Introduction
Follicular lymphoma (FL) is a highly heterogeneous,
indolent form of non-Hodgkin lymphoma (NHL). In China, FL
represents 10–20% of all new NHL diagnoses (1). The majority of patients with FL have a
favorable outcome; however, ~20% of patients still face the risk of
disease progression and adverse outcomes despite intensive
treatment (2–4). Given the heterogeneity of the disease,
several models have been proposed to predict treatment outcomes
(5,6). However, identifying high-risk patients
at the time of FL diagnosis remains challenging. Thus, mechanistic
studies into FL development and progression are vital for
identifying more prognostic predictors.
It has become increasingly evident that the survival
of indolent lymphoma cells is highly dependent upon their ability
to escape host immunity (7);
however, the immune evasion strategies remain unclear. Growing
evidence has suggested that the gut microbiota is associated with
immune cell dynamics and immune homeostasis in humans (8–10).
Recent studies have provided strong evidence of the association
between the gut microbiota and human diseases, including cancer
(11–13). Studies have also demonstrated the
prognostic roles of the gut microbiota in immunotherapy and
allogeneic stem cell transplantation (14–16).
These studies have garnered interest in other diseases where a
connection to the gut microbiota is suspected. Recent studies that
have assessed the gut microbiota in patients with NHL, especially
diffuse large B-cell lymphoma, have revealed an association between
gut microbiota composition and disease outcomes (17–19).
However, research on the gut microbiota in FL remains limited. To
the best of our knowledge, only one study has previously examined
the gut microbiota composition in a small cohort of patients with
primary gastrointestinal FL (20).
The present study aimed to investigate the characteristics of the
gut microbiota in patients with FL, and explored the prognostic
value of the gut microbiota composition and the abundance of
specific taxa.
Materials and methods
Patient enrollment and sample
collection
Patients diagnosed with FL (n=28), and age- and
sex-matched healthy controls (n=18) were enrolled from the
Hematology Department, Peking Union Medical College Hospital
(Beijing, China) between June 2021 and June 2022. All patients were
enrolled before initiating treatment or during monitoring of FL.
The exclusion criteria were as follows: i) History of chronic
gastrointestinal inflammatory diseases; ii) coexistence of other
types of tumors; iii) history of diarrhea within 2 weeks; and iv)
history of antibiotic use within 4 weeks (17,18,21,22).
On their first visit to the clinic, fresh fecal samples were
collected from the patients and stored at −20°C. Patients were
assessed by hematologists to determine their Ann Arbor stage
(23), FL International Prognostic
Index (FLIPI) score (24),
pathological grade and tumor burden according to the modified
Groupe d'Etude des Lymphomes Folliculaires (GELF) criteria
(25,26). Patient data, including age, sex, Ann
Arbor stage, World Health Organization pathological grade (25), FLIPI score, extranodal involvement,
tumor burden and laboratory findings [IL-6, IL-8, IL-10 and lactate
dehydrogenase (LDH)] were collected.
16S ribosomal RNA (rRNA) sequencing
and statistical analysis
The microbiota diversity and composition of the
samples were assessed using 16S rRNA gene sequencing. Briefly,
fecal samples stored at −20°C were processed within 24 h of
collection. Sample DNA was extracted using the PowerSoil DNA
Isolation Kit (Mo Bio Laboratories, Inc.). The V3-4 hypervariable
regions of the bacterial 16S rRNA gene were amplified with the
forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG) and reverse primer
806R (5′-GGACTACHVGGGTWTCTAAT) using polymerase chain reaction
(PCR) (27). The PCR products were
purified using Agencourt AMPure XP Kit (Beckman Coulter, Inc.) and
quantified by Nanodrop (Thermo Fisher Scientific, Inc.). The
quality of the amplicons was assessed using the ABI StepOnePlus
Real Time PCR system (Applied Biosystems, Inc.) and Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.). Deep sequencing was
performed on the MiSeq platform using the MiSeq Reagent Kit v2
(cat. no. MS-102-2003; Illumina, Inc.) at Allwegene Technology
(paired-end sequencing, raw read length, 300 bp; loading dose, ~1.6
pmol). After the run, image analysis, base calling and error
estimation were performed using Illumina Analysis Pipeline Version
2.6 (Illumina, Inc.). The raw data were first screened and
sequences were removed from consideration if they were shorter than
230 bp, had a low quality score (≤20), contained ambiguous bases or
did not exactly match to primer sequences and barcode tags.
Qualified reads were separated using the sample-specific barcode
sequences and trimmed with Illumina Analysis Pipeline Version 2.6.
The dataset was then analyzed using QIIME1 (v1.8.0). The sequences
were clustered into operational taxonomic units at a similarity
level of 97% to generate rarefaction curves, and to calculate the
richness and diversity indices (data not shown). The Ribosomal
Database Project Classifier tool was used to classify all sequences
into different taxonomic groups (28). QIIME1 (v1.8.0) software was used to
calculate the chao1 index, observed species and phylogenetic
diversity whole tree (29).
Principal coordinate analysis was used to analyze β-diversity using
R (v3.6.0) (30). The differences
between groups were analyzed using Mothur software (v.1.34.4)
(31), and linear discriminant
analysis effect size (LEfSe) was analyzed by Python (V2.7)
(32). The PICRUSt2 was used to
predict the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment from 16S sequence results (33).
The χ2 test, Fisher's exact test, and
Mann-Whitney U test were applied, where appropriate, to compare the
baseline demographics. Receiver operating characteristic (ROC)
curve analysis, logistic regression, and simple linear regression
analyses were conducted to analyze the association between
microbiota and tumor burden. Statistical analysis was conducted
using R (v3.6.0) and data were visualized using GraphPad Prism 9
(Dotmatics). P<0.05 was considered to indicate a statistically
significant difference.
Results
Baseline characteristics
Among the 28 patients, the median age was 51 years
(range, 30–70 years). The majority of patients had low-grade
advanced-stage disease (Ann Arbor stage III–IV, pathological grade
1–2). Most patients were at low risk as defined by the FLIPI score
(24). The extranodal involvement
sites included the gastrointestinal tract, bone marrow and skin.
According to the modified GELF criteria (26), 11 (39.3%) patients were classified
as having a high tumor burden. There was no significant difference
in age or sex between the patient and control groups (age: P=0.57,
sex: P=0.11). Baseline characteristics are listed in Table I.
 | Table I.Characteristics of patients with FL
and healthy controls. |
Table I.
Characteristics of patients with FL
and healthy controls.
| Characteristic | Patients with FL
(n=28) | Healthy controls
(n=26) | P-value |
|---|
| Median age, years
(range) | 51 (30–70) | 56 (35–64) | 0.57 |
| Sex, n (%) |
|
| 0.11 |
|
Male | 9 (32.1) | 14 (53.8) |
|
|
Female | 19 (67.9) | 12 (46.2) |
|
| Ann Arbor stage, n
(%) |
|
|
|
| I | 0 (0) |
|
|
| II | 3 (10.7) |
|
|
|
III | 16 (57.1) |
|
|
| IV | 9 (32.2) |
|
|
| WHO pathological
grade, n (%) |
|
|
|
| 1 | 18 (64.3) |
|
|
| 2 | 7 (25.0) |
|
|
| 3a | 3 (10.7) |
|
|
| 3b | 0 (0) |
|
|
| FLIPI score, n
(%) |
|
|
|
| Low
risk | 17 (60.7) |
|
|
|
Intermediate risk | 5 (17.9) |
|
|
| High
risk | 6 (21.4) |
|
|
| Extranodal site, n
(%) |
|
|
|
|
None | 17 (60.7) |
|
|
|
Gastrointestinal tract | 6 (21.4) |
|
|
| Bone
marrow | 5
(17.9)a |
|
|
|
Skin | 1 (3.6) |
|
|
| Tumor burden, n
(%) |
|
|
|
|
Low | 17 (60.7) |
|
|
|
High | 11 (39.3) |
|
|
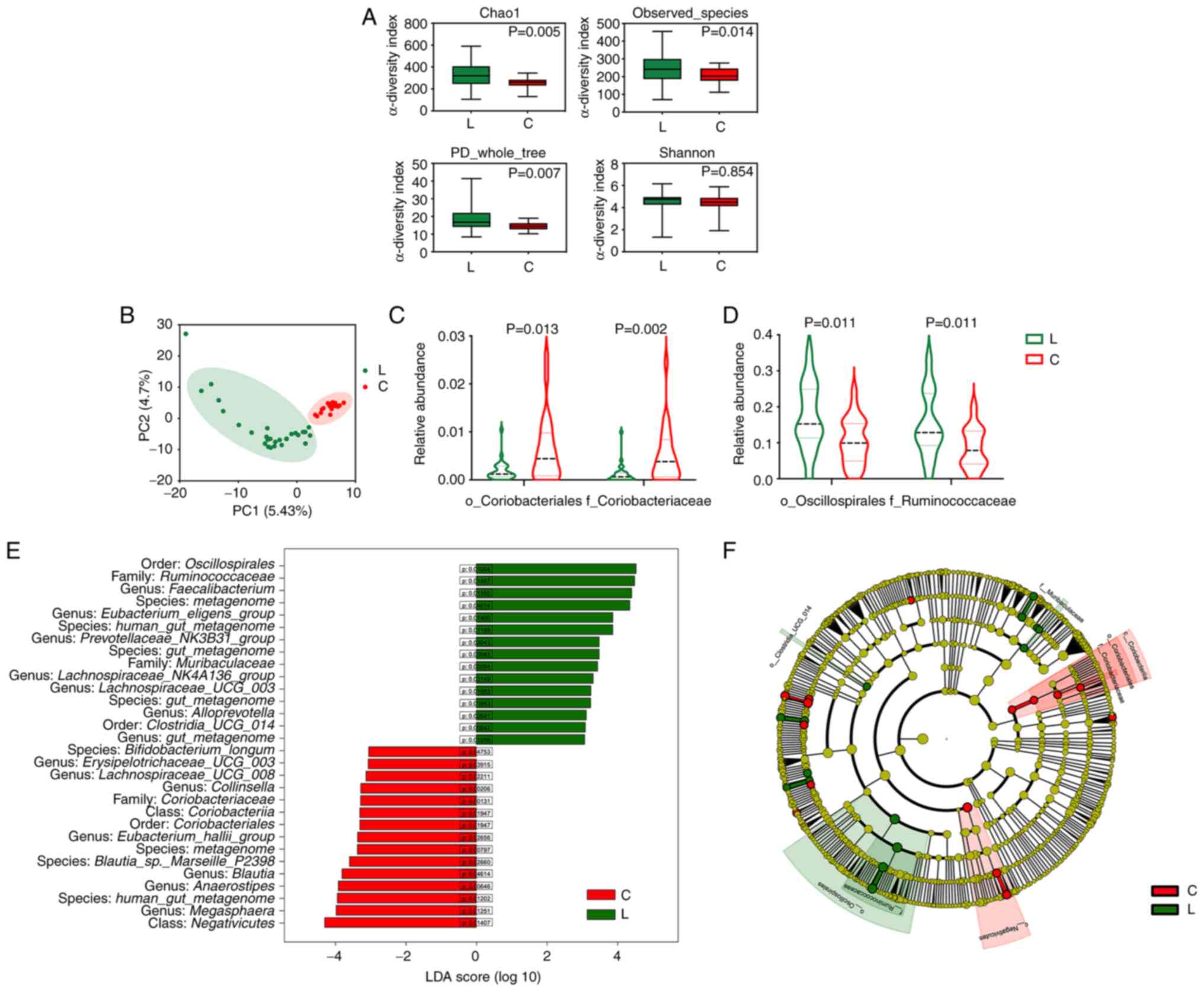
Patients with FL have an altered gut
microbiota
To investigate whether the gut microbiota was
altered in treatment-naïve patients with FL, patients with FL were
compared to healthy controls. The α-diversity analysis showed that
patients with FL had higher taxonomic diversity of gut microbiota
(Fig. 1A). The β-diversity analysis
showed a significant difference between patients with FL and
healthy controls regarding microbiota composition (Fig. 1B). The LEfSe analysis revealed that
the main difference in β-diversity was related to the overabundance
of Ruminococcaceae (family of the Oscillospirales order) in
patients with FL (Fig. 1D and E). A
decrease in Coriobacteriaceae abundance at the family level
were also observed in patients with FL (Fig. 1C and E). The cladogram shows the
relationships of the enriched taxa (Fig. 1F). To further investigate the
functional alteration of gut microbiota, PICRUSt2 was used to
predict the KEGG pathway enrichment based on the 16S rRNA
sequencing data. The activity of the ‘bacterial secretion system’
was significantly enriched in patients with FL (P=0.005), with a
concomitant decrease in nutrient metabolism function, including
‘carbohydrate metabolism’ (P=0.003), ‘fructose and mannose
metabolism’, and ‘thiamine metabolism’ (P=0.01) (Table SI). These results suggested that
the composition and function of the gut microbiota were altered in
patients with FL prior to treatment.
Gut microbiota is associated with
tumor burden
To explore the prognostic value of the gut
microbiota in FL, patients were grouped into a low tumor burden
group (B0, n=17) and a high tumor burden group (B1, n=11). Although
the α-diversity did not differ significantly (Fig. 2A), the β-diversity differed between
patients with low and high tumor burden (Fig. 2B). In the taxonomic comparison of
gut microbial composition, an overabundance of Ruminococcus
(genus of the Ruminococcaceae family) was observed in
patients with high tumor burden (Fig.
2C and E). The present study then specifically investigated the
predictive value of the abundance of Ruminococcus on tumor
burden. The ROC showed that the abundance of Ruminococcus
could be used to distinguish high and low tumor burdens in patients
(area under the curve=0.87, P=0.001; Fig. 2D). As determined by logistic
regression analysis, it was revealed that the high abundance of
Ruminococcus (defined by a relative abundance ≥0.8%; the
cut-off value defined by ROC curve analysis) was associated with an
increased risk of high tumor burden (OR 3.2, 95% CI 1.5–10.5,
P=0.0003) (Table SII).
Additionally, as determined by linear regression analysis, the
abundance of Ruminococcus was associated with LDH levels of
the patients (Fig. 2F). Baseline
cytokine concentrations (IL-6, IL-8 and IL-10) were also assessed;
however, no significant association was found between ILs and
Ruminococcus (data not shown). In addition, the clinical
characteristics of the patients with high or low
Ruminococcus were compared. Although not statistically
significant, patients with a high abundance of Ruminococcus
tended to have a higher pathological grade (P=0.16) and FLIPI score
(P=0.09) (Table II).
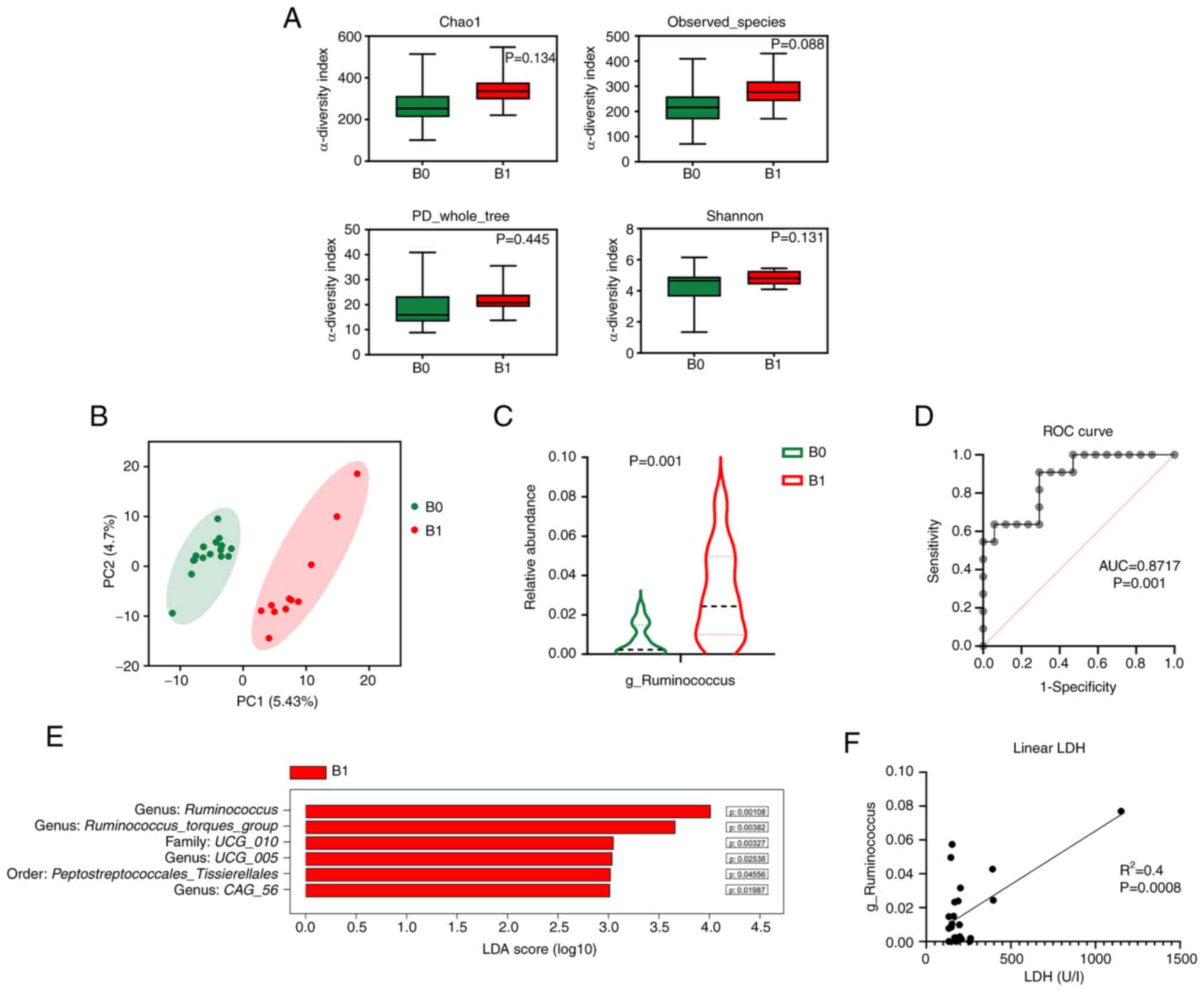 | Figure 2.Gut microbiota is associated with
tumor burden. (A) Bar plots of α-diversity indexes: Chao1, observed
species, phylogenetic diversity whole tree and Shannon. (B)
Principal coordinate analysis of β-diversity. (C) Violin plots of
the relative abundance of the genus Ruminococcus (Wilcoxon
rank-sum test). (D) ROC curve for predicting high tumor burden
based on the relative abundance of Ruminococcus. (E) Bar
plots of the differentially abundant taxa defined by LDA score
>3.0. (F) Estimated regression line obtained by linear
regression model linking LDH levels to the relative abundance of
Ruminococcus. AUC, area under the curve; B0, low
tumor-burden group; B1, high tumor-burden group; C, control; FLIPI,
Follicular Lymphoma International Prognostic Index; L, lymphoma;
LDA, linear discriminant analysis; LDH, lactate dehydrogenase; ROC,
receiver operating characteristic. |
 | Table II.Characteristics of patients with FL
and high or low Ruminococcus abundance. |
Table II.
Characteristics of patients with FL
and high or low Ruminococcus abundance.
| Characteristic | High
Ruminococcus (n=15) | Low
Ruminococcus (n=13) | P-value |
|---|
| Median age, years
(range) | 43 (30–70) | 51 (41–64) | 0.32 |
| Sex, n (%) |
|
| >0.99 |
|
Male | 5 (33.3) | 4 (30.8) |
|
|
Female | 10 (66.7) | 9 (69.2) |
|
| Ann Arbor stage, n
(%) |
|
| 0.87 |
| I | 0 (0) | 0 (0) |
|
| II | 1 (6.7) | 2 (15.4) |
|
|
III | 9 (60.0) | 7 (53.8) |
|
| IV | 5 (33.3) | 4 (30.8) |
|
| WHO pathological
grade, n (%) |
|
| 0.27 |
| 1 | 8 (53.3) | 10 (76.9) |
|
| 2 | 4 (26.7) | 3 (23.1) |
|
| 3a | 3 (20.0) | 0 (0) |
|
| 3b | 0 (0) | 0 (0) |
|
| FLIPI score, n
(%) |
|
| 0.18 |
| Low
risk | 7 (46.7) | 10 (76.9) |
|
|
Intermediate risk | 3 (20.0) | 2 (15.4) |
|
| High
risk | 5 (33.3) | 1 (7.7) |
|
| Extranodal site, n
(%) |
|
| 0.64 |
|
None | 9 (60.0) | 8 (61.5) |
|
|
Gastrointestinal tract | 2 (13.3) | 4 (30.8) |
|
| Bone
marrow | 3 (20.0) | 2
(15.4)a |
|
|
Skin | 1 (6.7) | 0 (0) |
|
| Tumor burden, n
(%) |
|
| 0.0003 |
|
High | 10 (66.7) | 1 (7.7) |
|
|
Low | 5 (33.3) | 12 (92.3) |
|
Discussion
In the present study, it was revealed that patients
with FL at diagnosis had an altered gut microbiota composition
compared with that in healthy individuals, characterized by an
overabundance of Ruminococcaceae and increased bacterial
secretion function. Furthermore, it was demonstrated that the
relative abundance of Ruminococcus was a significant
predictor of tumor burden.
Based on these findings, it was hypothesized that
microbial dysbiosis may be involved in the development and
progression of FL. It is generally believed that lymphoma cells
must evolve some immune escape strategy to develop from lymphoid
organs and survive in the periphery; however, the immune evasion
mechanisms remain poorly characterized (7). There is mounting evidence to support
the role of the microbiome in altering the immune system of the
host. The crosstalk between the gut microbiota and the immune
system influences localized mucosal immunity and has broader
effects, contributing to innate and adaptive immunity at multiple
levels (9,34). Mechanistic studies have shown that
gut microbial dysbiosis can coordinate helper T-cell responses via
the secretion of inflammatory molecules and induction of local
oxidative stress (35–37). As determined by PICRUSt2 prediction,
it was demonstrated in the present study that the bacterial
secretion system pathway was significantly enriched in patients
with FL, whereas the nutrient metabolism pathway was not. This
result indicated a functional shift in the gut microbiota toward a
pathological state. To determine the inflammatory status of the
patients, baseline cytokine concentrations (IL-6, IL-8, IL-10) and
LDH levels were measured in serum samples obtained along with the
fecal samples. Although no significant difference was determined in
cytokines due to the limited sample size, an association was
identified between Ruminococcus abundance and LDH levels.
However, increased LDH could be caused by various factors other
than the gut microbiome, and additional functional studies,
especially metagenomic studies, are warranted to elucidate the
mechanism underlying the interaction between the gut microbiota and
inflammation in patients with FL.
As determined by taxonomic analysis, it was revealed
that members of the Ruminococcaceae family, especially the
genus Ruminococcus, were associated with FL and a high tumor
burden. In the literature, Ruminococcaceae has been reported
to produce short-chain fatty acids that can induce the Wnt
pathway in intestinal stem cells to promote their proliferation
(38). Furthermore, high abundance
of Ruminococcus has been associated with several autoimmune
diseases, including inflammatory bowel disease and spondylarthritis
(39,40). This evidence, together with the
present findings, suggested a novel role for Ruminococcus in
the pathogenesis of several diseases.
To the best of our knowledge, the present study is
the first comprehensive microbiota analysis of treatment-naïve
patients with FL. The results suggested an association between gut
microbiota composition and the disease progression of FL. However,
the impact of these initial findings is limited by the small sample
size. Validation of these findings in larger clinical studies is
required. Moreover, long-term survival data will help to further
consolidate the prognostic value of gut microbiota. Future studies
should also investigate the regulatory mechanisms of the gut
microbiota in preclinical models to understand the interplay of
bacterial taxa and bacterial metabolites on the immune system.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Hongyun Chen, Dr
Jinrong Zhao and Dr Jinkai Lin (Peking Union Medical College
Hospital, Beijing, China) for coordinating and collecting patient
samples.
Funding
This study was funded by the National Natural Science Foundation
of China (NSFC) (grant no. 81970188) and the CAMS Innovation Fund
for Medical Sciences (CIFMS) (grant no. 2021-I2M-1-041). The
funders had no role in study design, data collection, analysis,
interpretation or report writing.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the figshare repository (http://doi.org/10.6084/m9.figshare.22559515).
Authors' contributions
ZFX, CW and WW participated in the patient data
acquisition. DQZ and YZ confirm the authenticity of all the raw
data. ZFX, DQZ and YZ performed statistical analyses. DBZ, DQZ, and
WZ designed the study. DBZ, WZ, and DQZ obtained funding for the
study. DBZ and WZ revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Peking Union Hospital (protocol code 202104293005224,
13rd May 2021; Beijing, China). All patients and healthy volunteers
provided informed written consent for the collection of
samples.
Patient consent for publication
All patients and healthy volunteers provided
informed written consent for the publication of this article.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chinese Society of Lymphoma, Chinese
Anti-cancer Association, Chinese Society of Hematology and Chinese
Medical Association, . Chinese guidelines for diagnosis and
treatment of follicular lymphoma (2020). Zhonghua Xue Ye Xue Za
Zhi. 41:537–44. 2020.(In Chinese). PubMed/NCBI
|
|
2
|
Bachy E, Seymour JF, Feugier P, Offner F,
López-Guillermo A, Belada D, Xerri L, Catalano JV, Brice P,
Lemonnier F, et al: Sustained progression-free survival benefit of
rituximab maintenance in patients with follicular lymphoma:
Long-term results of the PRIMA study. J Clin Oncol. 37:2815–2824.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Casulo C, Byrtek M, Dawson KL, Zhou X,
Farber CM, Flowers CR, Hainsworth JD, Maurer MJ, Cerhan JR, Link
BK, et al: Early relapse of follicular lymphoma after rituximab
plus cyclophosphamide, doxorubicin, vincristine, and prednisone
defines patients at high risk for death: An analysis from the
national lymphocare study. J Clin Oncol. 33:2516–2522. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zha J, Fan L, Yi S, Yu H, Zheng Z, Xu W,
Deng M, Lin Z, Li Z, Ping L, et al: Clinical features and outcomes
of 1845 patients with follicular lymphoma: A real-world multicenter
experience in China. J Hematol Oncol. 14:1312021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Silva A, Bassim S, Sarkozy C, Mottok A,
Lackraj T, Jurinovic V, Brodtkorb M, Lingjaerde OC, Sehn LH,
Gascoyne RD, et al: Convergence of risk prediction models in
follicular lymphoma. Haematologica. 104:e252–e255. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roschewski M, Staudt LM and Wilson WH:
Dynamic monitoring of circulating tumor DNA in non-Hodgkin
lymphoma. Blood. 127:3127–3132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laurent C, Charmpi K, Gravelle P, Tosolini
M, Franchet C, Ysebaert L, Brousset P, Bidaut A, Ycart B and
Fournié JJ: Several immune escape patterns in non-Hodgkin's
lymphomas. Oncoimmunology. 4:e10265302015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gopalakrishnan V, Helmink BA, Spencer CN,
Reuben A and Wargo JA: The influence of the gut microbiome on
cancer, immunity, and cancer immunotherapy. Cancer Cell.
33:570–580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Honda K and Littman DR: The microbiota in
adaptive immune homeostasis and disease. Nature. 535:75–84. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schluter J, Peled JU, Taylor BP, Markey
KA, Smith M, Taur Y, Niehus R, Staffas A, Dai A, Fontana E, et al:
The gut microbiota is associated with immune cell dynamics in
humans. Nature. 588:303–307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Okumura S, Konishi Y, Narukawa M, Sugiura
Y, Yoshimoto S, Arai Y, Sato S, Yoshida Y, Tsuji S, Uemura K, et
al: Gut bacteria identified in colorectal cancer patients promote
tumourigenesis via butyrate secretion. Nat Commun. 12:56742021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davar D, Dzutsev AK, McCulloch JA,
Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding
Q, Pagliano O, et al: Fecal microbiota transplant overcomes
resistance to anti-PD-1 therapy in melanoma patients. Science.
371:595–602. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Ma C, Duan Y, Heinrich B, Rosato
U, Diggs LP, Ma L, Roy S, Fu Q, Brown ZJ, et al: Gut microbiome
directs hepatocytes to recruit MDSCs and promote
cholangiocarcinoma. Cancer Discov. 11:1248–1267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matson V, Fessler J, Bao R, Chongsuwat T,
Zha Y, Alegre ML, Luke JJ and Gajewski TF: The commensal microbiome
is associated with anti-PD-1 efficacy in metastatic melanoma
patients. Science. 359:104–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Viaud S, Saccheri F, Mignot G, Yamazaki T,
Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ,
et al: The intestinal microbiota modulates the anticancer immune
effects of cyclophosphamide. Science. 342:971–976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peled JU, Gomes ALC, Devlin SM, Littmann
ER, Taur Y, Sung AD, Weber D, Hashimoto D, Slingerland AE,
Slingerland JB, et al: Microbiota as predictor of mortality in
allogeneic hematopoietic-cell transplantation. N Engl J Med.
382:822–834. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan L, Wang W, Zhang W, Zhang Y, Wei C,
Li J and Zhou D: Gut microbiota in untreated diffuse large B cell
lymphoma patients. Front Microbiol. 12:6463612021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Diefenbach CS, Peters BA, Li H, Raphael B,
Moskovits T, Hymes K, Schluter J, Chen J, Bennani NN, Witzig TE and
Ahn J: Microbial dysbiosis is associated with aggressive histology
and adverse clinical outcome in B-cell non-Hodgkin lymphoma. Blood
Adv. 5:1194–1198. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yoon SE, Kang W, Choi S, Park Y, Chalita
M, Kim H, Lee JH, Hyun DW, Ryu KJ, Sung H, et al: The influence of
microbial dysbiosis on immunochemotherapy-related efficacy and
safety in diffuse large B-cell lymphoma. Blood. 141:2224–2238.
2023.PubMed/NCBI
|
|
20
|
Zeze K, Hirano A, Torisu T, Esaki M,
Shibata H, Moriyama T, Umeno J, Fujioka S, Okamoto Y, Fuyuno Y, et
al: Mucosal dysbiosis in patients with gastrointestinal follicular
lymphoma. Hematol Oncol. 38:181–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jakobsson HE, Jernberg C, Andersson AF,
Sjölund-Karlsson M, Jansson JK and Engstrand L: Short-term
antibiotic treatment has differing long-term impacts on the human
throat and gut microbiome. PLoS One. 5:e98362010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rashidi A, Ebadi M, Rehman TU, Elhusseini
H, Nalluri H, Kaiser T, Holtan SG, Khoruts A, Weisdorf DJ and
Staley C: Gut microbiota response to antibiotics is personalized
and depends on baseline microbiota. Microbiome. 9:2112021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carbone PP, Kaplan HS, Musshoff K,
Smithers DW and Tubiana M: Report of the committee on Hodgkin's
disease staging classification. Cancer Res. 31:1860–1861.
1971.PubMed/NCBI
|
|
24
|
Solal-Céligny P, Roy P, Colombat P, White
J, Armitage JO, Arranz-Saez R, Au WY, Bellei M, Brice P, Caballero
D, et al: Follicular lymphoma international prognostic index.
Blood. 104:1258–1265. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alaggio R, Amador C, Anagnostopoulos I,
Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D,
Calaminici M, et al: The 5th edition of the World Health
Organization classification of haematolymphoid tumours: Lymphoid
neoplasms. Leukemia. 36:1720–1748. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brice P, Bastion Y, Lepage E, Brousse N,
Haïoun C, Moreau P, Straetmans N, Tilly H, Tabah I and
Solal-Céligny P: Comparison in low-tumor-burden follicular
lymphomas between an initial no-treatment policy, prednimustine, or
interferon alfa: A randomized study from the groupe d'etude des
lymphomes folliculaires. groupe d'etude des lymphomes de l'adulte.
J Clin Oncol. 15:1110–1117. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Caporaso JG, Lauber CL, Walters WA,
Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N and Knight R:
Global patterns of 16S rRNA diversity at a depth of millions of
sequences per sample. Proc Natl Acad Sci USA. 108 (Suppl
1):S4516–S4522. 2011. View Article : Google Scholar
|
|
28
|
Cole JR, Wang Q, Fish JA, Chai B,
McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR and Tiedje
JM: Ribosomal database project: Data and tools for high throughput
rRNA analysis. Nucleic Acids Res. 42:(Database Issue). D633–D642.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2022, URL. https://www.R-project.org/
|
|
31
|
Schloss PD, Westcott SL, Ryabin T, Hall
JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH,
Robinson CJ, et al: Introducing mothur: Open-source,
platform-independent, community-supported software for describing
and comparing microbial communities. Appl Environ Microbiol.
75:7537–7541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pilgrim M and Willison S: Dive into python
3.2. Springer; 2009
|
|
33
|
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel
SN, Brown JR, Taylor CM, Huttenhower C and Langille MGI: PICRUSt2
for prediction of metagenome functions. Nat Biotechnol. 38:685–688.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Johansson ME, Jakobsson HE, Holmén-Larsson
J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, Arike L, Wising C,
Svensson F, Bäckhed F and Hansson GC: Normalization of host
intestinal mucus layers requires long-term microbial colonization.
Cell Host Microbe. 18:582–592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gaboriau-Routhiau V, Rakotobe S, Lécuyer
E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M,
Brandi G, et al: The key role of segmented filamentous bacteria in
the coordinated maturation of gut helper T cell responses.
Immunity. 31:677–689. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fattizzo B, Cavallaro F, Folino F and
Barcellini W: Recent insights into the role of the microbiome in
malignant and benign hematologic diseases. Crit Rev Oncol Hematol.
160:1032892021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamamoto ML and Schiestl RH: Intestinal
microbiome and lymphoma development. Cancer J. 20:190–194. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie J, Li LF, Dai TY, Qi X, Wang Y, Zheng
TZ, Gao XY, Zhang YJ, Ai Y, Ma L, et al: Short-chain fatty acids
produced by Ruminococcaceae mediate α-linolenic acid promote
intestinal stem cells proliferation. Mol Nutr Food Res.
66:e21004082022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hall AB, Yassour M, Sauk J, Garner A,
Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et
al: A novel Ruminococcus gnavus clade enriched in
inflammatory bowel disease patients. Genome Med. 9:1032017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Breban M, Tap J, Leboime A, Said-Nahal R,
Langella P, Chiocchia G, Furet JP and Sokol H: Faecal microbiota
study reveals specific dysbiosis in spondyloarthritis. Ann Rheum
Dis. 76:1614–1622. 2017. View Article : Google Scholar : PubMed/NCBI
|