Introduction
Lung cancer is one of the most common malignant
tumors worldwide (1). The annual
global incidence of lung cancer is over 1,800,000 (2). Various factors, such as smoking,
habits and environment, contribute to the morbidity and mortality
of lung cancer (3). In developed
countries, lung cancer mortality has decreased since the 1990s
(4). However, in China, patients
with lung cancer have a higher mortality burden (5). Although great advances have been made
in lung cancer treatment, such as chemotherapy, radiotherapy and
surgery, the long-term overall survival rates remain unsatisfactory
(6) due to limited access to
diagnosis and treatment (7).
Therefore, the identification of novel diagnostic markers for lung
cancer is vital.
Forkhead box P2 (FOXP2), a transcription
factor (8), is located on
chromosome 7q31 and is a key regulator of metabolism, development
and differentiation (9).
FOXP2 is involved in embryonic and organ development,
including that of the heart, lungs and central nervous system
(10). Abnormal FOXP2
expression contributes to the pathogenesis of lung disorders,
including lung cancer. For instance, FOXP2 alleviates LPS-induced
apoptosis in human pulmonary alveolar epithelial cells and protects
against acute lung injury (11).
FOXP2 expression is decreased in patients with lung cancer
(12). Notably, FOXP2-mediated
upregulation of DNASE1L3 suppresses tumor cell proliferation
and angiogenesis in lung adenocarcinoma (13). However, its role in lung cancer
remains to be elucidated.
TGFβ regulates cell proliferation, migration and
differentiation (14). However,
TGFβ is frequently deregulated in carcinogenesis (15). TGFβ activation induced by tumor and
stromal cells promotes tumor growth and metastasis (16). In addition, TGFβ signaling drives
epithelial-to-mesenchymal transition processes (17), which may contribute to the
chemoresistance and immune surveillance of tumor cells (18). Activated TGFβ ligand coordinates
with its receptors to phosphorylate SMADs, which promotes nuclear
translocation of SMADS to regulate the expression of the TGFβ
target (19). In lung cancer,
hypoxia-inducible factor 1-α-mediated activation of TGFβ/SMAD
signaling accelerates tumor cell glycolysis and growth (20). Epigenetically stimulated
TGFβ2 transcription enhances the radioresistance of lung
cancer (21). However, the roles of
TGFβ/SMAD signaling in lung cancer are still not fully
understood.
The present study investigated the potential role of
FOXP2 in lung cancer. Gene and protein expression were
determined using RT-qPCR and western blotting, respectively.
Functional analysis was performed using the CCK-8, colony
formation, Transwell and TUNEL assays.
Materials and methods
Sampling
A total of 20 lung cancer tissues and adjacent
tissues (>5 cm away from the tumor) [10 males and 10 females; 6
patients aged <60 years old (45–59 years old) and 14 patients
aged ≥60 years old] were collected from patients hospitalized at
Changzhou First People's Hospital (Jiangsu, China). The tissues
were immediately frozen in liquid nitrogen and stored at −80°C for
further processing. All diagnoses of non-small cell lung cancer
were confirmed using pathological assays, including computed
tomography, nuclear magnetic resonance imaging and
immunohistochemistry. Patients who had previously received
chemotherapy or radiotherapy were excluded from the present study.
The present study was approved by the Ethics Committee of Changzhou
First People's Hospital [Jiangsu, China; approval no.
(2019)003].
Cell culture
Human lung cancer (A549, H1975 and H596) and human
bronchial epithelial (16HBE) cell lines were provided by ATCC.
Cells were cultured in RPMI-1640 medium (HyClone; Cytiva)
containing 10% FBS at 37°C in an incubator (Thermo Fisher
Scientific, Inc.) with 5% CO2. Cells were treated with
10 µM of SRI-011381 (MedChemExpress), an agonist of TGFβ
signaling.
Cell transfection
pcDNA3.1 and pcDNA3.1-FOXP2 were obtained from
Shanghai GenePharma Co., Ltd. A549 Cells with good growth state
were taken for seed plate and transfected when the cell density
reached 60%. The culture medium was replaced with a non-antibiotic
medium 12 h before transfection. Cells were divided into the
following groups: Control, untreated; pc-negative control (NC),
transfected with 5 µl of Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) and 20 µM pcDNA3.1 for 6 h at 37°C;
pc-FOXP2, transfected with 5 µl of Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) and 20 µM pcDNA3.1-FOXP2 for 6 h at
37°C and then replaced with complete medium and culture continued
for 24–48 h.
Reverse transcription-quantitative
(RT-q) PCR
All experimental operations were performed according
to the manufacturer's protocols. When the cell density reached
1×106, total RNA was extracted from the cells. cDNA was
synthesized using the RevertAid First Strand cDNA Synthesis Kit
(cat. no. K1622; Thermo Fisher Scientific, Inc.). The mRNA
expression was determined using PCR with the FastStart Universal
SYBR Green Master (Rox) Kit (cat. no. 04913914001; Roche
Diagnostics). mRNA expression was normalized to that of
GAPDH. Results were measured using the 2−ΔΔCq
method (22). The PCR conditions
were as follows: Pre-denaturation at 95°C for 1 min, followed by
denaturation at 95°C for 15 sec, and annealing and extension at
60°C for 30 sec, for 40 cycles. This was repeated three times for
each set. The primer sequences were: GAPDH, F: 5′
AGAAGGTGGTGAAGCAGGCGTC 3′ and R: 5′ AAAGGTGGAGGAGTGGGTGTCG 3′; and
FOXP2, F: 5′-GATGCAACAACTCCAGCAG-3′ and R:
5′-AGGACTTAAGCCAGCTTGAG-3′.
Western blotting
Cells in good condition were collected and the cell
culture medium was discarded. Thereafter, cells were washed twice
with PBS, RIPA lysis buffer (cat. no. BL504A; Biosharp Life
Sciences) was added and the cells were shaken on ice for 15 min.
The supernatant was centrifuged at 4°C, at 10,000 × g, for 5 min,
and then the sample was heated at 100°C for 10 min. The protein
concentration was determined using the BCA method. The protein (20
µg/lane) was isolated using SDS-PAGE (10%; 120 V) and transferred
onto PVDF membranes (MilliporeSigma). Thereafter, the PVDF
membranes were treated with 5% non-fat milk at room temperature to
block them for 30 min, and incubated with primary antibodies at 4°C
overnight: FOXP2 (cat. no. ab16046; 1:1,000), BAX (cat. no.
ab32503; 1:2,500), BCL-2 (cat. no. ab182858; 1:2,000), p-SMAD3
(cat. no. ab52903; 1:2,000), SMAD3 (cat. no. ab208182; 1:1,000),
SMAD4 (cat. no. ab40759; 1:5,000), TGFβR1 (cat. no. ab235578;
1:1,000), zinc finger E-box binding homeobox 1 (ZEB1; cat. no.
ab203829; 1:500), zinc finger protein SNAI1 (SNAIL; cat. no.
ab216347; 1:1,000) and GAPDH (cat. no. ab181602; 1:5,000) and then
with HRP-labeled secondary antibody incubated at room temperature
for 1 h. (cat. no. ab205718; 1:10,000). All antibodies were
provided by Abcam. Proteins were visualized using an enhanced
chemiluminescence kit (Tanon Science & Technology Co., Ltd.).
Finally, ImageJ (National Institutes of Health) was used to analyze
the gray value of the images.
CCK-8 assay
The cells were plated in 24-well plates at a density
of 1×105 cells per well and incubated for 0, 24, 48 and
72 h after transfection at 37°C. The cells were then cultured with
CCK-8 (10 µl; Beijing Solarbio Science & Technology Co., Ltd.)
and cultured for another 4 h at 37°C. Subsequently, absorbance
values were determined using a microplate reader at a wavelength of
450 nm.
Colony formation assay
Cells were plated in a 96-well plate precoated with
soft agar and cultured at 37°C in a 5% CO2 incubator for
14 days. After fixing with 100% methanol at room temperature for 30
min, cells were stained with 1% crystal violet at room temperature
for 15 min. Images were captured under a microscope (Leica
Microsystems GmbH). Colonies with diameters >2 mm in the
predetermined fields of interest were counted (magnification,
×200).
Transwell assay
Transwell chambers were pre-coated with Matrigel (BD
Biosciences) and placed on ice for 30 min to form an even coating.
Homogeneous serum-free cell suspensions (5×105
cells/well) were added to the upper chambers and the lower chambers
were supplemented with 10% FBS. Transwell culture dishes were
placed in a 5% CO2 cell incubator at 37°C for 24 h.
Cells in the lower chamber were fixed and stained with 1% crystal
violet (Beyotime Institute of Biotechnology). Finally, the number
of migrated or invaded cells in predetermined fields of interest
was calculated based on images captured using a microscope (CKX53;
Olympus Corporation; magnification, ×400).
TUNEL assay
Cells were harvested, fixed in 4% paraformaldehyde
(MilliporeSigma) at room temperature for 15 min and permeabilized
with 0.25% Triton-X100 (Dalian Meilun Biotechnology Co., Ltd.).
Thereafter, cells were stained using an in situ cell death
detection kit (MilliporeSigma). Images were visualized using a
fluorescence microscope (Nikon Corporation). The cell death rate
was calculated as TUNEL-positive cells/total cells ×100.
Statistical analysis
Statistical analyses were performed using GraphPad
software, version 9.5.1 (GraphPad; Dotmatics). Data were presented
as mean ± standard deviation. Student's t-test was performed to
analyze the differences between two groups, whereas one-way
analysis of variance and Tukey's post hoc test were applied for
multigroup analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
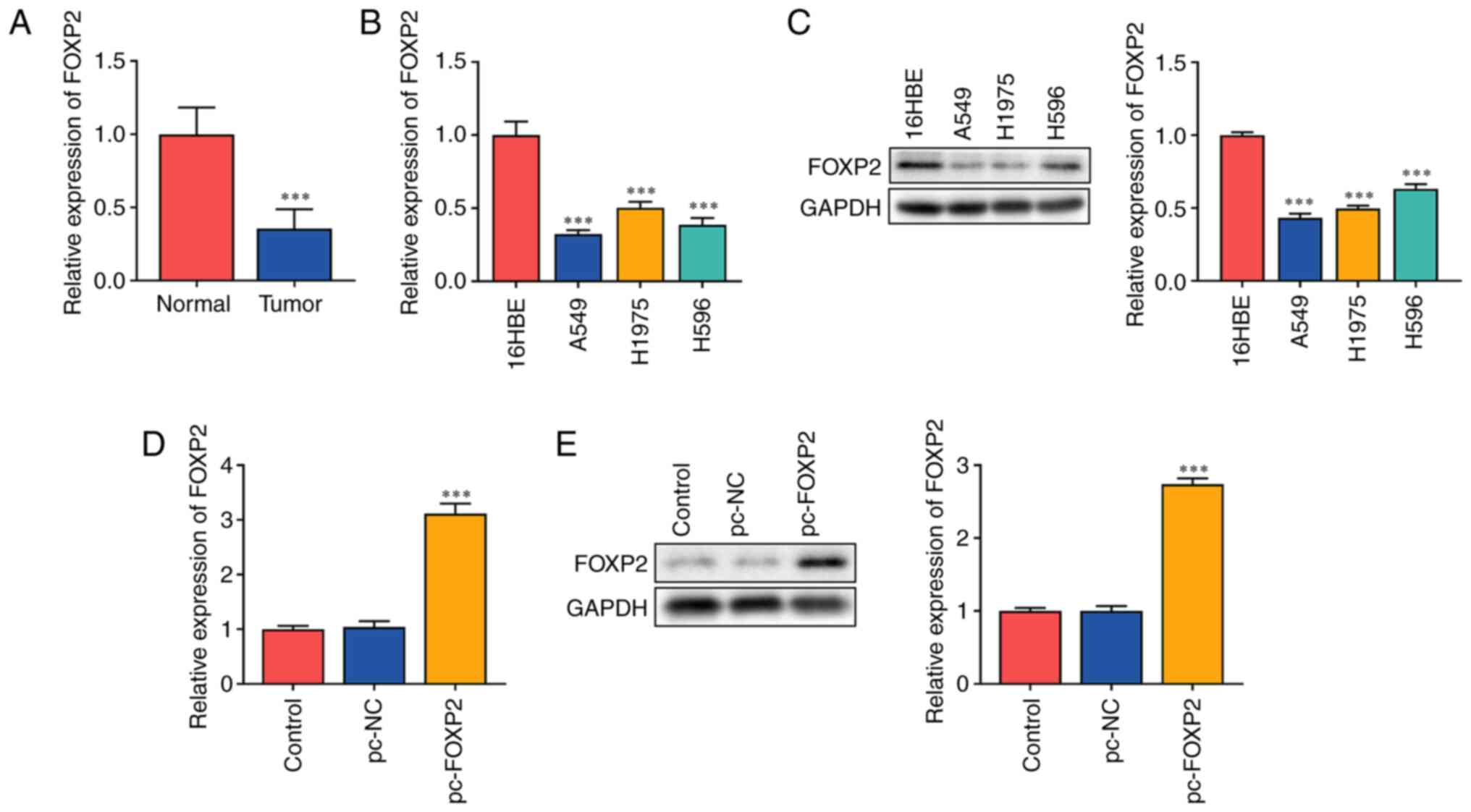
FOXP2 is downregulated in lung
cancer
The present study hypothesized that FOXP2
functions as an antitumor gene in lung cancer. It was found that
FOXP2 mRNA expression significantly decreased in patients
with lung cancer (Fig. 1A). In
addition, mRNA (Fig. 1B) and
protein (Fig. 1C) expressions of
FOXP2 were markedly decreased in lung cancer cells. A549 cells with
a significant difference in FOXP2 expression were used for
subsequent experiments. The potential role of FOXP2 in lung
cancer was further investigated. A549 cells were transfected with
FOXP2 overexpression plasmids. FOXP2 expression in the
pc-FOXP2 group was significantly increased at both the mRNA and
protein levels (Fig. 1D and E),
suggesting that the cells were successfully transfected.
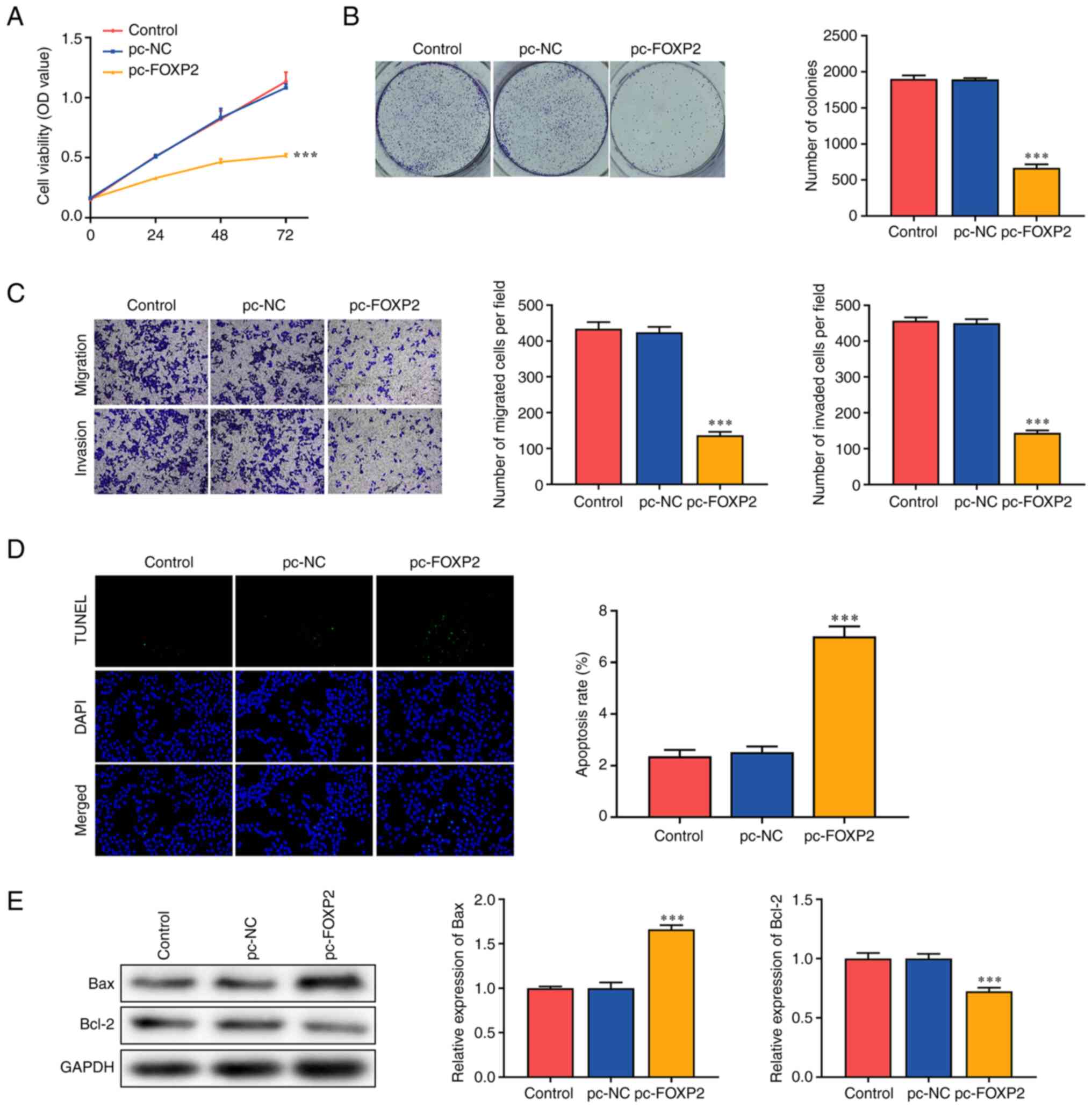
FOXP2 overexpression suppresses the
aggressiveness of A549 cells
Functional assays were performed to determine the
effects of FOXP2 on lung cancer cell function. The
overexpression of FOXP2 significantly suppressed the
viability of A549 cells compared with that in the pc-NC group
(Fig. 2A). In addition,
FOXP2 overexpression markedly inhibited colony formation in
A549 cells (Fig. 2B). Migratory and
invasive abilities were significantly suppressed in the pc-FOXP2
group (Fig. 2C). FOXP2
overexpression significantly increased the TUNEL-positive cells
(Fig. 2D). In addition,
FOXP2 overexpression increased BAX expression and suppressed
BCL-2 expression (Fig. 2E). These
findings suggested that FOXP2 overexpression suppressed the
malignant behavior of lung cancer cells.
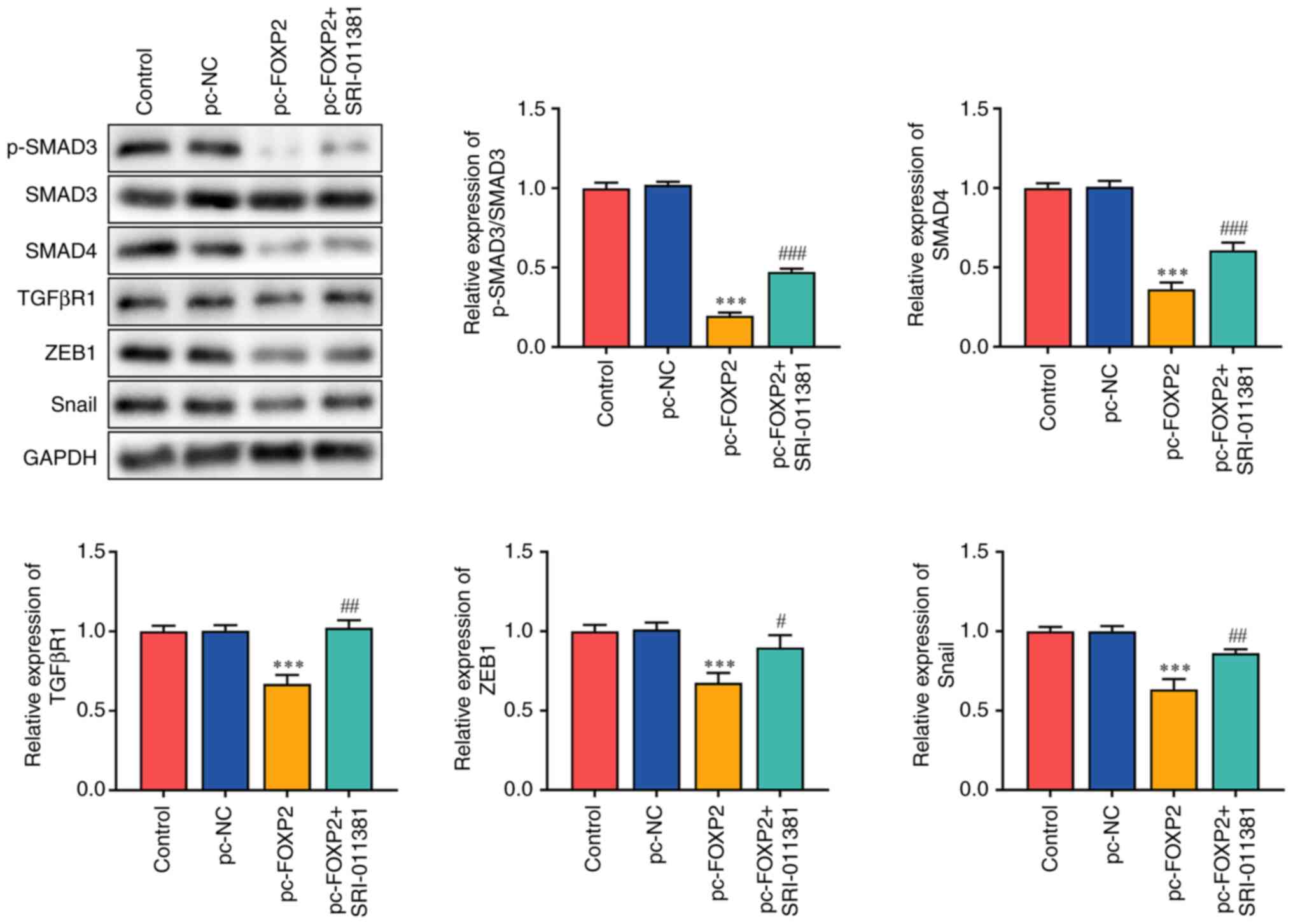
FOXP2 blocks TGFβ/SMAD signaling in
lung cancer
TGFβ/SMAD is involved in the carcinogenesis of lung
cancer (23–25). FOXP2 overexpression suppresses tumor
aggressiveness. Therefore, it was hypothesized that FOXP2
may inhibit the development of lung cancer by targeting TGFβ
signaling. It was found that FOXP2 overexpression
significantly inhibited SMAD3 phosphorylation and suppressed the
protein expression of SMAD4, TGFβR1, ZEB1 and SNAIL. However, the
TGFβ/SMAD signaling agonist SRI-011381 reversed the effects of
pc-FOXP2 (Fig. 3).
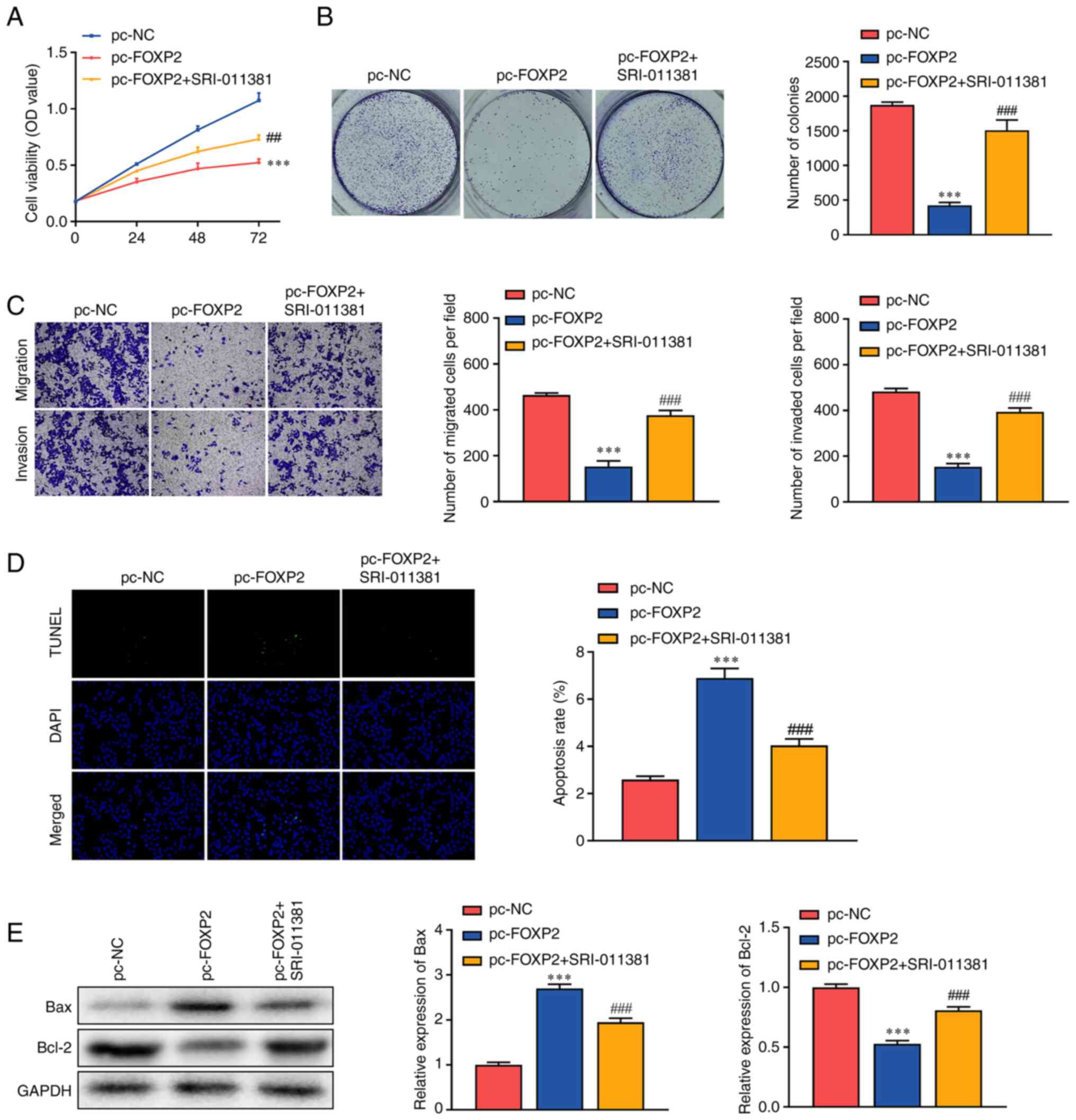
Activation of TGFβ/SMAD signaling
promotes the malignant behaviors of lung cancer
Rescue assays were conducted to verify the role of
FOXP2 and TGFβ/SMAD in lung cancer. Following exposure to
SRI-011381, a TGFβ/SMAD signaling agonist, the inhibition of cell
viability induced by FOXP2 overexpression was alleviated
(Fig. 4A). In addition, SRI-011381
treatment significantly abrogated the effects of FOXP2
overexpression and increased the number of tumor cell clones
(Fig. 4B). This was consistent with
the results of the Transwell assay. In addition, the inhibition of
tumor cell migration and invasion induced by FOXP2
overexpression was markedly abrogated by SRI-011381 (Fig. 4C). In addition, SRI-011381 treatment
significantly alleviated the effects of FOXP2 overexpression
and suppressed apoptosis in A549 cells (Fig. 4D). SRI-011381 treatment
significantly dampened the effects of FOXP2 overexpression,
increased BCL-2 protein expression and downregulated BAX. These
findings suggested that FOXP2 may suppress the aggressiveness of
lung cancer by targeting TGFβ/SMAD signaling.
Discussion
In the present study, FOXP2 expression was
downregulated in lung cancer. Notably, FOXP2 overexpression
suppressed the proliferative, migratory and invasive abilities of
lung cancer cells and promoted tumor cell apoptosis. In addition,
FOXP2 blocked TGFβ signaling, the activation of which enhances
malignant behaviors in tumor cells.
Increasing evidence indicates that FOXP2
functions as an oncogene in various types of cancer. For instance,
circST3GAL6-mediates upregulation of FOXP2 promotes
apoptosis and autophagy in gastric cancer (26). FOXP2 overexpression inhibits
the migration of colon cancer (27). However, the role of FOXP2 in
cancer remains unclear. Activation of HN1L/FOXP2 signaling-mediated
stemness promotes tumor growth and migration in prostate cancer
(28). In addition, FOXP2
overexpression promotes the migration and invasion of colorectal
cancer cells (29), suggesting that
it may also function as an oncogene. Therefore, identifying the
exact role of FOXP2 in lung cancer is vital. In the present
study, FOXP2 expression was decreased in lung cancer cells.
In addition, FOXP2 overexpression suppressed the
proliferative, migratory and invasive abilities of lung cancer
cells, suggesting that FOXP2 may function as an antitumor
gene in lung cancer. These findings were consistent with those of
previous studies (12,13).
FOXP2 alters cellular functions by regulating
the expression of its targets (30). For instance, FOXP2 epigenetically
activates RPS6KA6 to enhance tumor cell apoptosis in thyroid cancer
(31). FOXP2 interacts with
caspase-1 to drive tumor cell pyroptosis in colorectal cancer
(23). In the present study, FOXP2
blocked TGFβ signaling, which plays a key role in the pathogenesis
of cancers. However, its role varies with the stages of tumors. At
the early stages, TGFβ signaling functions as a tumor suppressor
and promotes cell cycle arrest (24). However, the enrichment of
proinflammatory TGFβ induces the degradation of epithelial
functions and the acquisition of mesenchymal features (25), promoting tumor cell migration and
invasion. In addition, the continuous release of TGFβ contributes
to the immune evasion of tumor cells by recruiting macrophages,
cancer-associated fibroblasts and neutrophils (32). In the present study,
SRI-011381-mediated activation of TGFβ/SMAD signaling promoted the
proliferation, migration and invasion of lung cancer cells and
suppressed tumor cell apoptosis. These findings suggested that
FOXP2 suppressed the aggressiveness of lung cancer cells by
targeting TGFβ/SMAD signaling.
The present study had some limitations. First, it
included only 20 participants and did not distinguish the malignant
degree of lung cancer. Future studies with a larger sample size are
needed to confirm the results and further studies are needed to
investigate the correlation between FOXP2 expression and malignant
degree of lung cancer. Second, avoiding recollection bias when
obtaining past information was difficult. In addition, some of the
clinical data were missing. Therefore, well-designed studies are
warranted in the future.
In conclusion, FOXP2 functions as an
antitumor gene in lung cancer. FOXP2 suppressed the proliferation,
migration and invasion of lung cancer cells and promoted apoptosis
in lung cancer cells by blocking TGFβ/SMAD signaling. Therefore,
FOXP2/TGFβ/SMAD signaling may be a potential target for lung
cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by Changzhou Key Medical
Discipline (grant no. CZXK202205).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZYL conceived and designed the study. WYS, SH and LZ
performed the literature search, performed the experiments and data
extraction. HB analyzed and interpreted the data. WYS and HB
drafted the manuscript. ZYL, WYS and SH confirm the authenticity of
all the raw data and revised the final version of the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Changzhou First People's Hospital [Jiangsu, China;
approval no. (2019)003]. All patients signed informed consent
forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Hamann HA, Ver Hoeve ES, Carter-Harris L,
Studts JL and Ostroff JS: Multilevel opportunities to address lung
cancer stigma across the cancer control continuum. J Thorac Oncol.
13:1062–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhuo C, Zhuang H, Gao X and Triplett PT:
Lung cancer incidence in patients with schizophrenia:
Meta-analysis. Br J Psychiatry. 215:704–711. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nooreldeen R and Bach H: Current and
Future development in lung cancer diagnosis. Int J Mol Sci.
22:86612021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen
W, Li X, Wang L, Wang L, Liu Y, et al: Mortality, morbidity, and
risk factors in China and its provinces, 1990–2017: A systematic
analysis for the Global Burden of Disease Study 2017. Lancet.
394:1145–1158. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang D, Liu Y, Bai C, Wang X and Powell
CA: Epidemiology of lung cancer and lung cancer screening programs
in China and the United States. Cancer Lett. 468:82–87. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang M, Herbst RS and Boshoff C: Toward
personalized treatment approaches for non-small-cell lung cancer.
Nat Med. 27:1345–1356. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zugazagoitia J and Paz-Ares L:
Extensive-Stage small-cell lung cancer: First-Line and second-line
treatment options. J Clin Oncol. 40:671–680. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nudel R and Newbury DF: Foxp2. Wiley
Interdiscip Rev Cogn Sci. 4:547–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
den Hoed J, Devaraju K and Fisher SE:
Molecular networks of the FOXP2 transcription factor in the brain.
EMBO Rep. 22:e528032021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Furlong R: FOXP2 tells a cautionary tale.
Nat Rev Genet. 19:592–593. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nan CC, Zhang N, Cheung KCP, Zhang HD, Li
W, Hong CY, Chen HS, Liu XY, Li N and Cheng L: Knockdown of lncRNA
MALAT1 Alleviates LPS-Induced acute lung injury via inhibiting
apoptosis through the miR-194-5p/FOXP2 Axis. Front Cell Dev Biol.
8:5868692020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren T, Liu C, Hou J and Shan F:
Hsa_circ_0043265 suppresses proliferation, metastasis, EMT and
promotes apoptosis in non-small cell lung cancer through
miR-25-3p/FOXP2 pathway. Onco Targets Ther. 13:3867–3880. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meng F, Yang X and Xiao P: DNASE1L3
regulation by transcription factor FOXP2 affects the proliferation,
migration, invasion and tube formation of lung adenocarcinoma. Exp
Ther Med. 25:722022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng D, Fu M, Wang M, Wei Y and Wei X:
Targeting TGF-β signal transduction for fibrosis and cancer
therapy. Mol Cancer. 21:1042022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hao Y, Baker D and Ten Dijke P:
TGF-beta-mediated epithelial-mesenchymal transition and cancer
metastasis. Int J Mol Sci. 20:27672019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Syed V. TGF-beta signaling in cancer. J
Cell Biochem. 117:1279–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mortezaee K, Majidpoor J and Kharazinejad
E: Epithelial-mesenchymal transition in cancer stemness and
heterogeneity: Updated. Med Oncol. 39:1932022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giannelli G, Koudelkova P, Dituri F and
Mikulits W: Role of epithelial to mesenchymal transition in
hepatocellular carcinoma. J Hepatol. 65:798–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou JY, Zheng SR, Liu J, Shi R, Yu HL and
Wei M: MiR-519d facilitates the progression and metastasis of
cervical cancer through direct targeting Smad7. Cancer Cell Int.
16:212016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang Y, Chen Z, Lu T, Bi G, Li M, Liang
J, Hu Z, Zheng Y, Yin J, Xi J, et al: HIF-1alpha switches the
functionality of TGF-β signaling via changing the partners of smads
to drive glucose metabolic reprogramming in non-small cell lung
cancer. J Exp Clin Cancer Res. 40:3982021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jie X, Fong WP, Zhou R, Zhao Y, Zhao Y,
Meng R, Zhang S, Dong X, Zhang T, Yang K, et al: USP9X-mediated
KDM4C deubiquitination promotes lung cancer radioresistance by
epigenetically inducing TGF-β2 transcription. Cell Death Differ.
28:2095–2111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao P, Huang WH, Cao L, Wang T and Chen
LM: Low expression of FOXP2 predicts poor survival and targets
caspase-1 to inhibit cell pyroptosis in colorectal cancer. J
Cancer. 13:1181–1192. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Colak S and Ten Dijke P: Targeting TGF-β
signaling in cancer. Trends Cancer. 3:56–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JH and Massague J: TGF-beta in
developmental and fibrogenic EMTs. Semin Cancer Biol. 86((Pt 2)):
136–145. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu P, Zhang X, Cao J, Yang J, Chen Z, Wang
W, Wang S, Zhang L, Xie L, Fang L, et al: The novel role of
circular RNA ST3GAL6 on blocking gastric cancer malignant
behaviours through autophagy regulated by the FOXP2/MET/mTOR axis.
Clin Transl Med. 12:e7072022. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao Q, Liu C, Cui Q, Luan X, Wang Q and
Zhou C: miR-190b promotes colorectal cancer progression through
targeting forkhead box protein P2. Exp Ther Med. 19:79–84.
2020.PubMed/NCBI
|
|
28
|
Nong S, Wang Z, Wei Z, Ma L, Guan Y and Ni
J: HN1L promotes stem cell-like properties by regulating TGF-β
signaling pathway through targeting FOXP2 in prostate cancer. Cell
Biol Int. 46:83–95. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang R, Xiang G, Duan X, Wang H, He K and
Xiao J: MiR-132-3p inhibits proliferation, invasion and migration
of colorectal cancer cells via down-regulating FOXP2 expression.
Acta Biochim Pol. 69:371–377. 2022.PubMed/NCBI
|
|
30
|
Vernes SC, Newbury DF, Abrahams BS,
Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE,
Geschwind DH, et al: A functional genetic link between distinct
developmental language disorders. N Engl J Med. 359:2337–2345.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang F, Xiao Z and Zhang S: FOXP2
regulates thyroid cancer cell proliferation and apoptosis via
transcriptional activation of RPS6KA6. Exp Ther Med. 23:4342022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tauriello DVF, Sancho E and Batlle E:
Overcoming TGFβ-mediated immune evasion in cancer. Nat Rev Cancer.
22:25–44. 2022. View Article : Google Scholar : PubMed/NCBI
|