1. By contrast, cell lines with MLL-rearrangement
such as NOMO-1, MOLM13 and MV4-11 exhibited high levels of CYTH4
expression. This suggests that the regulation of CYTH4 may be
influenced by fusion proteins and their associated signaling
pathways. This hypothesis is in line with the study by Stengel
et al, which identified CYTH4 as a target regulated by the
fusion protein RUNX1-RUNX1T1 (38).
As for clinical characteristics, high CYTH4
expression was significantly correlated with high WBC, higher risk
status and RUNX1 mutation. These unfavorable factors are known to
adversely affect the prognosis of patients with AML (39,40),
indicating that CYTH4 might act as a negative prognostic factor.
The survival analysis provided ultimate evidence that high
expression of CYTH4 was associated with poor survival, validated in
three different datasets. The multivariate analysis also confirmed
the adverse prognostic effect of CYTH4. Additionally, it was
observed that in patients with high CYTH4 expression, those who
received chemotherapy plus transplantation had better survival
outcomes than those who received chemotherapy alone. It suggested
that CYTH4 might serve as an indicator to guide therapy, and
transplantation could potentially overcome the adverse effect of
high CYTH4 expression. BM evaluation is used through the diagnosis,
management and follow-up of AML (41). However, in the present univariate
and multivariate analysis, results showed that BM blast was not a
risk factor for AML. The reason might be that ~80% (n=119/151) of
the patients had a high percentage (>50%) of BM blasts at
diagnosis. Therefore, the prognostic value of BM blasts was not
significant in that particular cohort. In vitro functional
analysis further confirmed that CYTH4 exerted oncogenic effects on
AML cell lines, thereby underscoring its potential value as a
prognostic biomarker in AML. These findings were consistent with
previous studies that reported cytohesins have a variety of
biological activities and are involved in cell proliferation
(16), migration (18) and invasion (15) during carcinogenesis. Ren et
al (21) reported that
inhibiting CYTH1 could reduce the expression of the anti-apoptotic
protein MCL1. Due to their identical structural organization
(21), CYTH4 may also play a role
in leukemogenesis by regulating essential molecules and pathways
associated with cell proliferation (16,18)
and apoptosis (21). Moreover, GO
analysis and GSEA revealed that genes associated with CYTH4 were
involved in cell defense response, signal transduction and
apoptosis. Therefore, the present study suggests that CYTH4 is
upregulated in AML and may play a crucial role in AML
leukemogenesis. However, the specific mechanism by which CYTH4
contributes to leukemogenesis requires further investigation.
In AML, the data of the current study showed that
CYTH4-associated genes were largely involved in the immune response
such as antigen processing and presentation, and positive
regulation of T cell proliferation and differentiation. This is
consistent with the study by Wang et al (10) which demonstrated CYTH2 participated
in immunoregulation. The KEGG analysis and GSEA also proved that
CYTH4 was involved in immune response, but the exact mechanism
needs to be further explored. Immunotherapy, including checkpoint
inhibitors and chimeric antigen receptor-T therapy, is playing an
increasingly important role in the treatment of leukemia (42). Further investigation of the role of
CYTH4 in immunoregulation might provide novel insight into
improving therapeutic efficacy and overcoming obstacles encountered
in immunotherapy. Apart from the immunoregulation effect, the KEGG
result showed that CYTH4 was related to allograft rejection and
involved in GVHD in patients with AML undergoing transplantation.
Further investigation of CYTH4 might help us improve the success
rate of transplantation. However, this hypothesis requires further
study.
The current study has some limitations that need to
be addressed. Firstly, the results were mainly based on
bioinformatics analysis of public datasets; additional experimental
validations, especially in vivo, are required to confirm the
findings. Secondly, it was suggested that CYTH4 expression might be
used as a prognostic biomarker in AML. However, compared with gene
mutation and cytogenetic abnormities, there is more to consider
before the gene expression profile could be used as a biomarker.
For instance, the optimal cut-off value between high and low
expression of CYTH4 is difficult to determine, because expression
is a relative concept. The accuracy, feasibility and clinical
utility of the CYTH4 expression need to be well demonstrated in
larger patient cohorts. Nonetheless, the present study provides
valuable insights into the potential role of CYTH4 as a prognostic
biomarker in AML and prompts further investigations into its
clinical relevance and therapeutic potential.
CYTH4 is upregulated in AML, and the high expression
of CYTH4 is associated with poor survival. CYTH4 can potentially be
used as a prognostic marker in AML.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant nos. 82070161 and
81870134), the China Postdoctoral Science Foundation (grant no.
2022TQ0217), the Natural Science Foundation of Guangdong Province
(grant no. 2022A1515012465), the Natural Foundation of Shenzhen
Science and Technology Innovation Commission (grant no.
JCYJ20220531103003007), the Natural Science Foundation of Shenzhen
University General Hospital (grant no. SUGH2020QD008), the Shenzhen
Key Laboratory Foundation (grant no. ZDSYS20200811143757022), the
Sanming Project of Shenzhen (grant no. SZSM202111004) and the
Medical-Engineering Interdisciplinary Research Foundation of
ShenZhen University (grant no. 00000305).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HW and YL designed the research. HW, YX and WZ
acquired and analyzed the data, and performed statistical analysis.
HW and YX performed in vitro function validation. HW drafted
the manuscript. YX, WZ and YL revised the manuscript. HW and YL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
CYTH4
|
cytohesin-4
|
|
HPA
|
Human Protein Atlas
|
|
CCLE
|
Cancer Cell Line Encyclopedia
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
|
FAB
|
French-American-British
|
|
ROC
|
Receiver Operating Characteristic
curve
|
|
OS
|
overall survival
|
|
EFS
|
event-free survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
FC
|
fold-change
|
|
FDR
|
false discovery rate
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
BM
|
bone marrow
|
|
WBC
|
white blood cell
|
|
PB
|
peripheral blood
|
References
|
1
|
Döhner H, Wei AH, Appelbaum FR, Craddock
C, DiNardo CD, Dombret H, Ebert BL, Fenaux P, Godley LA, Hasserjian
RP, et al: Diagnosis and management of AML in adults: 2022 ELN
recommendations from an international expert panel. Blood.
140:1345–1377. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
SEER*Explorer, . An interactive website
for SEER cancer statistics. Surveillance Research Program, National
Cancer Institute. 2023 Apr 19;Available from:. https://seer.cancer.gov/statistics-network/explorer/
|
|
3
|
Guan W, Zhou L, Li Y, Yang E, Liu Y, Lv N,
Fu L, Ding Y, Wang N, Fang N, et al: Profiling of somatic mutations
and fusion genes in acute myeloid leukemia patients with FLT3-ITD
or FLT3-TKD mutation at diagnosis reveals distinct evolutionary
patterns. Exp Hematol Oncol. 10:272021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Menezes AC, Jones R, Shrestha A, Nicholson
R, Leckenby A, Azevedo A, Davies S, Baker S, Gilkes AF, Darley RL
and Tonks A: Increased expression of RUNX3 inhibits normal human
myeloid development. Leukemia. 36:1769–1780. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan F, Li J, Milosevic J, Petroni R, Liu
S, Shi Z, Yuan S, Reynaga JM, Qi Y, Rico J, et al: KAT6A and ENL
form an epigenetic transcriptional control module to drive critical
leukemogenic gene-expression programs. Cancer Discov. 12:792–811.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li XP, Zhang WN, Mao JY, Zhao BT, Jiang L
and Gao Y: Integration of CD34+CD117dim
population signature improves the prognosis prediction of acute
myeloid leukemia. J Transl Med. 20:3592022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duncavage EJ, Schroeder MC, O'Laughlin M,
Wilson R, MacMillan S, Bohannon A, Kruchowski S, Garza J, Du F,
Hughes AEO, et al: Genome sequencing as an alternative to
cytogenetic analysis in myeloid cancers. N Eng J Med. 384:924–935.
2021. View Article : Google Scholar
|
|
8
|
Letai A: Precision medicine in AML:
Function plus-omics is better than either alone. Cancer Cell.
40:804–806. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi C, Cai C, Cheng Z, Zhao Y, Yang X, Wu
Y, Wang X, Jin Z, Xiang Y, Jin M, et al: Genome-wide CRISPR-Cas9
screening identifies the CYTH2 host gene as a potential therapeutic
target of influenza viral infection. Cell Rep. 38:1105592022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Çil Ç, Harnett MM and Pineda MA:
Cytohesin-2/ARNO: A novel bridge between cell migration and
immunoregulation in synovial fibroblasts. Front Immunol.
12:8098962022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ratcliffe CDH, Siddiqui N, Coelho PP,
Laterreur N, Cookey TN, Sonenberg N and Park M: HGF-induced
migration depends on the PI(3,4,5)P3-binding
microexon-spliced variant of the Arf6 exchange factor cytohesin-1.
J Cell Biol. 218:285–298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyamoto Y, Torii T, Homma K, Oizumi H,
Ohbuchi K, Mizoguchi K, Takashima S and Yamauchi J: The adaptor
SH2B1 and the phosphatase PTP4A1 regulate the phosphorylation of
cytohesin-2 in myelinating schwann cells in mice. Sci Signal.
15:eabi52762022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Donaldson JG and Jackson CL: ARF family G
proteins and their regulators: Roles in membrane transport,
development and disease. Nat Rev Mol Cell Biol. 12:362–375. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee YJ, Bae JH, Kim WI, Kim JH, Cho SW,
Lee SH, Jeon JS, Nam HS, Lee SH, Lee SH and Cho MK: Cytohesin-2 is
upregulated in malignant melanoma and contributes to tumor growth.
Ann Dermatol. 31:93–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan T, Sun J, Hu J, Hu Y, Zhou J, Chen Z,
Xu D, Xu W, Zheng S and Zhang S: Cytohesins/ARNO: The function in
colorectal cancer cells. PloS One. 9:e909972014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu K, Gao J, Yang X, Yao Y and Liu Q:
Cytohesin-2 as a novel prognostic marker for hepatocellular
carcinoma. Oncol Rep. 29:2211–2218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan T, Sun J, Zhou J, Fu Z, Hu Y, Zheng S
and Zhang S: Function and mode of action of cytohesins in the
epidermal growth factor pathway in colorectal cancer cells. Oncol
Lett. 5:521–526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu Y, Li J, Feng MX, Yang XM, Wang YH,
Zhang YL, Qin W, Xia Q and Zhang ZG: Cytohesin-3 is upregulated in
hepatocellular carcinoma and contributes to tumor growth and
vascular invasion. Int J Clin Exp Pathol. 7:2123–2132.
2014.PubMed/NCBI
|
|
19
|
Bill A, Schmitz A, König K, Heukamp LC,
Hannam JS and Famulok M: Anti-proliferative effect of cytohesin
inhibition in gefitinib-resistant lung cancer cells. PloS One.
7:e411792012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Q, Wang Q, Wu S and Zhang J:
Clinical implication and immunological characterisation of the
ARF-GEF family member CYTH4 in ovarian cancer. Autoimmunity.
53:434–442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren WX, Guo H, Lin SY, Chen SY, Long YY,
Xu LY, Wu D, Cao YL, Qu J, Yang BL, et al: Targeting cytohesin-1
suppresses acute myeloid leukemia progression and overcomes
resistance to ABT-199. Acta Pharmacol Sin. Aug 29–2023.(Epub ahead
of print).
|
|
22
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cancer Genome Atlas Research Network, .
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A,
Hoadley K, Triche TJ Jr, Laird PW, et al: Genomic and epigenomic
landscapes of adult de novo acute myeloid leukemia. N Engl J Med.
368:2059–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Jonge HJ, Woolthuis CM, Vos AZ, Mulder
A, van den Berg E, Kluin PM, van der Weide K, de Bont ES, Huls G,
Vellenga E and Schuringa JJ: Gene expression profiling in the
leukemic stem cell-enriched CD34+ fraction identifies target genes
that predict prognosis in normal karyotype AML. Leukemia.
25:1825–1833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tomasson MH, Xiang Z, Walgren R, Zhao Y,
Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, et
al: Somatic mutations and germline sequence variants in the
expressed tyrosine kinase genes of patients with de novo acute
myeloid leukemia. Blood. 111:4797–4808. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wouters BJ, Löwenberg B,
Erpelinck-Verschueren CA, van Putten WL, Valk PJ and Delwel R:
Double CEBPA mutations, but not single CEBPA mutations, define a
subgroup of acute myeloid leukemia with a distinctive gene
expression profile that is uniquely associated with a favorable
outcome. Blood. 113:3088–3091. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sherman BT, Hao M, Qiu J, Jiao X, Baseler
MW, Lane HC, Imamichi T and Chang W: DAVID: A web server for
functional enrichment analysis and functional annotation of gene
lists (2021 update). Nucleic Acids Res. 50:W216–W221. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Newman AM, Steen CB, Liu CL, Gentles AJ,
Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA,
Steiner D, et al: Determining cell type abundance and expression
from bulk tissues with digital cytometry. Nat Biotechnol.
37:773–782. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Short NJ and Kantarjian H: Choosing
between intensive and less intensive front-line treatment
approaches for older patients with newly diagnosed acute myeloid
leukaemia. The Lancet Haematology. 9:e535–e545. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ito A, Fukaya M, Okamoto H and Sakagami H:
Physiological and pathological roles of the cytohesin family in
neurons. Int J Mol Sci. 23:50872022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stengel KR, Ellis JD, Spielman CL, Bomber
ML and Hiebert SW: Definition of a small core transcriptional
circuit regulated by AML1-ETO. Mol Cell. 81:530–545.e5. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gatua M, Navari M, Ong'ondi M, Onyango N,
Kaggia S, Rogena E, Visani G, Abinya NA and Piccaluga PP: Molecular
profiling of kenyan acute myeloid leukemia patients. Front Genet.
13:8437052022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang H, Li Y, Lv N, Li Y, Wang L and Yu L:
Predictors of clinical responses to hypomethylating agents in acute
myeloid leukemia or myelodysplastic syndromes. Ann Hematol.
97:2025–2038. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Percival ME, Lai C, Estey E and Hourigan
CS: Bone marrow evaluation for diagnosis and monitoring of acute
myeloid leukemia. Blood Rev. 31:185–192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chergui A and Reagan JL: Immunotherapy in
acute leukemias: Past success paves the way for future progress.
Cancers (Basel). 15:41372023. View Article : Google Scholar : PubMed/NCBI
|