Introduction
Breast cancer (BRCA) has become the most prevalent
cancer in women, with ~20,000 new cases reported each year, making
it the primary cause of cancer deaths in women worldwide (1–3). Based
on the International Agency for Research on Cancer reports, BRCA
has accounted for 31% of all malignancies in women by 2023
(4). The available clinical
strategies for the treatment of BRCA include surgical excision,
radiotherapy, endocrine therapy, immunotherapy and targeted
therapy, however, due to residual tumors and drug resistance,
patients with BRCA still generally have a poor prognosis and the
rate of relapse remains high (5).
Therefore, it is crucial to explore the molecular mechanisms of
BRCA development to identify significantly effective therapeutic
targets and prognostic indicators.
Autophagy is a conserved intracellular, degrative
process that is activated in response to various stressors and
regulated by evolutionarily conserved autophagy-related genes
(ATGs) (6,7). Under conditions of nutrient
starvation, cells undergo a lysosomal-dependent self-digestive
process in which cytoplasmic components, including damaged proteins
and organelles, are hydrolyzed to produce nutrients and energy
necessary for cell maintenance (8).
Previous studies reported that autophagy plays a dual role in
cancer development whereby it can inhibit metastasis in the early
stage of tumorigenesis, while promoting tumor progression by
increasing cell growth, proliferation and survival in the late
stage (6,9,10).
Therefore, the function of autophagy may depend on the oncogenic
driving factor.
Recent studies have revealed a reciprocal interplay
between epithelial-mesenchymal transition (EMT) and
autophagy-related signaling pathways (11–13).
EMT-induced autophagy is considered a novel mechanism that
regulates the cytotoxic activity of T-lymphocytes and tumors in
BRCA, and the tumor cells undergoing EMT display tumor resistance
associated with autophagy induction (14). A previous study reported that
autophagy induced by heat treatment upregulated TGF-β signaling
activity and promoted the EMT phenotype, thereby enhancing the
metastasis ability in BRCA (15).
Thus, the key factors that are associated with EMT and autophagic
processes could be utilized as prognostic markers or drug targets
for BRCA therapy.
The Human Autophagy Database (HADb; http://www.autophagy.lu/index.html) showed that
neuroregulatory protein 2 (NRG2) is an autophagy-related gene, and
plays an critical role in malignant tumorigenesis of various human
cancer types, such as breast (16),
prostate (17), and lung (18) cancer. NRGs activate the ErbB
tyrosine kinase receptor family members, which can initiate a
variety of downstream signal transduction pathways related to cell
proliferation and differentiation, apoptosis, migration and
adhesion (19). Zhao et al
(20) reported that NRG2 was highly
expressed in glioma tissues of different grades, which may
partially regulate the expression of GFAP in glioma cells through
Akt signaling, thus affecting the survival rate of patients.
Previous studies investigated that NRG2 participated in the
development of ATGs involved in the prognostic signature for
gastric cancer and prostate cancer (21,22).
However, there is a lack of research focusing on the role of NRG2
in BRCA.
In the present study, an autophagy-related
prognostic model was constructed utilizing The Cancer Genome Atlas
(TCGA) database (https://portal.gdc.cancer.gov/) and a hub gene was
selected for further study. The biological function and immune cell
infiltration of the NRG2 gene were analyzed by bioinformatics and
the ability of NRG2 to regulate autophagy and EMT was determined by
in vitro experiments.
Materials and methods
Data
Through the HADb, 222 ATGs were found. The dataset
from TCGA-BRCA (phs000178) was used for the analysis. The ‘DESeq2’
package in R (version 4.1.1, http://www.r-project.org/) (23) was employed to conduct differentially
expressed gene (DEG) analysis in both groups. A total of 10,935
DEGs with significant statistical differences were selected when
the threshold value was set at |log2FC|>1 and P<0.01.
Definition of the autophagy-related
prognostic model
Univariate Cox regression analysis was performed
using survival (version 3.2.10; http://CRAN.R-project.org/package=survival) and rms
(version 6.3–0; http://hbiostat.org/R/rms/), and the futime and fustat
of two cohorts of patients in all BRCA samples were compared. From
31 intersecting genes, 10 prognostic ATGs were identified
(P<0.2).
Construction of autophagy-related
prognostic model in BRCA
Lasso regression was used to create a prognostic
signature utilizing the samples from the TCGA cohort. Multiple Cox
regression analysis was performed using survival (version 3.2.10)
and rms (version 6.3–0) to identify if the marker genes may
function as stand-alone predictors of patient survival. The
regression coefficients (β) from the multivariate Cox regression
model were combined with the relevant gene expression levels to
generate a multi-gene marker-based predictive risk score. TCGA data
was used to train the risk score model, which was built using
glmnet (version 4.1.7) as follows: Risk score=expression level of
interferon-γ (IFNG) × (−0.17421391598924) + expression level of
neuregulin 1 (NRG1) × (−0.110265298892829) + expression level of
NRG2 × (0.0562837346428505) + expression level of c-FOS ×
(−0.014540998384746) + expression level of eukaryotic translation
initiation factor 4E-binding protein 1 (EIF4EBP1) ×
(0.124693974397309). The median risk score was regarded as the
cut-off value to partition the cohort of patients with BRCA from
TCGA into high-risk and low-risk groups. Kaplan-Meier (KM) survival
curves were conducted with a two-sided log-rank test, and P<0.05
was considered to indicate a significant difference. Time-dependent
receiver operating characteristic (ROC) curve analyses were
conducted to evaluate the predictive capability of the model; the
closer the AUC was to 1, the better the diagnosis. AUC values of
0.5–0.7 represented a low accuracy, 0.7–0.9 represented a moderate
accuracy and >0.9 represented a high accuracy.
Identification of hub gene
Protein-protein interaction (PPI) analysis of 31
differentially expressed autophagy-related genes (DEATGs) was
performed using STRING database (https://cn.string-db.org/). The Cytoscape (version
3.9.1) (24) plugin CytoHubba
(25) was used to identify hub
genes in the module subnet, while Clustering Coefficient methods
were utilized to identify hub genes.
Gene expression and
clinicopathological character analysis
BRCA in TCGA [paraneoplastic (n=113); tumor
(n=1,113)], GSE26304 [normal (n=6); cancer (n=109)] (26) and GSE45827 [normal (n=11); cancer
(n=144)] (27) from the Gene
Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) public database were
selected for differential analysis. Clinical and gene expression
data [including T-classification, M-classification,
N-classification, age, ethnicity, estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor 2 (HER2) status of patients] were extracted from the TCGA
database to explore NRG2 expression in different clinical subgroups
of BRCA.
Survival analysis
Kaplan-Meier survival analysis was performed using
the survival (version 3.2.10) and survminer (version 0.4.9;
http://CRAN.R-project.org/package=survminer).
TCGA-BRCA data (removing normal samples and samples without
clinical information) was categorized into two groups based on high
and low NRG2 expression levels. The prognostic impact of NRG2 was
also assessed based on overall survival (OS), progression-free
interval (PFI) and disease-specific survival (DSS). The diagnostic
value of NRG2 was assessed using the pROC (version 1.18.0;
http://rdocumentation.org/packages/pROC/versions/1.18.5)
to generate ROC curves. Cox regression analysis was performed using
the survival package and forest plot was visualized using ggplot2
(version 3.3.3; http://ggplot2.tidyverse.org).
Functional clustering analysis
Spearman correlation analysis of NRG2 was performed
using the LinkedOmics database (http://www.linkedomics.org/login.php) (28), a total of 13,455 genes were screened
according to P<0.01. Genes with absolute values of correlation
coefficients >0.3 were subjected to Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using
the clusterProfiler (version 4.4.4) (29) and visualized using the ggplot2
package. The BRCA data in TCGA were divided into high and low
expression groups based on NRG2 level, and the low expression group
was used as the control for difference analysis, and the genes with
P<0.01 were selected as the analysis list for Gene Set
Enrichment Analysis (GSEA) enrichment analysis. GSEA was performed
using c2.cp.reactome.v2022.1.Hs.symbols.gmt (Reactome Pathway
Database) and h.all.v7.5.1.symbols.gmt (Hallmarks). Genes with
P<0.01 and |log2FC|>2 were selected as the analysis list for
GO-KEGG enrichment analysis associated with log2FC which was
performed with the clusterProfiler. Log2FC was used to calculate
the Zscore value corresponding to each enriched pathway via the
GOplot (version1.0.2) (30).
Immunity analysis
Correlation between NRG2 and immune infiltration
matrix data of 24 immune cells (31) was assessed by the ssGSEA (Single
Sample Gene Set Enrichment Analysis) of the R package GSVA (version
1.46.0) (32). Spearman correlation
analysis was used to identify the relationship between NRG2
expression and the expression of immune checkpoint genes (PDCD1,
CD274, HAVCR2, TIGIT, SIGLEC15, CTLA4, LAG3 and PDCD1LG2). The
chemokine and receptor related with NRG2 was analyzed by the tumor
and immune system interaction database (TISIDB; http://cis.hku.hk/TISIDB/) (33).
Cell culture
MDA-MB-231 and 293T cells were obtained from the
American Type Culture Collection. Cells were cultured in Dulbecco's
Modified Eagle Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Every Green; Zhejiang Tianhang
Biotechnology Co., Ltd.) and 1% penicillin/streptomycin (Dalian
Meilun Biology Technology Co., Ltd.), and incubated at 37°C and 5%
CO2 in a humidified equipment.
Stable cell line screening
The short hairpin RNA-mediated RNA interference
assay was established to induce knockdown of NRG2 in MDA-MB-231
cells. First, three small interfering (si) RNA oligonucleotides
targeting the NRG2 coding sequence were designed with the following
sequences: siRNA-1: 5′-GCCGAGACATTCGCATCAAAT-3′, siRNA-2:
5′-GCAGCGGCTCGGGCGGCGGCT-3′, siRNA-3: 5′-TCGGCGTCGGACGACGACGCG-3′,
and the scrambled negative control: 5′-TCGTGATCAATCACAGGCACA-3′.
The siRNA sequences were synthesized by Beijing Tsingke Biotech
Co., Ltd. Each of the siRNAs (50 nM) was transiently transfected
into MDA-MB-231 cells in 6-well plate using Lipofectamine
2000® (Thermo Fisher Scientific, Inc.; cat. no.
11668019) at 37°C for 24 h to determine its knockdown efficiency.
Then, the lentivirus vector plko.1 (Promega Corporation) with siRNA
sequences was constructed (plko.1-shNRG2 or plko.1-sh-control), and
prepared for lentivirus packaging. The plasmid (plko.1-shNRG2 or
plko.1-sh-control), pCMV–VSV-G (Promega Corporation) and
pCMV-Gag-Pol (Promega Corporation) was used at a 4:3:1 ratio (12 µg
total DNA in a 10-cm dish) were cotransfected into 293T cells using
PEI reagent (Polyplus-transfection SA). After 72 h of incubation at
37°C, viral supernatant was collected and filtered through a
0.22-µm filter. The virus titer was determined using a Lenti-Pac™
HIV qRT-PCR Titration Kit (GeneCopoeia, Inc.), and then the virus
was used to infect MDA-MB-231 cells at the multiplicity of
infection of 5. Puromycin (1 µg/ml) was used to screen stable
MDA-MB-231 cells with NRG2 silence or the control group after
lentiviral infection. The efficiency of NRG2 silencing was
evaluated using inverted fluorescence microscopy (GFP+)
and reverse transcription-quantitative PCR (RT-qPCR) 5 days after
infection.
RT-qPCR
Total RNA was extracted using Ultrapure RNA Kit
(CWBio) from cells. Reverse Hifair® III 1st Strand cDNA
Synthesis SuperMix (Shanghai Yeasen Biotechnology Co., Ltd.) was
used to reverse transcribe the total RNA into complementary DNA
(cDNA) according to manufacturer's protocol. After which, the qPCR
assay was performed using 2× Universal SYBR Green (ABclonal Biotech
Co., Ltd.). The instrument for qPCR was CFX Connect Real-Time PCR
Detection System (Bio-Rad) with the following conditions: 40 cycles
of 95°C for 10 sec, 60°C for 20 sec and 72°C for 15 sec were
performed after 10 min at 95°C. The 2−ΔΔCq method was
used to calculate the relative expression (34). GAPDH was regarded as the internal
control to standardize the results. Primer sequences were as
follows: NRG2-forward (F): 5′-ACAGCGGAAGCAGATGCAC-3′, reverse (R):
5′-GTTTCTCTCCTGATGACATGGTC-3′; GAPDH-F: 5′-TGCACCACCAACTGCTTAGC-3′,
R: 5′-GGCATGGACTGTGGTCATGAG-3′.
Cell proliferation assays
Cell counting kit-8 (CCK-8) assay was used to assess
cell proliferation. Cells were attached to 96-well plate at
indicated times and cell viability was measured by adding CCK8
reagents (Dalian Meilun Biology Technology Co., Ltd.) with
incubation for 2 h. The OD450 value of each sample was quantified
using a spectrophotometer. Cell proliferation was also evaluated by
a colony formation assay. MDA-MB-231 cells were plated at a low
density (2×103 cells/well) in 6-well plate, and culture
media with 30% serum (Every Green; Zhejiang Tianhang Biotechnology
Co., Ltd. was replaced every 3 days. Following 2 weeks of
incubation, 4% paraformaldehyde (Dalian Meilun Biology Technology
Co., Ltd.), was used to fix the cells at room temperature for 30
min. After staining with 0.1% crystal violet (Servicebio) for 15
min at room temperature, excess crystal violet was washed away with
PBS, and the plaques were imaged and analyzed using the ImageJ
software (version 1.54f; National Institutes of Health).
Transwell assay
First, 50 µl of DMEM-diluted Ceturegel®
Matrix LDEV-Free (Shanghai Yeasen Biotechnology Co., Ltd.) was
added to each well of the Transwell inserts (Labselect; Beijing
Lanjieke Technology Co., Ltd.) and incubated at 37°C for 1 h. A
volume of 500 µl of complete medium (DMEM supplemented with 10%
FBS) was plated to the bottom chamber as a chemoattractant, and
1×105 cells/well were seeded in the upper chamber in 100
µl of serum-free medium. After incubation at 37°C for 12 h, the
cells remaining at the upper membrane were removed by washing with
PBS, and the cells on the lower membrane were fixed with 4%
paraformaldehyde at room temperature for 30 min and stained with 1X
Modified Giemsa stain (Beyotime Institute of Biotechnology) at room
temperature for 45 min, and then images were captured using the
inverted fluorescence microscope.
Cell wound healing assay
A total of 1×105 cells/well were grown
with complete medium in a 6-well plate. Upon reaching 75%
confluence, the cell layers were scratched using a sterile pipette
tip, and washed with PBS to remove cell debris. After which, the
cells were cultured at 37°C for 72 h in serum-free medium, and the
healing process was recorded using an inverted fluorescent
microscope. The wound healing was assessed using the closure rate,
which was calculated as follows: [(Wound area at 0 h-wound area at
indicated h)/wound area at 0 h] ×100%.
Western blotting
Total cell proteins were extracted from the
harvested cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology) and PMSF (Beyotime Institute of Biotechnology). The
protein concentration in the supernatants was quantified by BCA
detection assay (cat. no. P0010S; Beyotime Institute of
Biotechnology). The Omni-Easy™ One-step Color PAGE Gel Rapid
Preparation Kit (Epizym, Inc.; cat. nos. PG211 and PG213) was used
to prepare 7.5 and 12.5% gels. Aliquots containing 20 µg total
protein were fractionated by SDS-PAGE gel electrophoresis and
blotted onto a PVDF membrane (cat. no. IPVH00010; MilliporeSigma).
The membrane was blocked for 1 h at room temperature with 5%
non-fat milk, and was incubated with primary antibodies at 4°C
overnight. The membrane was washed and incubated for 1 h at room
temperature with secondary antibodies, and the expression of
proteins was visualized using the ECL developing solution (Biosharp
Life Sciences; cat. no. BL520A). Primary antibodies used in the
present study were as follows: mouse anti-GAPDH (1:5,000; ABclonal
Biotech Co., Ltd.; cat. no. AC033), rabbit anti-LC3B (1:1,000;
ABclonal Biotech Co., Ltd.; cat. no. A11282), rabbit anti-P62
(1:2,000; ABclonal Biotech Co., Ltd.; cat. no. A19700), rat
anti-E-cadherin (1:500; Santa Cruz Biotechnology, Inc.; cat. no.
sc-59778) and mouse anti-Vimentin (1:500; Santa Cruz Biotechnology,
Inc.; cat. no. sc-373717); and the secondary antibodies were as
follows: HRP Goat Anti-Rat IgG (H+L; 1:10,000; ABclonal Biotech
Co., Ltd.; cat. no. AS028), HRP Goat Anti-Mouse IgG (H+L; 1:10,000;
ABclonal Biotech Co., Ltd.; cat. no. AS003) and HRP Goat
Anti-Rabbit IgG (H+L; 1:10,000; ABclonal Biotech Co., Ltd.; cat.
no. AS014). The gray value was measured and quantified by ImageJ
software (version 1.54f; National Institutes of Health).
Immunofluorescence
Cells were grown on cell climbing slices until
reaching ~80% confluence, after which 4% paraformaldehyde was used
to fix the cells at room temperature for 30 min. The fixed cells
were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room
temperature. Non-specific binding was reduced by incubating the
cells with 5% BSA at room temperature for 1 h, and cells were with
rabbit anti-LC3B (ABclonal Biotech Co., Ltd.; cat. no. A11282) and
p62 (ABclonal Biotech Co., Ltd.; cat. no. A19700) antibodies
(1:200) overnight at 4°C. The next day, cells were incubated with
secondary anti-rabbit-Cy3 conjugated antibody (1:200; Wuhan
Servicebio Technology Co., Ltd.; cat. no. GB21303) at room
temperature for 1 h. Nuclei were stained using
4′,6-diamidino-2-phenylindole (DAPI; Beyotime Institute of
Biotechnology) at room temperature for 10 min. Finally, slides were
cover-slipped by water-soluble glycerol-based mounting medium and
viewed on an inverted fluorescent microscope. The results of
immunofluorescence assay were analyzed by ImageJ software (version
1.54f; National Institutes of Health).
Statistical analysis
All data analysis was performed using GraphPad Prism
software (version 8.0.1; Dotmatics). The statistical differences in
experimental groups were analyzed using unpaired student's t-test
or one-way ANOVA with Tukey's post hoc test. Each treatment of cell
samples was replicated at least three times. Data are presented as
mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Construction of autophagy-related
prognostic model for breast invasive carcinoma
Different expressions of ATGs were analyzed using
P<0.01 and FC>1 thresholds (Fig.
1A and B). According to the cox regression analysis results,
among 31 DEATGs, 10 genes associated with the prognosis of patients
were identified (P<0.2; Fig.
1C). The LASSO logistic regression indicated an autophagy risk
score model composed of seven genes: TP63, PTK6, IFNG, NRG1, NRG2,
FOS and EIF4EBP1 (Fig. 1D and E).
In addition, the model was examined using ROC curves to assess the
diagnostic utility of the autophagy model in the TCGA-BRCA cohort
(Fig. 1F). The distributions of the
risk scores, survival time, survival status and expression patterns
of 7 genes are displayed in Fig.
1G. Subsequently, a PPI network of 31 DEATGs was constructed
(Fig. 1H), and the hub genes were
screened by the Cytohubba plugin of Cytoscape (Table SI). The results showed that NRG2
was the gene with higher clustering coefficient scores in the
autophagy risk model than the scores of PTK6 and NRG1, suggesting
that it may play a significant role in the nosogenesis of BRCA
associated with autophagy.
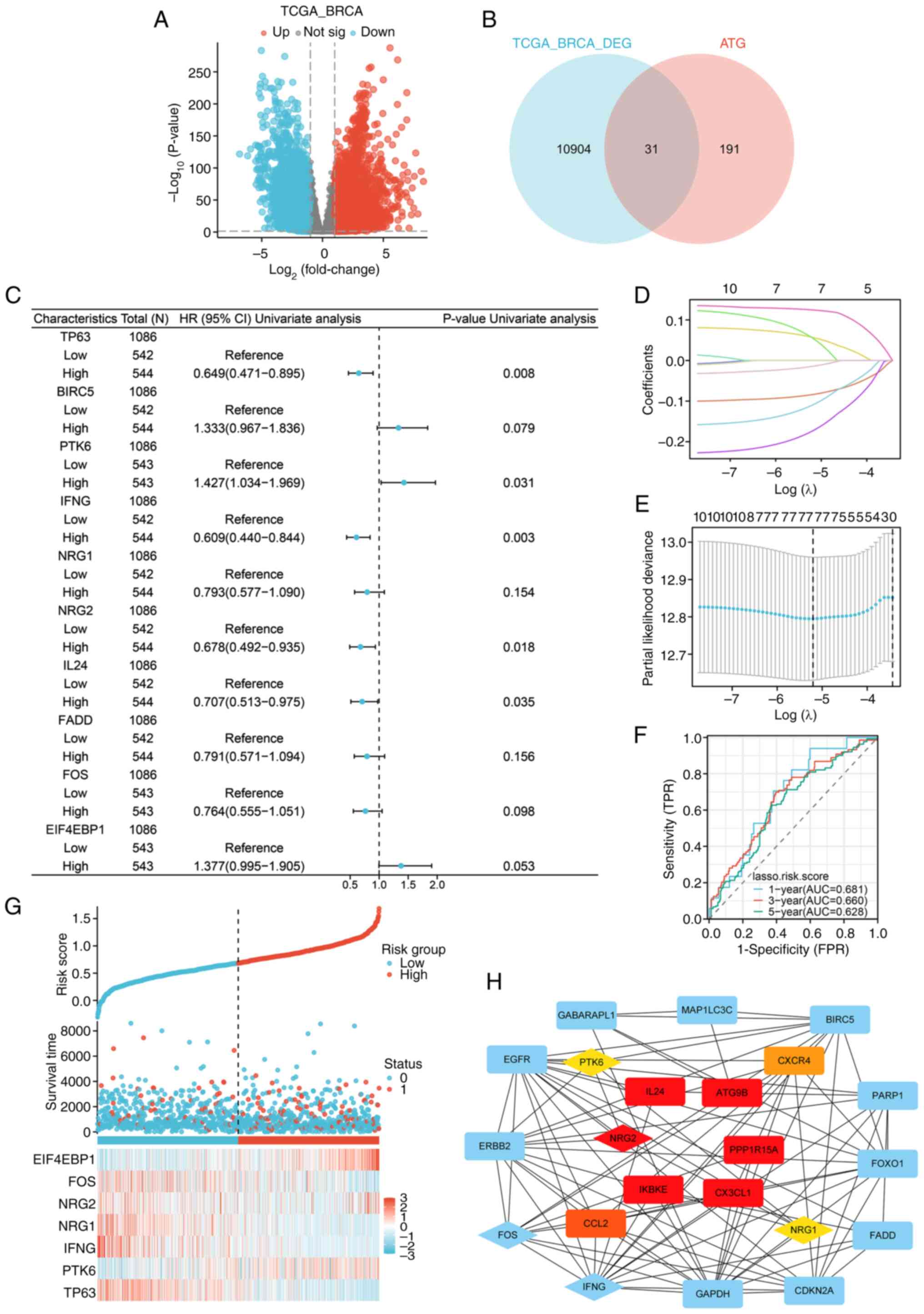 | Figure 1.Construction of a prognostic
autophagy model for BRCA. (A) Volcano map of the DEGs in TCGA-BRCA
(P<0.01, |log2FC|>1). (B) Venn diagram of DEGs and ATGs. (C)
Forest map of univariate cox regression analysis (P<0.2). (D)
Lasso coefficient spectrum of seven autophagy-related genes. (E)
Cross-validation of adjustment parameter selection in a
proportional hazards model. (F) ROC curves of the autophagy risk
model from the TCGA-BRCA cohort. (G) Distribution of the risk
score, survival status and expression profiles of seven genes in
TCGA set. (H) PPI network of 31 DEATGs, with the genes in the
autophagy-related prognostic model displayed by diamonds, and the
top ten genes with the highest clustering coefficient scores were
displayed in red (high), orange (moderate) and yellow (low) based
on their scores, respectively. BRCA, breast cancer; DEGs,
differential expressed genes ATGs, autophagy-related genes; ROC,
receiver operating characteristic; TCGA, The Cancer Genome Atlas;
PPI, protein-protein interaction; DEATGs, different expression of
autophagy-related genes. |
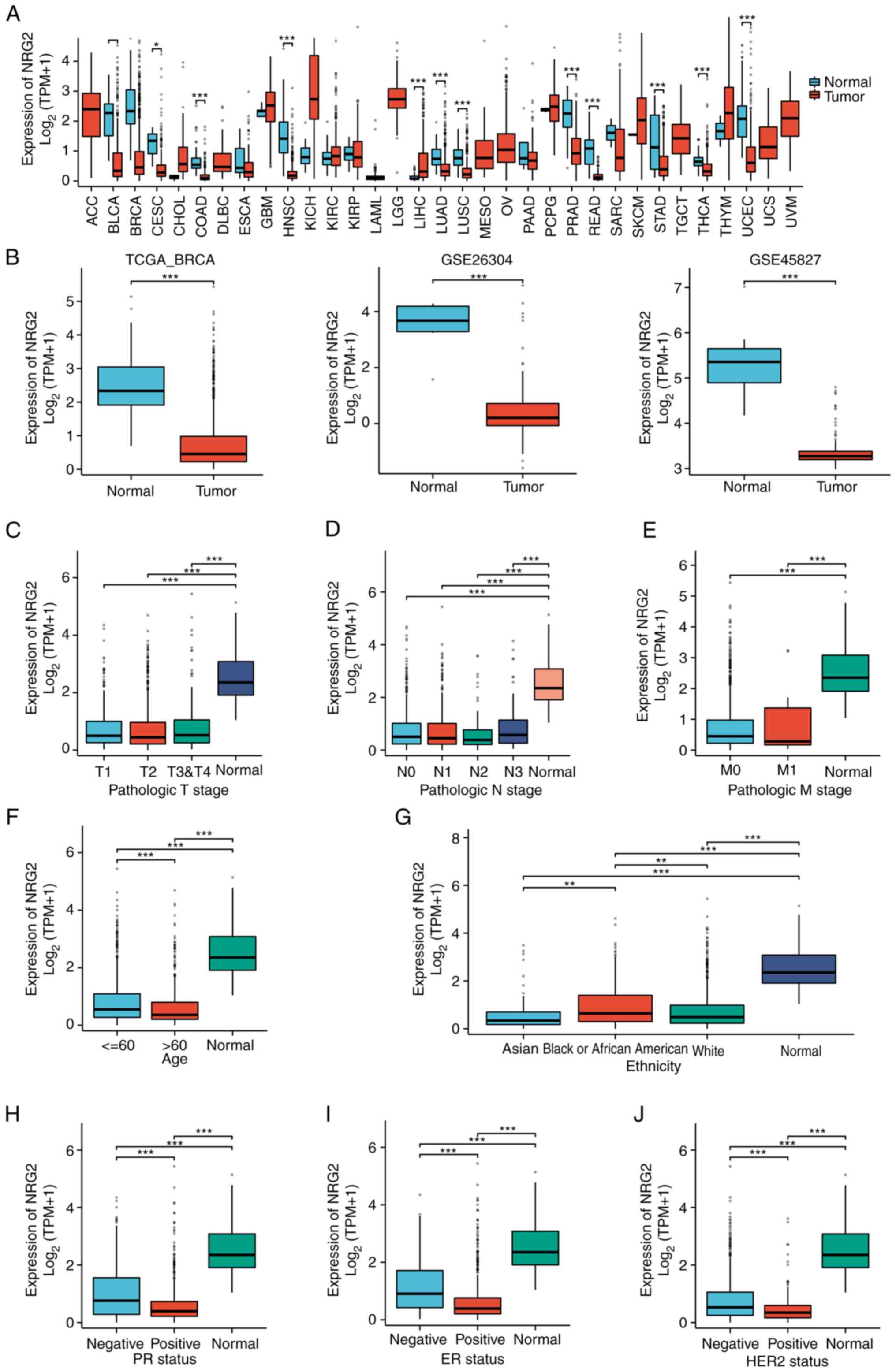
Expression of NRG2 in pan-cancer and
BRCA
In light of the significance of NRG2 in the
autophagy-related prognosis model of BRCA, a comprehensive
investigation of NRG2 was conducted. The pan-cancer expression
analysis showed that NRG2 expression was significantly lower than
normal tissue in cancers such as BLCA, BRCA, CESC and COAD
(P<0.01; Fig. 2A). The
transcriptional mRNA level of NRG2 in BRCA was analyzed based on
TCGA and GEO data, with a significant down-regulation in BRCA
tissues compared with normal tissues found (P<0.001; Fig. 2B). Clinical (including
T-classification, M-classification, N-classification, age,
ethnicity, PR, ER and HER2 status of patients) and gene expression
data were extracted from TCGA to explore the expression of NRG2 in
different clinical subgroups of BRCA (Table SII). The results indicated
significant differences in the expression of NRG2 among age and
ethnicity, while no significant variance was observed across stages
of TMN classification (Fig. 2C-G).
In addition, it was found that the expression of NRG2 was
negatively associated with the status of PR, ER and HER2 (Fig. 2H-J). Based on the results, further
analyses were conducted to determine the expression level of NRG2
among distinct subtypes of BRCA, and the significant prognostic
(P<0.05) and diagnostic (AUC=0.972) value of NRG2 was found,
specifically in Luminal B subtype of BRCA comparing with other BRCA
types (Fig. S1).
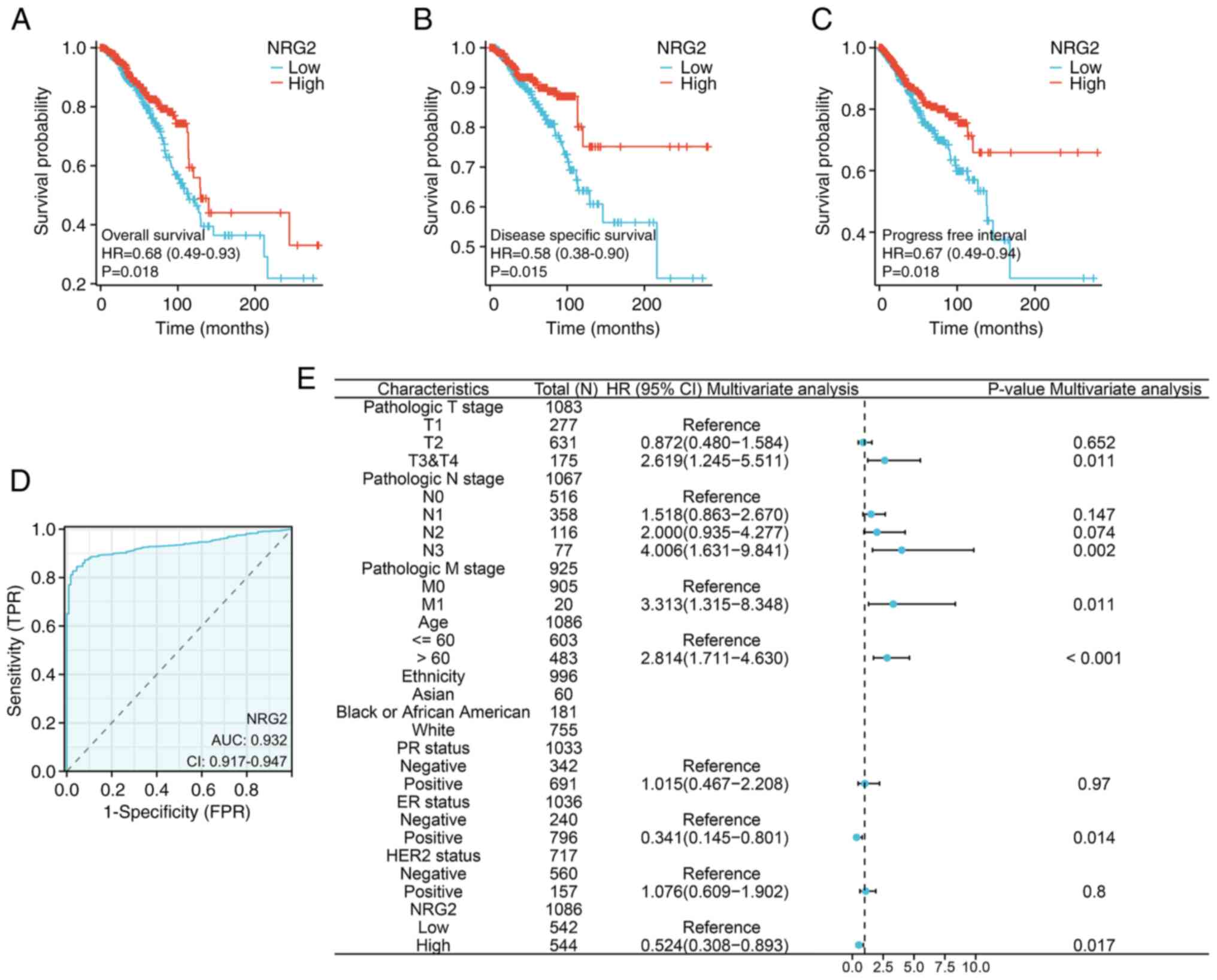
Association between NRG2 expression
and prognosis of patients with BRCA
To determine whether the NRG2 level impacted the
clinical outcomes of patients with BRCA, a prognostic model was
constructed using KM survival curves. The results revealed that
patients with low NRG2 expression had significantly poor OS, PFI
and DSS compared with patients with high NRG2 levels (Fig. 3A-C; P<0.05). Additionally, the
diagnostic value of NRG2 expression was evaluated using a ROC
curve, and the AUC was calculated as 0.932, indicating that NRG2
had high accuracy in predicting outcomes (Fig. 3D). In univariate and multivariate
Cox regression analyses, risk scores were found to be associated
with OS. The multivariable Cox analysis revealed that advanced
tumor stages (T3 & T4), lymph node involvement (N3), distant
metastasis (M1), older age, ER-negative status and low expression
of NRG2 were independent risk factors influencing OS in BRCA
(P<0.05; Fig. 3E).
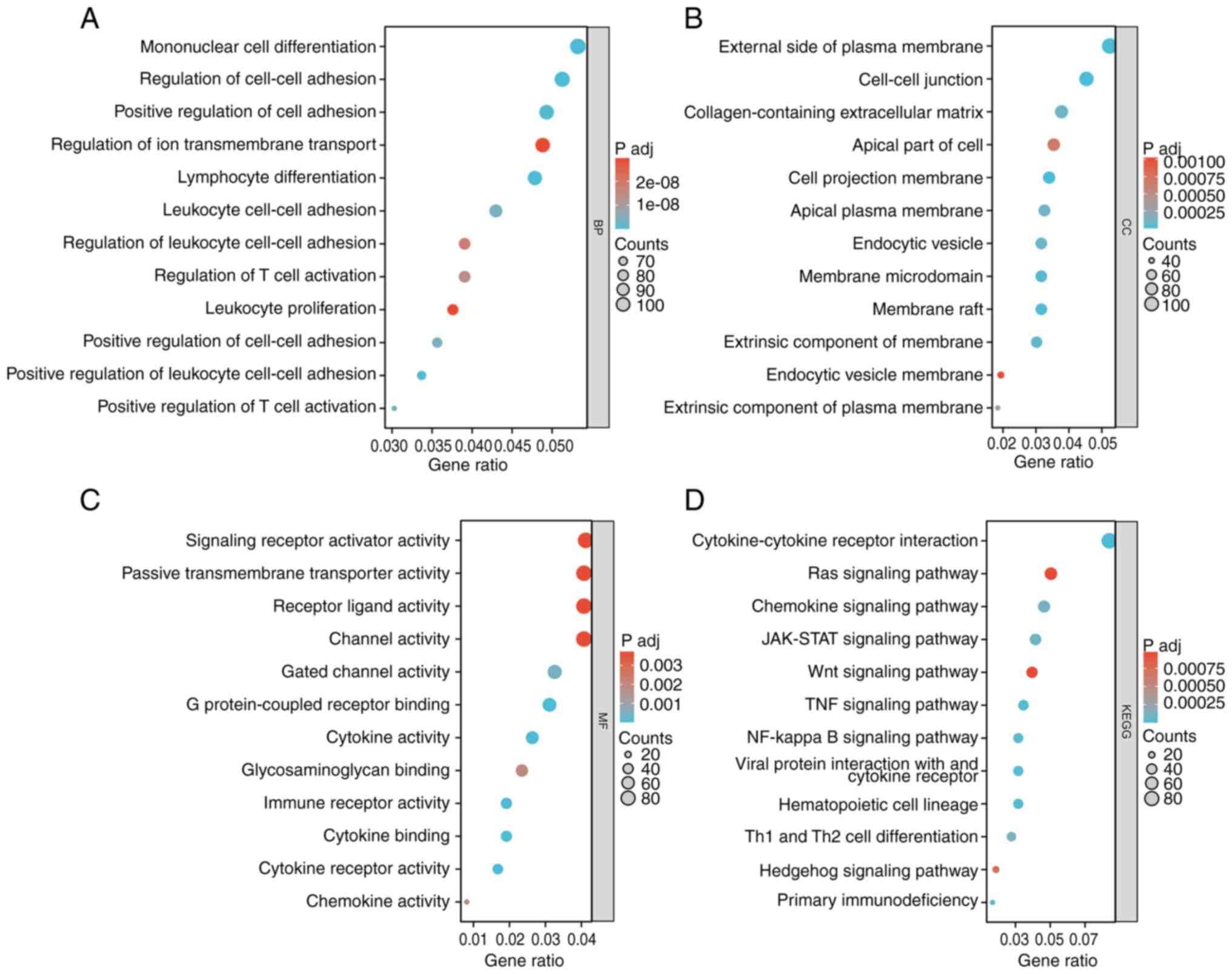
Gene set enrichment analysis of NRG2
related and co-expressed genes
To clarify the function of NRG2, related analysis of
NRG2 was performed using the LinkedOmics database, and a total of
13,455 genes were detected (P<0.01; Fig. S2, Fig.
S3, Fig. S4), in which 833
biological processes (BP), 43 cellular components (CC), 108
molecular functions (MF) and 48 KEGG signaling pathways were
obtained in GO-KEGG enrichment analysis. The enrichment analysis
showed that NRG2 related genes were significantly enriched in BP
such as mononuclear cell differentiation, lymphocyte
differentiation and leukocyte cell-cell adhesion (Fig. 4A); for CC analysis, NRG2 and its
related genes were located in regions such as the external side of
the plasma membrane and cell-cell junctions (Fig. 4B); for MF analysis, NRG2 related
genes were significantly enriched in several functions including
signaling receptor activator activity and passive transmembrane
transport activity (Fig. 4C); KEGG
enrichment analysis showed that NRG2 and its related genes were
mainly enriched in the Ras, JAK-STAT, Wnt, TNF and NF-κB signaling
pathways (Fig. 4D).
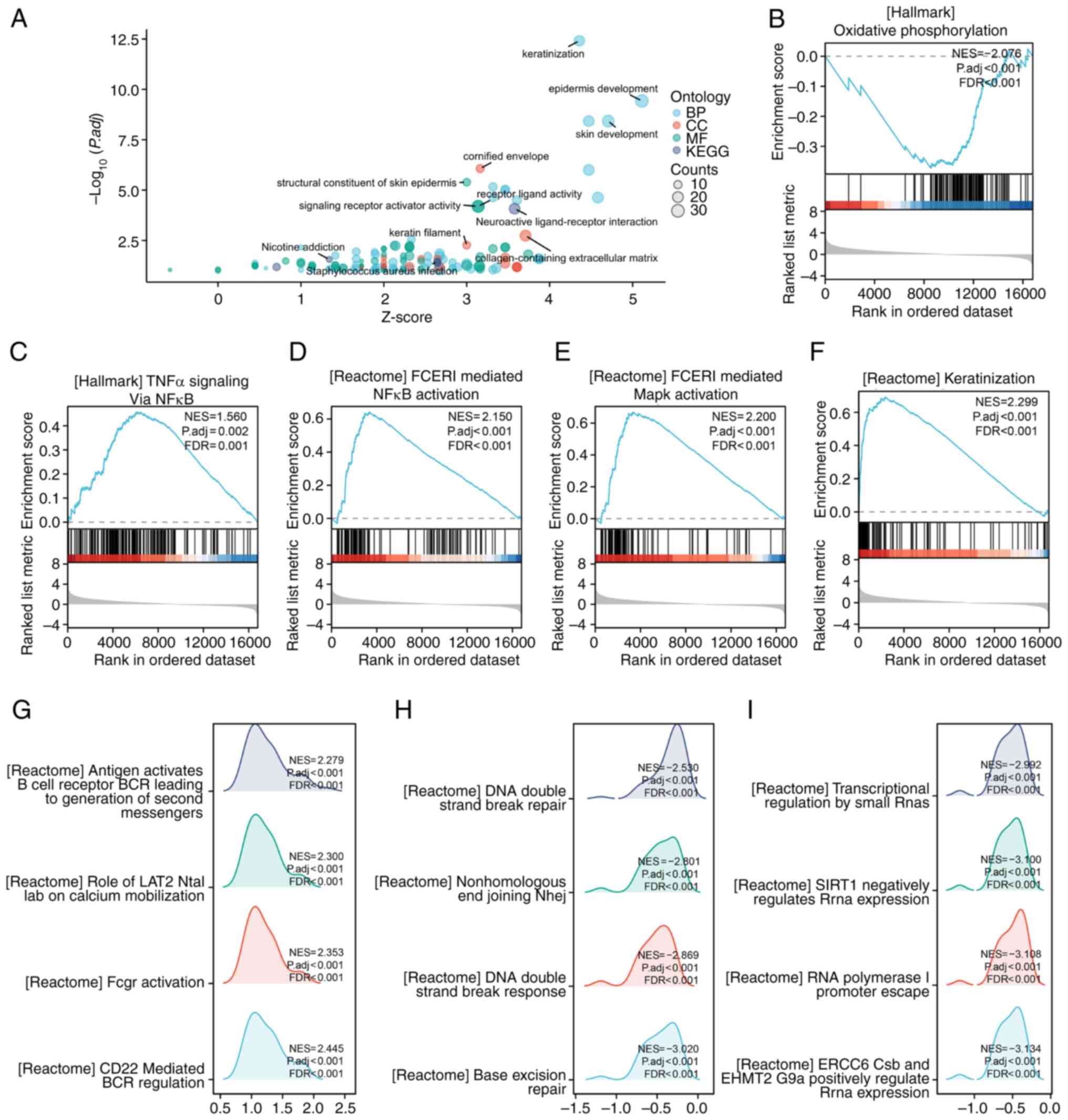
Gene set GO-KEGG enrichment analysis showed that
keratinization, epidermis development and neuroactive
ligand-receptor interaction were significantly enriched in the NRG2
high-expression group (Fig. 5A).
The enriched signaling pathways in the NRG2 high/low expression
group were identified using the GSEA assay. GSEA analysis revealed
a significant enrichment of keratinization, TNFα signaling via
NF-κB, FCERI mediated MAPK activation and FCERI mediated NF-κB
activation in the NRG2 high expression group, while oxidative
phosphorylation was enriched in the NRG2 low expression group
(Fig. 5B-F). Several representative
pathways with high GSEA scores were selected and it was found that
co-expression genes of NRG2 were involved in relevant processes
such as immune response, nuclear DNA repair and
transcription-related pathways (Fig.
5G-I).
Relationship between NRG2 expression
and immune indices
Given the association between NRG2 and immunity,
correlation analyses between the expression levels of NRG2 and
immune infiltration matrix data in BRCA were performed. The
majority of immune cell infiltration was positively related with
NRG2 expression, while Th2 and Th17 cells were negatively related
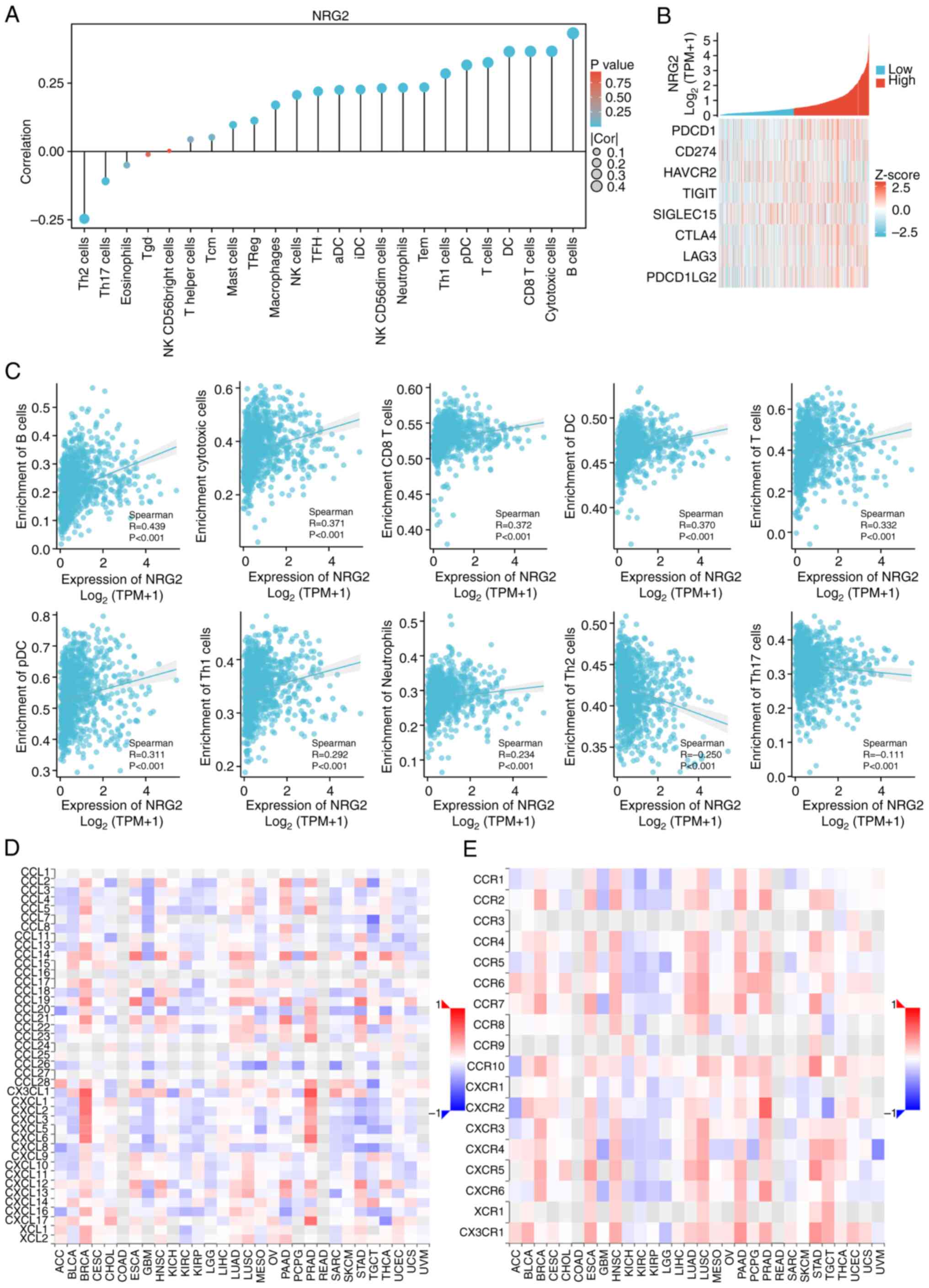
with NRG2 levels (Fig. 6A and C).
Considering that NRG2 may act as a potential tumor suppressor gene
in BRCA, the relationship between NRG2 and immune checkpoints
(PDCD1, CD274, HAVCR2, TIGIT, SIGLEC15, CTLA4, LAG3 and PDCD1LG2)
were further evaluated. NRG2 was found to be positively correlated
with the expression levels of the majority of immune checkpoints
(P<0.001; Fig. 6B). Moreover, it
was further validated that NRG2 may positively regulate the
chemokine-like CX3CL1 and CXCL family, specifically CXCL1-6, and
may be positively associated with molecules including CCR2, CCR7
and CCR10 (Fig. 6D-E). These
findings suggested that anti-tumoral immunity and immune escape may
be involved in NRG2-mediated BRCA carcinogenesis.
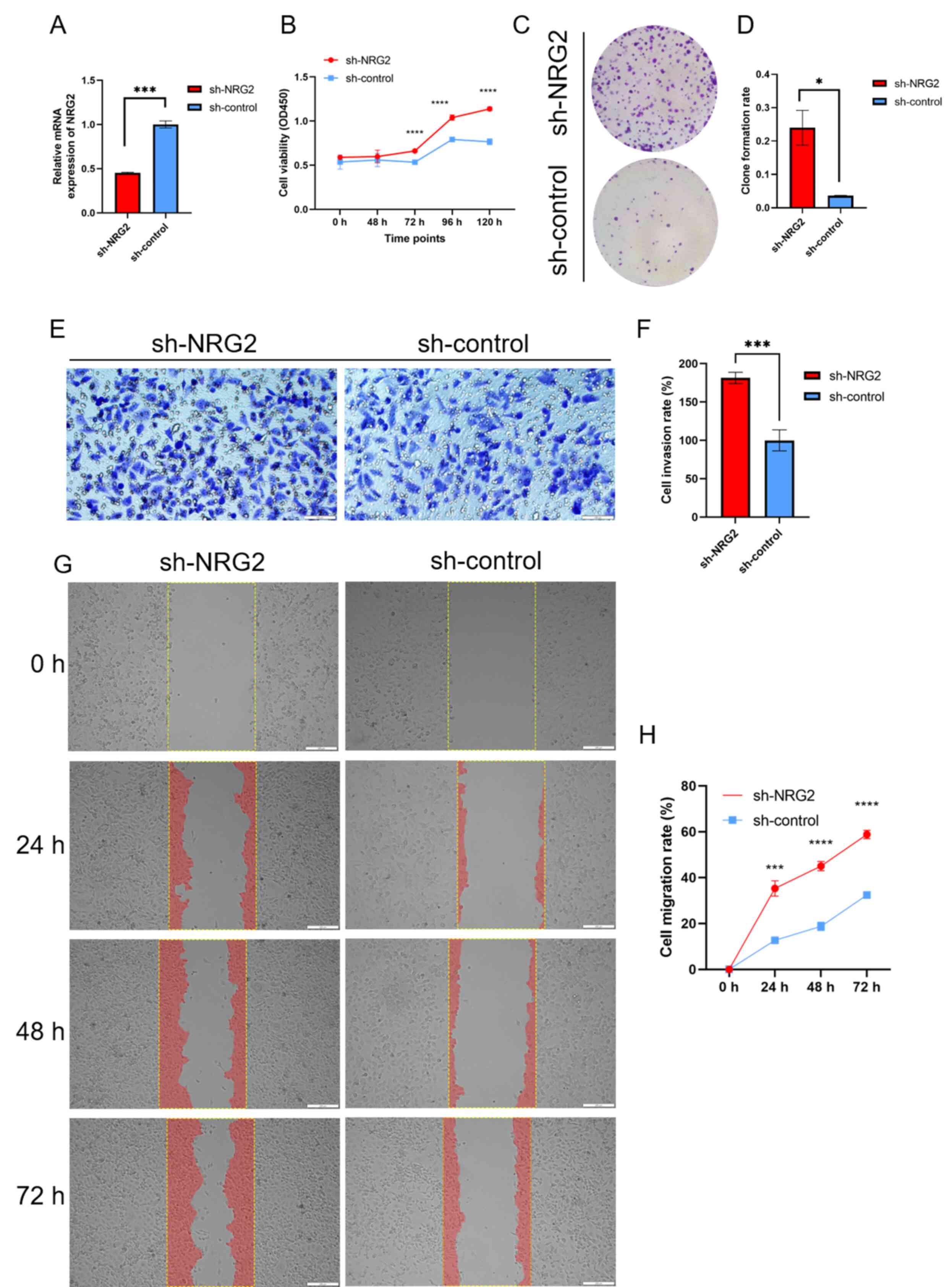
NRG2 knockdown facilitates cell
proliferation, invasion and migration of MDA-MB-231 cells
Based on bioinformatics analysis of the NRG2 gene,
in vitro experiments were conducted to investigate its tumor
inhibition effect in BRCA cells. The results of siRNA transient
transfection revealed that siRNA-1 showed the best knockdown
efficiency compared with the other two sequences, and was thus
selected for subsequent stable cell screening (data not shown). The
stable MDA-MB-231 cells with sh-NRG2 or sh-control were validated
by RT-qPCR assay (Fig. 7A). CCK8
and colony formation assay showed that silencing of NRG2
significantly enhanced the proliferation ability of MDA-MB-231
cells (Fig. 7B-D). Transwell assay
demonstrated a significant promotion in invasion capacity when the
expression of NRG2 was decreased compared with the control group
(Fig. 7E and F). Migration
capability was quantified by calculating the area of cells
migrating into the scratched part at 0, 24, 48 and 72 h (Fig. 7G-H), and the results revealed that
the migration ability was significantly enhanced in the NRG2
knockdown group compared with the sh-control cells.
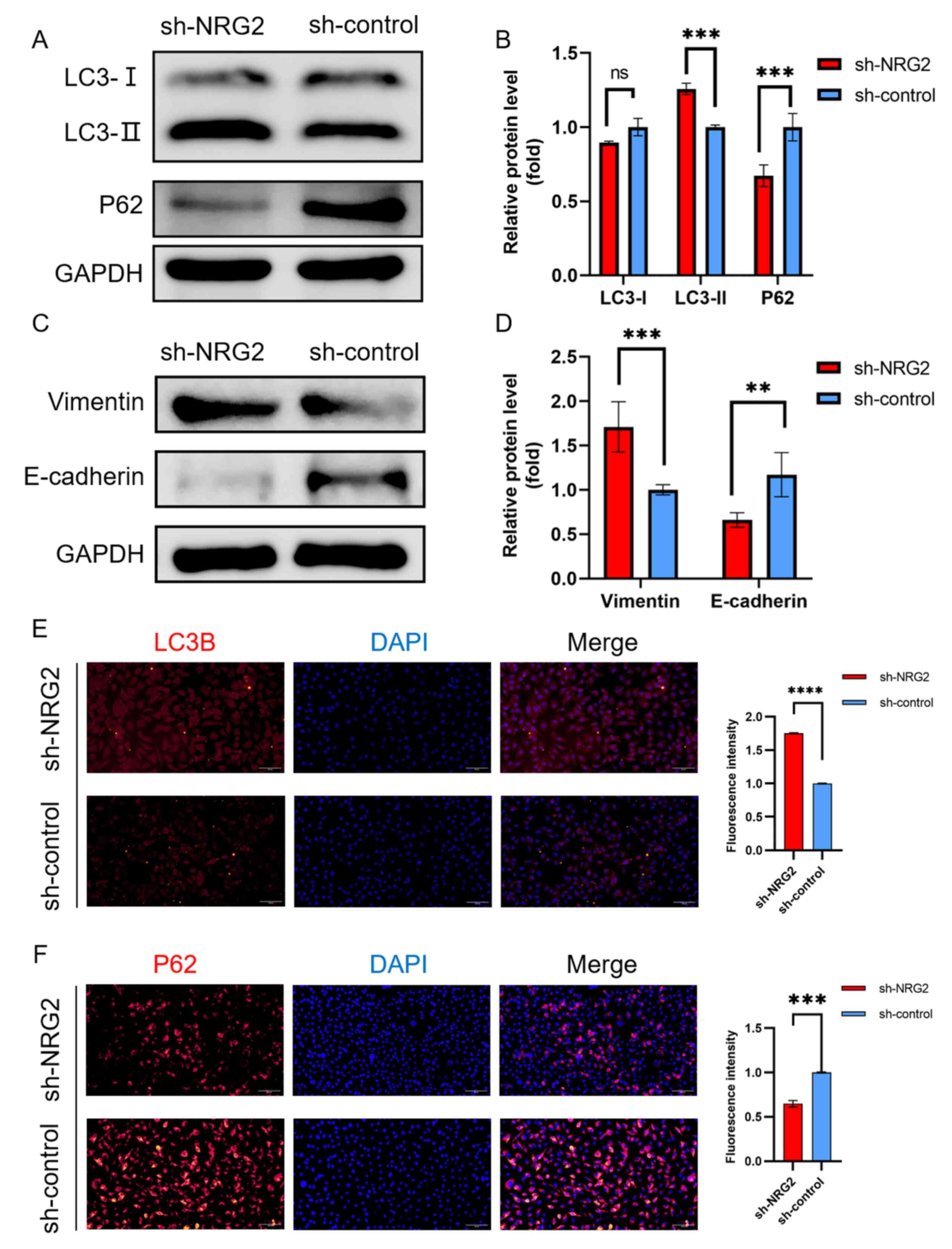
NRG2 knockdown promotes autophagy and
EMT
Autophagic activity was evaluated by measuring
autophagic protein expression (LC3 and P62) using western blotting
and immunofluorescence. Elevated LC3-II and consumed P62 indicated
an increased initiation of autophagy in response to NRG2 knockdown
(Fig. 8A, B, E and F). Mesenchymal
and epithelial state markers of EMT, vimentin and E-cadherin
expression levels were detected to assess the EMT process. The
results showed that upregulated vimentin and downregulated
E-cadherin were identified in MDA-MB-231cells with sh-NRG2
transfection compared with the sh-control cells (Fig. 8C and D).
Discussion
BRCA is the predominant malignancy affecting women
globally and exhibits the highest fatality rate in female patients.
ATGs exert a dual influence on BRCA, as shown in previous studies
where certain ATGs have been implicated in inhibiting
tumorigenesis, whereas others have been found to promote tumor
characteristics in malignant mammary cells (35–38).
Considering the important role of autophagy in BRCA, an
autophagy-related prognosis model of BRCA in TCGA dataset was
constructed, and it was found that NRG2 served as a hub gene
through the PPI network of DEATGs.
In the present study, a comprehensive bioinformatics
analysis was performed to explore the potential regulatory pathways
and biological functions of NRG2 in BRCA. Additionally, the effect
of NRG2 on the malignant characteristics of BRCA was verified by
experiments. According to the findings, NRG2 was significantly
under-expressed in patients with BRCA and may change during the
early stages of BRCA. Survival analysis results revealed that the
OS, PFI and DSS rate of patients with high NRG2 expression was
significantly greater than that of low levels. The ROC curve
indicated that low NRG2 expression has a good diagnostic value.
Finally, the results of the multiple Cox regression analyses showed
that low NRG2 expression is an independent risk factor for OS. The
aforementioned findings indicated that NRG2 may serve as a
prognostic and diagnostic biomarker for BRCA. Furthermore, the
lowest expression of NRG2 was observed in the luminal B subtype
comparing with luminal A, Her2 and basal subtypes, and the overall
survival and ROC curve analysis revealed compelling evidence
supporting the significant diagnostic and prognostic value of NRG2
specifically in the luminal B subtype.
NRG2 expression was positively correlated with most
immune cells such as B cells, cytotoxic cells, CD8+ T
cells, dendritic cells, T cells and neutrophils, which were reduced
in tumor immune microenvironments when NRG2 expression decreased.
The inhibition of CD22 mediated B-cell receptor (BCR) regulation
and antigen activated BCR leading to generation of second
messengers in GSEA enrichment analysis may cause decrease B cell
activation, B cell antigen receptor-induced proliferation and B
cell turnover rates (39,40). GO-KEGG analysis also showed that the
NRG2 related genes were enriched in regulation of T cell activation
and chemokine activity. The TISIDB demonstrated that NRG2 might
positively regulate the CX3CL1 and CXCL family especially CXCL1-6.
The main function of CXCL1-6 is recruitment of immunocytes,
especially neutrophils (41). The
decrease of recruitment of immunocytes induced by NRG2 knockdown in
BRCA also reduced immune infiltration level. Inhibiting of
anti-tumoral immunity and enhancing of immune escape may be
strongly correlated with oncogenic processes mediated by NRG2.
In the present study, the functional analysis in
MDA-MB-231 cells demonstrated that NRG2 silencing significantly
contributed to the malignant characteristics of tumor cells
including cell proliferation, migration and invasion. Both the
processes of autophagy and EMT are crucially involved in the
invasion and metastasis of cancer cells. On one hand, autophagy
provides energy vital to cancer cells and can widely modulate the
EMT process. On the other hand, EMT can regulate autophagy via
pathways such as WNT and NF-κB (11). The occurrence of autophagy was
observed through the conversion of LC3-I to LC3-II, as well as the
degradation of p62. EMT was indicative by decreased epithelial
indicator E-cadherin and increased mesenchymal marker vimentin.
From the enrichment results of GO-KEGG in the
present study, various pathways that are highly related to the
occurrence and development of cancer were detected, such as Ras,
JAK-STAT, Wnt, TNF, NF-κB and Hedgehog signaling pathway. Several
investigations have demonstrated an elevated level of autophagy in
cells with RAS-activating mutations which promote tumor growth,
survival and oncogenesis, and are linked to the progression of
certain lethal cancers (42–44).
The cross-regulatory relationships between Hedgehog, Wnt and NF-κB
pathways regulate the expression and function of EMT-inducing
transcription factors, and in turn, affect basic cellular
mechanisms such as proliferation, differentiation and survival
(45–47).
GSEA enrichment results showed that transcription
and DNA repair were enriched in the low-NRG2 expression group while
the MAPK and NF-κB signaling pathways were enriched in high-NRG2
expression group. Copetti et al (48) showed that NF-κB can induce autophagy
by transactivating Beclin-1, while autophagy regulation by MAPK has
similarities with NF-κB. Additionally, Xu et al (49) proved that NF-κB and MAPK inhibitors
upregulated LC3-II mRNA expression and sustained autophagy.
Therefore, the autophagy mediated by knockdown of NRG2 may be
caused by inhibiting the MAPK and NF-κB signaling pathway.
Although the present study has made noteworthy
contributions in elucidating the role of NRG2 in BRCA, it does have
the following limitations. First, the data used for analysis was
from public databases. As such, although the available data was
meaningful and contributes to the knowledge of the biofunction of
NRG2, its function still needs further verification through in
vitro and in vivo experiments. Furthermore, although the
present study proved that NRG2 can promote autophagy and EMT in
BRCA through bioinformatics combined with experiments, its exact
mechanism and its role in other cell lines needs to be investigated
in further studies.
In summary, the present study found that NRG2 is an
autophagy-related prognostic biomarker, which is significantly
associated with an improved prognosis. Downregulation of NRG2 may
be strongly associated with oncogenic processes involving the
inhibition of anti-tumor immunity and enhancement of immune
evasion. The NRG2 gene may act as a tumor suppressor factor that
inhibits cell proliferation, invasion and migration by regulating
the pathological process of autophagy and EMT, suggesting that NRG2
could be used as a prognostic marker for clinical therapy of
BRCA.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Yuan Cao at The
Medicine & Sciences Analysis Center of Wuhan University of
Science and Technology (Wuhan, China) for their help with
immunofluorescence imaging and analysis. The authors would also
like to thank Dr Hui Li at Tianyou Hospital (Wuhan, China) for
assisting in the preparation of materials for ethics approval.
Funding
This study was supported by the Foundation of Hubei Province
Supporting Enterprise Technology Innovation Development (grant no.
2021BAB126), Wuhan East Lake High-tech Zone ‘JieBangGuaShuai’
Project (grant no. 2022KJB113) and Foundation of Wuhan University
of Science and Technology (grant no. 2016×z036).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RJZ and JJD executed the project, analyzed the
bioinformatics data and wrote the original manuscript draft; RLZ
carried out the data curation; MYW and XTD were responsible for
methodology optimization, the analysis of experimental data and
construction of figures; QZ, ZRW and FL performed the in
vitro experiments using cell lines; DY assisted with the design
of clinical experiment and revision of the manuscript; YX designed
the whole project and critically revised the manuscript. RJZ, JJD
and YX confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Islami F, Siegel RL, Ward EM and
Jemal A: Global Cancer in Women: Burden and Trends. Cancer
Epidemiol Biomarkers Prev. 26:444–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang L, Chen W, Liu S and Chen C:
Targeting breast cancer stem cells. Int J Biol Sci. 19:552–570.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yun CW and Lee SH: The roles of autophagy
in cancer. Int J Mol Sci. 19:34662018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levine B and Kroemer G: Biological
functions of autophagy genes: A disease perspective. Cell.
176:11–42. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jain V, Singh MP and Amaravadi RK:
Amaravadi, Recent advances in targeting autophagy in cancer. Trends
Pharmacol Sci. 44:290–302. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Debnath J, Gammoh N and Ryan KM: Autophagy
and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol.
24:560–575. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gundamaraju R, Lu W, Paul MK, Jha NK,
Gupta PK, Ojha S, Chattopadhyay I, Rao PV and Ghavami S: Autophagy
and EMT in cancer and metastasis: Who controls whom? Biochim
Biophys Acta Mol Basis Dis. 1868:1664312022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Si L and Yang Z: Regulatory effects of
lncRNAs and miRNAs on the crosstalk between autophagy and EMT in
cancer: A new era for cancer treatment. J Cancer Res Clin Oncol.
148:547–564. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Babaei G, Aziz SG and Jaghi NZZ: EMT,
cancer stem cells and autophagy; The three main axes of metastasis.
Biomed Pharmacother. 133:1109092021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akalay I, Janji B, Hasmim M, Noman MZ,
Thiery JP, Mami-Chouaib F and Chouaib S: EMT impairs breast
carcinoma cell susceptibility to CTL-mediated lysis through
autophagy induction. Autophagy. 9:1104–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Lu C, Wang F, Guo H, Wang Z, Yin H
and Li J: Heat treatment-induced autophagy promotes breast cancer
cell invasion and metastasis via TGF-β2-mediated
epithelial-mesenchymal transitions. PeerJ. 11:e146402023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marshall C, Blackburn E, Clark M,
Humphreys S and Gullick WJ: Neuregulins 1–4 are expressed in the
cytoplasm or nuclei of ductal carcinoma (in situ) of the human
breast. Breast Cancer Res Treat. 96:163–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karthaus WR and Hofree M: Regenerative
potential of prostate luminal cells revealed by single-cell
analysis. Science. 368:497–505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trombetta D, Sparaneo A, Fabrizio FP, Di
Micco CM, Rossi A and Muscarella LA: NRG1 and NRG2 fusions in
non-small cell lung cancer (NSCLC): Seven years between lights and
shadows. Expert Opin Ther Targets. 25:865–875. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao WJ, Yi SJ, Ou GY and Qiao XY:
Neuregulin 2 (NRG2) is expressed in gliomas and promotes migration
of human glioma cells. Folia Neuropathol. 59:189–197. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li F, Shang Y, Zhang H, She J, Wang G and
Sun Q: Development of a novel autophagy-related gene prognostic
signature for gastric cancer. Transl Cancer Res. 10:2790–2800.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu D, Jiang L, Luo S, Zhao X, Hu H, Zhao G
and Tang W: Development of an autophagy-related gene expression
signature for prognosis prediction in prostate cancer patients. J
Transl Med. 18:1602020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sepulveda JL: Using R and bioconductor in
clinical genomics and transcriptomics. J Mol Diagn. 22:3–20. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muggerud AA, Hallett M, Johnsen H, Kleivi
K, Zhou W, Tahmasebpoor S, Amini RM, Botling J, Børresen-Dale AL,
Sørlie T and Wärnberg F: Molecular diversity in ductal carcinoma in
situ (DCIS) and early invasive breast cancer. Mol Oncol. 4:357–368.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gruosso T, Mieulet V, Cardon M, Bourachot
B, Kieffer Y, Devun F, Dubois T, Dutreix M, Vincent-Salomon A,
Miller KM and Mechta-Grigoriou F: Chronic oxidative stress promotes
H2AX protein degradation and enhances chemosensitivity in breast
cancer patients. EMBO Mol Med. 8:527–549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.PubMed/NCBI
|
|
30
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ru B, Wong CN, Tong Y, Zhong JY, Zhong
SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, et al: TISIDB: An
integrated repository portal for tumor-immune system interactions.
Bioinformatics. 35:4200–4202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niklaus NJ, Tokarchuk I, Zbinden M,
Schläfli AM, Maycotte P and Tschan MP: The multifaceted functions
of autophagy in breast cancer development and treatment. Cells.
10:14472021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin L, Chen Y, Cheng D, He Z, Shi X, Du B,
Xi X, Gao Y and Guo Y: YAP inhibits autophagy and promotes
progression of colorectal cancer via upregulating Bcl-2 expression.
Cell Death Dis. 12:4572021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu X, Ma B, Chen M, Zhang Y, Ma Z and
Chen H: Prognostic Autophagy-Related genes of gastric cancer
patients on chemotherapy. Front Genet. 12:7208492021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ding J, Wang C, Sun Y, Guo J, Liu S and
Cheng Z: Identification of an Autophagy-Related signature for
prognosis and immunotherapy response prediction in ovarian cancer.
Biomolecules. 13:3392023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Poe JC, Fujimoto Y, Hasegawa M, Haas KM,
Miller AS, Sanford IG, Bock CB, Fujimoto M and Tedder TF: CD22
regulates B lymphocyte function in vivo through both
ligand-dependent and ligand-independent mechanisms. Nat Immunol.
5:1078–1087. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Harwood NE and Batista FD: Early events in
B cell activation. Annu Rev Immunol. 28:185–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou C, Gao Y, Ding P, Wu T and Ji G: The
role of CXCL family members in different diseases. Cell Death
Discov. 9:2122023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gonçalves PR, Rocha-Brito KJ, Fernandes
MR, Abrantes JL, Durán N and Ferreira-Halder CV: Violacein induces
death of RAS-mutated metastatic melanoma by impairing autophagy
process. Tumour Biol. 37:14049–14058. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su H, Yang F, Fu R, Li X, French R, Mose
E, Pu X, Trinh B, Kumar A, Liu J, et al: Cancer cells escape
autophagy inhibition via NRF2-induced macropinocytosis. Cancer
Cell. 39:678–693.e11. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee JJ and Jain V: Clinical translation of
combined MAPK and autophagy inhibition in RAS mutant cancer. Int J
Mol Sci. 22:124022021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Malla RR and Kiran P: Tumor
microenvironment pathways: Cross regulation in breast cancer
metastasis. Genes Dis. 9:310–324. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mirzaei S, Saghari S, Bassiri F, Raesi R,
Zarrabi A, Hushmandi K, Sethi G and Tergaonkar V: NF-κB as a
regulator of cancer metastasis and therapy response: A focus on
epithelial-mesenchymal transition. J Cell Physiol. 237:2770–2795.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Copetti T, Bertoli C, Dalla E, Demarchi F
and Schneider C: p65/RelA modulates BECN1 transcription and
autophagy. Mol Cell Biol. 29:2594–2608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu K, Chen W, Wang X, Peng Y, Liang A,
Huang D, Li C and Ye W: Autophagy attenuates the catabolic effect
during inflammatory conditions in nucleus pulposus cells, as
sustained by NF-κB and JNK inhibition. Int J Mol Med. 36:661–668.
2015. View Article : Google Scholar : PubMed/NCBI
|