Introduction
Gliomas are aggressive brain tumors that vary in
incidence based on patient demographics and location, typically
affecting 2–10 individuals per 100,000 in the population. Their
malignant and invasive nature poses significant health challenges
as high recurrence rates after radiotherapy and chemotherapy can
result in high mortality rates (1,2).
Although early diagnosis and surgery can improve the survival rate
of patients (3), high-grade
gliomas, especially glioblastomas (GBM, Grade IV), have low
survival rates (4). Despite
considerable treatment efforts, GBM remains the most common and
aggressive form of brain tumor in humans (5). Considering that GBM is associated with
high incidence, recurrence and mortality rates (6) there is a clear need for increased
research efforts to identify prognostic markers and effective drugs
that can improve early diagnosis and treatment.
F-box and leucine-rich repeat protein 6 (FBXL6) is a
member of the leucine-rich protein family. FBXL6 serves a crucial
role in phosphorylation-dependent ubiquitination, which is
essential for cell development and differentiation (7). The ubiquitin-proteasome system (UPS)
is an essential component of post-translational modifications and
serves a critical role in various cellular activities, such as the
cell cycle (8), apoptosis (9), DNA damage repair (10), immune response (11) and tumor development (12). Consequently, the study of the UPS in
tumors has received increasing attention (13,14).
Ubiquitination involves a three-enzyme cascade consisting of E1
(Ub-activating), E2 (Ub-conjugating) and E3 (Ub-ligase) enzymes
(15). Notably, a number of F-box
proteins, which serve as substrate-recognition subunits within
Skp1-cullin-F-box protein E3 ligase complexes, serve a crucial role
in various cellular processes (16,17).
Specifically, they mediate the ubiquitination and subsequent
degradation of target proteins (18), primarily influencing tumor
development through substrate turnover (19,20).
FBXL6 activates the estrogen receptor (ER) by promoting
transcription and mediating protein degradation. This highlights
the significance of FBXL6 in the modulation of ER activity and the
potential for targeted FBXL6-based strategies in the treatment of
ER-related cancers (21). Chan
et al (16) previously
reported that FBXL6 serves a critical role in human tumor
development and acts as a distinct prognostic marker for malignant
progression in renal cell carcinoma (22,23).
In liver cancer, the accumulation of FBXL6 promotes the
stabilization and activation of c-Myc by preventing the degradation
of HSP90AA1. Activated c-Myc, in turn, binds directly to the
promoter region of FBXL6, inducing its mRNA expression (24). In colorectal cancer, FBXL6 is highly
expressed and is associated with poor prognosis. It interacts with
phospho-p53 (S315), facilitating polyubiquitination at K291/292 and
consequently inhibiting the signal transduction of P53 (22). Despite these aforementioned
findings, there are few reports on the impact of FBXL6 expression
on gliomas and its biological function remains largely unexplored.
Therefore, further investigation is necessary to determine the
prognostic potential of FBXL6 and drug sensitivity in patients with
glioma.
Building on previous studies, we investigated the
specific role of FBXL6 in gliomas. By utilizing The Cancer Genome
Atlas (TCGA), Gene Expression Profiling Interactive Analysis (GEPI)
and Chinese Glioma Genome Atlas (CGGA) databases, the present study
comprehensively analyzed the relationship between prognostic value,
drug sensitivity and FBXL6 expression in glioma. In addition, the
expression and prognostic significance of FBXL6 in gliomas was
examined using various methods such as western blot, reverse
transcription-quantitative PCR and immunohistochemistry (IHC). The
present study may offer novel insights to guide future clinical
approaches to treating human gliomas.
Materials and methods
Data and preprocessing
Gene expression data and complete clinical
annotations were obtained from the CGGA (cgga.org.cn/) and TCGA
(https://portal.gdc.cancer.gov/)
databases. Patient data included the age, sex, radiotherapy and
chemotherapy statuses of patients, complete follow-up information,
histopathological classification and primary/recurrent status of
World Health Organization (WHO) malignant gliomas. In the present
study, the mRNAseq_693 and mRNAseq_325 glioma cohorts were
analyzed. Tumor-immune system interactions and drug bank database
(https://www.drugbank.ca/) analysis was used to
determine the correlation between target genes and lymphocytes,
immune regulators and immune checkpoints in gliomas.
Differential expression and prognostic
analysis of FBXL6 in gliomas
The expression levels of FBXL6 in gliomas were
analyzed using data from the TCGA database. The ‘limma’ package in
the R platform was used to analyze the differential expression of
FBXL6 between normal and tumor tissues in gliomas. Low-grade
gliomas typically refer to WHO grade I or II tumors, which tend to
grow slowly, have distinct borders and invade surrounding tissues
to a lesser extent. Patients with low-grade gliomas generally have
a better prognosis (25).
Conversely, high-grade gliomas usually refer to WHO grade III or IV
tumors and are characterized by rapid growth, indistinct borders
and high invasiveness. Patients with high-grade gliomas typically
have a poorer prognosis (26).
Visualization analysis was performed using the ‘ggplot2’ and
‘ggpubr’ packages in R. Additionally, the prognostic value of FBXL6
in gliomas was analyzed using the TCGA and CGGA databases.
Clinical feature correlation analysis
and nomogram construction
The CGGA database provided detailed clinical data,
including patient age, sex, radiotherapy and chemotherapy status,
complete follow-up information, histological classification and
primary/recurrent status of WHO malignant gliomas. TCGA clinical
data included sex, age and tumor grade. Therefore, a detailed
analysis of the correlation between FBXL6 expression and clinical
features using was performed using bioinformatic tools. Receiver
operating characteristic (ROC) curves were used to determine the
accuracy of FBXL6 expression in predicting 1-, 3- and 5-year
survival rates in patients with glioma. To construct a nomogram
using both FBXL6 expression data and clinical data, the ‘rms,’
‘survival,’ and ‘regplot’ packages in R were used to assess the
impact of age, sex and tumor grade on patients with glioma.
Clinical decision curve analysis was used to evaluate the accuracy
of the 1-, 3- and 5-year survival rate predictions in patients with
glioma. Additionally, the reliability of the nomogram was assessed
using calibration curves.
Correlation between genes and
enrichment analyses
To investigate the specific mechanisms underlying
FBXL6 expression in gliomas, gene correlation and enrichment
analyses were performed. For the correlation analysis, the
filtering criteria were R≥0.6 and P<0.001. To explore the
biological processes and pathways related to the correlated genes
in gliomas, enrichment analyses were performed using the Gene
Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway and Gene Set Enrichment Analysis (GSEA) databases.
Correlation between immune
microenvironment, immune cell infiltration and FBXL6
expression
The role of FBXL6 expression in immune cell
infiltration was investigating by using the CiberSort algorithm to
study the distribution of 22 tumor-infiltrating immune cells in
high and low FBXL6 expression groups. Furthermore, Spearman
correlation analysis was conducted to explore the strength of the
association between FBXL6 expression and the 22 types of
immune-infiltrating cells. Immune and stromal scores were
calculated using estimation methods to reflect the relationship
between FBXL6 expression and the immune microenvironment. TIMER2
(http://timer.cistrome.org/) analysis
FBXL6 expression in different correlation between immune cells and
glioma subtypes.
Drug sensitivity analysis
With the advancement of precision medicine, the
demand for personalized treatment has increased. The sensitivity of
gene expression and drugs is a critical factor in personalized
treatment (27). Robust prediction
of in vivo chemotherapy responses by collecting pretreatment
baseline gene expression levels and drug sensitivity data from
cancer cell lines has been a long-standing and controversial issue
in pharmacogenomics. Cancer cell lines in labs may not accurately
reflect patient tumor complexity, leading to possible mismatches
between predicted and real responses to chemotherapy.
Patient-specific genetic variations and tumor environments also
affect treatment outcomes, challenging the applicability of cell
line data to predict patient responses effectively. Drug response
information and drug-targeting pathways were collected from the
Genomics of Drug Sensitivity in Cancer database (https://www.cancerrxgene.org/). Spearman correlation
analysis was performed to identify drugs related to the risk
score.
Cell culture, western blot and
RT-qPCR
The U251 cell (cat. no. TCHu 58) line was obtained
from the China Center for Typical Culture Collection. LN229 (cat.
no. iCell-h124) and HEB (cat. no. XY-XB-1640) cell lines were
purchased from Shanghai Xuan Ya Biotechnology Co., Ltd. U251 and
LN229 are high-grade glioma cell lines with overexpression of the
TP53 and EGFR genes, which serve crucial roles in the pathogenesis
and progression of gliomas. Additionally, U251 cells also exhibit
specific gene expressions such as neurofibromin, cyclin-dependent
kinase inhibitor 2A and phosphatidylinositol 3,4,5-triphosphate
3-phosphatase and dual-specificity protein phosphatase, which are
associated with the pathogenesis of gliomas. HEB is a normal human
brain glial cell line that exhibits a polygonal shape when adhered
to the matrix, with a more uniform size and a patchy growth pattern
(28). Compared to HEB cells, U251
cells have a higher apoptosis rate and a faster proliferation rate.
Cells were cultured in DMEM supplemented with 10% FBS (Gibco;
Thermo Fisher, Inc.), 100 units/ml penicillin and 100 mg/ml
streptomycin (Gibco; Thermo Fisher, Inc.). To prepare cell samples
for western blot, cells were lysed with RIPA lysis buffer (Beyotime
Institute of Biotechnology). The mass of protein/lane is 150 µg.
Proteins were separated by SDS-PAGE using a XCell SureLock
Mini-Cell (Invitrogen; Thermo Fisher Scientific, Inc.) and
transferred to a PVDF membrane (Invitrogen; Thermo Fisher, Inc.).
The concentration of the separating gel was between 6% and 8%.
Membranes were blocked in 5% non-fat milk in PBS (Invitrogen;
Thermo Fisher, Inc.) at room temperature for 1 h before incubation
with primary antibodies overnight at 4°C. The primary antibodies
used were anti-FBXL6 antibodies (cat. no. PA5-64927; 1:200; Thermo
Fisher, Inc). The membranes were then incubated with the
appropriate HRP-conjugated secondary antibodies (cat. no. C510051;
1:5,000; Shanghai Shenggong Biology Engineering Technology Service,
Ltd.) at 37°C on a shaker for 2 h. Anti-GAPDH antibodies (cat. no.
b181602; 1:2,000; Abcam) were used for the reference protein.
Immunoreactive bands (cat. no. WBKLS0500; Millipore Immobilon
Western, ECL; EMD Millipore, Billerica, MA, USA) were quantified
using ImageJ software (v.1.48; National Institutes of Health,
Bethesda, MD, USA).
RNA was extracted from glioma cells using
TRIzol® reagent (Invitrogen; Thermo Fisher, Inc.) and
quantified using a 725 spectrophotometer (Shanghai Sunny Hengping
Scientific Instrument Co., Ltd.). Oligo dT (Roche Applied Science,
10814270001) was used to prime cDNA synthesis. RT-qPCR was
performed using the SYBR Green Premix Ex Taq (Takara Biotechnology
Co., Ltd) on an Illumina Eco (Illumina, Inc.). Total RNA was
extracted from tissue and cells using Tripure Isolation reagent
(Roche Applied Science; Penzberg; Germany), according to the
manufacturer's instructions. The Transcription First Strand cDNA
Synthesis kit (Roche Applied Science) was used to synthesize cDNA
from 1 µg total RNA at room temperature at 24°C for 50 min.
Differences in gene expression were calculated using the
2−ΔΔCq method (29). The
thermocycling conditions were as follows: Initial denaturation at
94°C for 5 min, followed by 30 cycles at 94°C for 45 sec, 59°C for
45 sec and 72°C for 45 sec, followed by 72°C for 45 sec and final
extension at 72°C for 10 min. Experiments were performed in
triplicate with SYBR Green I Master mix (Roche Applied Science);
GAPDH was used as the internal control. The primers used were as
follows: FBXL6 forward (F), 5′-CATCAACCGTAATAGCATTCCCC-3′ and
reverse (R), 5′-CACATCAGGTTCAACAGCCG-3′ and GAPDH F,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and R,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
IHC
In the present study, paraffin-embedded glioma
specimens and matched adjacent non-tumor tissues were surgically
excised from patients at the Department of Neurosurgery and
forwarded to the Department of Pathology for the pathological
diagnosis of glioma. Sample collection took place from January 2020
to June 2023. Upon confirmation of glioma diagnosis,
paraffin-embedded specimens were stored within the Department of
Pathology. This was a retrospective study and informed consent for
this study was waived because the patients had previously provided
written informed consent for the use of their post-operative
samples and information in future clinical research. The study
protocol, including the use of these tissue samples, was approved
by the Ethics Committee of the Ninth People's Hospital affiliated
with Shanghai Jiao Tong University School of Medicine (approval no.
H9H-2023-T489-1; Shanghai, China). Clinical samples were fixed in
4% paraformaldehyde at room temperature for 24 h, subsequently
embedded in paraffin, and sectioned to a uniform thickness of 3
micrometers. Immunostaining was performed using the two-step
Elivision Plus kit system (Dako; Agilent Technologies, Inc.). The
sections were dewaxed in xylene, rehydrated with a series of
ethanol solutions (100, 95, 80 and 70%), and then boiled in
ethylenediaminetetraacetic acid buffer (pH 9.0) for 20 min in an
autoclave. Next, 0.3% H2O2 was used to block
endogenous peroxidase activity at room temperature for 15 min, and
the sections were incubated with normal goat serum (1:20; Beyotime
Institute of Biotechnology) for 20 min at room temperature to
reduce non-specific binding. Tissue sections were incubated with
the anti-FBXL6 (cat. no. PA5-64927; 1:200; Thermo Fisher) for 50
min at room temperature. The secondary antibody was applied using
the Envision Detection kit (SM802; Dako; Agilent Technologies;
Ready-to-use type) for 20 min at room temperature. Slides were
stained for 2 min with diaminobenzidine tetrahydrochloride (DAB)
and then counterstained 2 min with hematoxylin at room temperature.
The completed sections were mounted using a neutral resin (K-0212,
Shanghai Jiehao Biotechnology Co., Ltd.). The stained tumor cells
were assessed with a Nikon conventional optical microscope in 10
independent fields at magnification, ×400. Immunohistochemical
image results were quantified using ImageJ software (v.1.48;
National Institutes of Health, Bethesda, MD, USA). All the sections
were scored by two independent pathologists who were blinded to the
type of sample. Specifically, the intensity of positive cytoplasmic
staining was rated on a scale of 0–3 (0, negative; 1, light brown;
2, medium brown; and 3, dark brown). The corresponding percentages
of positively stained cells were set as: 1, 1–25%; 2, 26–50%; 3,
51–75%; and 4, 76–100%.
Statistical analysis
All statistical analyses were performed using R
(version 4.1.2; RStudio, Inc.). The results are presented as the
median ± interquartile range or mean ± standard deviation (SD).
Western blot data were analyzed using GraphPad Prism (version 5;
Dotmatics). One way ANOVA was employed to compare the means ± SD
between two groups followed by the Least Significant Difference
posts hoc test. The Chi-square test and Fisher's exact test were
utilized for analyzing qualitative data. For comparisons between
two groups, the Mann-Whitney U test (non-parametric) was used,
while the Kruskal-Wallis test followed by Dunn's test was applied
for comparisons involving >2 groups. The log-rank test and
Kaplan-Meier survival analysis were used to compare prognosis among
the different risk groups. Univariate and multivariate Cox
regression analyses were performed to evaluate the prognostic value
of risk models. The significance of the correlation was assessed
using the Spearman's rank correlation test. All experiments were
validated with three repeated trials. P<0.05 was considered to
indicate a statistically significant difference.
Results
High FBXL6 expression was associated
with poor prognosis in glioma
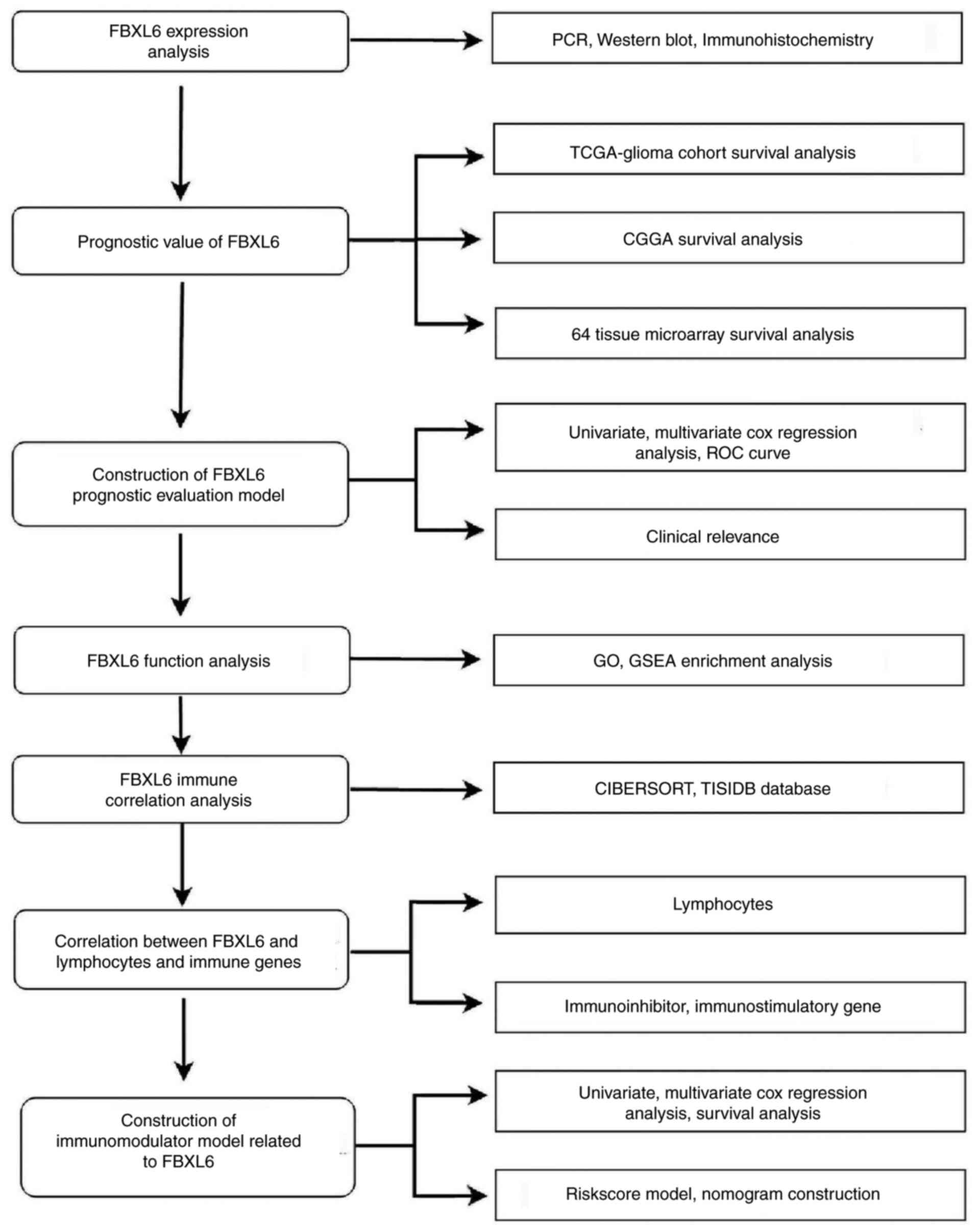
The data collection and analysis performed in the
present study (Fig. 1) identified
key biomarkers in glioma. Multicenter screening and validation were
performed and FBXL6 was identified as a key marker. Compared with
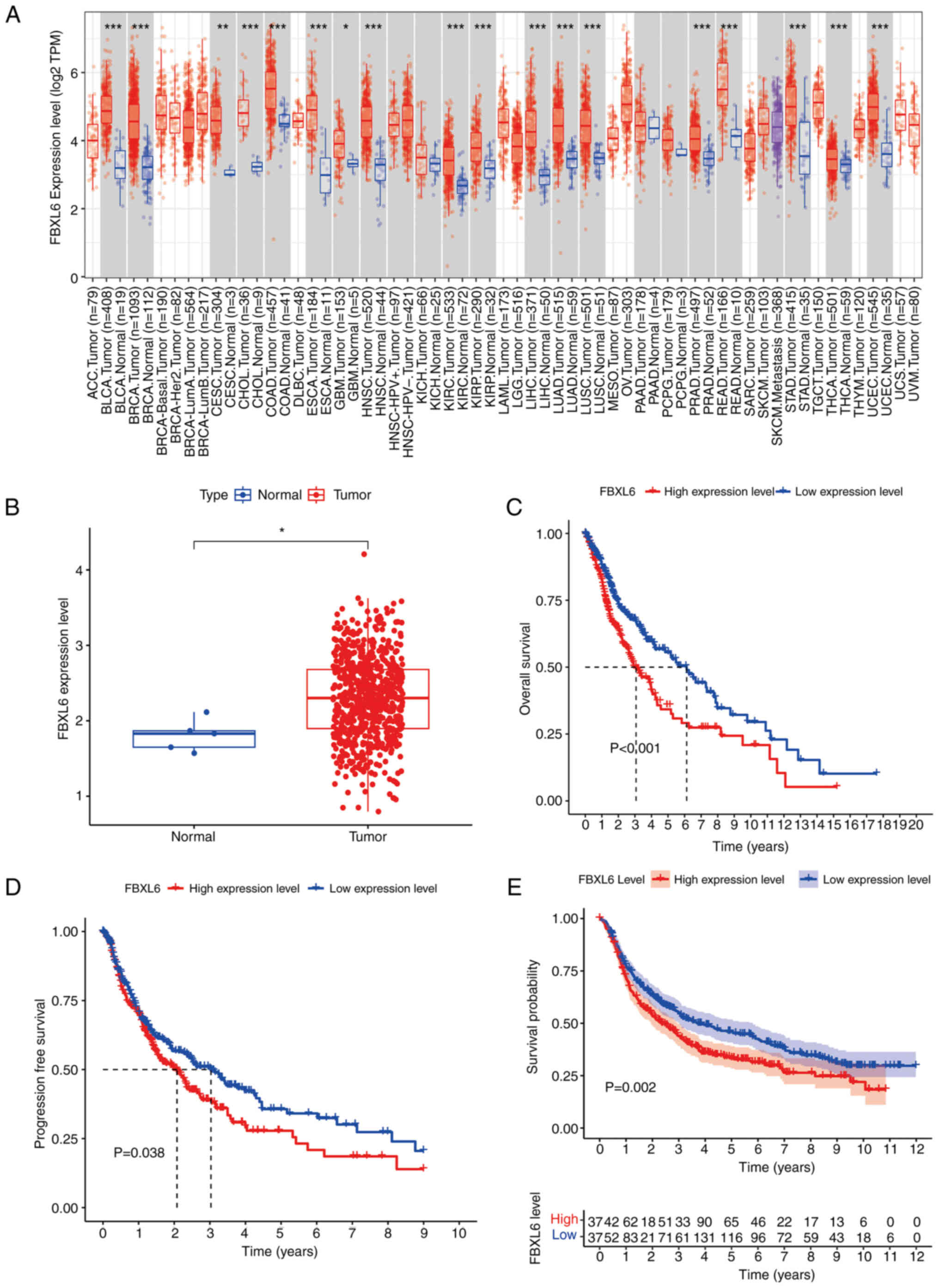
normal tissues, pan-cancer analysis demonstrated that FBXL6 was
highly expressed not only in GBM, but also in certain other
cancers, including bladder, breast and colorectal cancers (Fig. 2A). TCGA-glioma analysis demonstrated
consistently higher FBXL6 expression in tumor tissues compared with
normal tissues in the TIME2 database (Fig. 2B). Kaplan-Meier curve analysis of
TCGA-glioma data demonstrated that high FBXL6 expression was
associated with decreased overall survival and progression-free
survival compared with low FBXL6 expression, an unfavorable outcome
for patients with glioma (Fig.
2C-D). Moreover, the CGGA database showed the same relationship
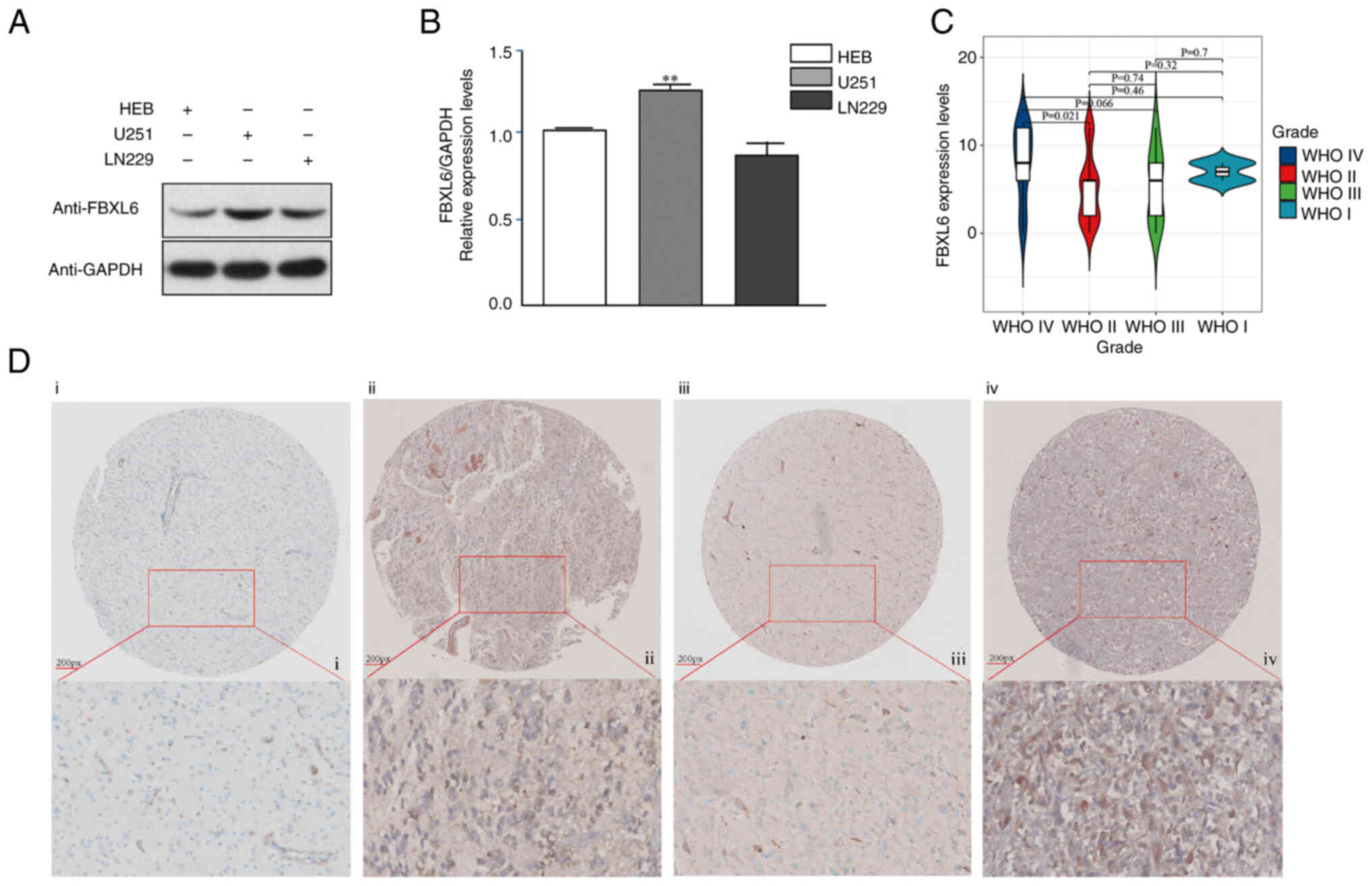
between FBXL6 expression and prognosis (Fig. 2E). Western blot was performed to
examine the FBXL6 protein levels in glioma U251 and normal HEB
cells. The protein expression level of FBXL6 was higher in U251
cells compared with HEB cells (Fig.
3A). The RT-qPCR results demonstrated that FBXL6 mRNA
expression levels were significantly higher in U251 compared with
HEB cells (Fig. 3B). Given the
crucial role of FBXL6 in cancer (22,30),
its expression levels were investigated across different stages of
glioma. A tissue microarray comprising samples from 64 glioma
patients was constructed for this purpose. Patients were stratified
into two groups based on their IHC Score (IRS): i) Low-expression
group, IRS≤7; ii) and High-expression group, IRS≥8. Subsequently,
the expression level of FBXL6 in the glioma tissue microarray was
assessed via IHC. Compared with low-grade glioma, the expression of
FBXL6 protein increases in high-grade gliomas. Furthermore, the
protein expression level of FBXL6 was increased in patients with
glioma at WHO Grade IV compared with patients at WHO Grade II stage
(Fig. 3C,D; Table I). These findings suggested that the
high expression of FBXL6 in glioma may affect its biological
function.
 | Table I.Tissue microarray clinical features
from 64 patient samples. |
Table I.
Tissue microarray clinical features
from 64 patient samples.
|
|
| FBXL6 expression
level |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Total patients, n
(%) | Low, n (%) | High, n (%) | P-value |
|---|
| Number of patients,
n | 64 | 33 | 31 |
|
| Sex |
|
|
| 0.825 |
|
Male | 36 (56.3%) | 19 (57.6%) | 17 (54.8%) |
|
|
Female | 28 (43.7%) | 14 (42.4%) | 14 (45.2%) |
|
| Age, years |
|
|
| <0.01 |
|
≥45 | 38 (59.4%) | 13 (39.4%) | 25 (80.6%) |
|
|
<45 | 26 (40.6%) | 20 (60.6%) | 6 (19.6%) |
|
| Grade |
|
|
|
|
| I | 3 (4.7%) | 3 (9.1%) | 0 (0.0%) | 0.2388 |
| II | 24 (37.5%) | 19 (57.6%) | 5 (16.1%) | 0.0008 |
|
III | 17 (26.6%) | 6 (18.2%) | 11 (35.5%) | 0.1594 |
| IV | 20 (31.2%) | 5 (15.1%) | 15 (48.4%) | 0.0065 |
FBXL6 expression correlated with
clinical features of glioma
CGGA, univariate (Fig.
4A) and multivariate (Fig. 4B)
COX regression analyses demonstrated that FBXL6 expression was
associated with poor survival and was considered an independent
prognostic factor in each dataset. FBXL6 was also associated with
IDH mutations, Primary, Recurrent and Secondary status, WHO grade,
chemotherapy regime and 1p19q co-deletion. Therefore, FBXL6 may
have a high predictive value in patients with gliomas and could
potentially be used as a prognostic factor for gliomas. In
addition, the expression level of FBXL6 was significantly higher in
patients with glioma undergoing chemotherapy compared with those
not undergoing chemotherapy (Fig.
4C). According to the WHO classification, the higher the glioma
grade, the higher the expression level of FBXL6 (Fig. 4D). In various subtypes of
pathological tissues, the expression of FBXL6 was correlated with
multiple subtypes of glioma and compared with other subtypes, FBXL6
had the highest expression level in glioblastoma (Fig. 4E). There was no statistical
significance observed between FBXL6 expression levels and the IDH1
mutation or 1p19q codeletion status (Fig. S1A-B). These results suggested that
FBXL6 expression promoted the occurrence and development of
glioblastoma and could potentially serve as a potential molecular
marker.
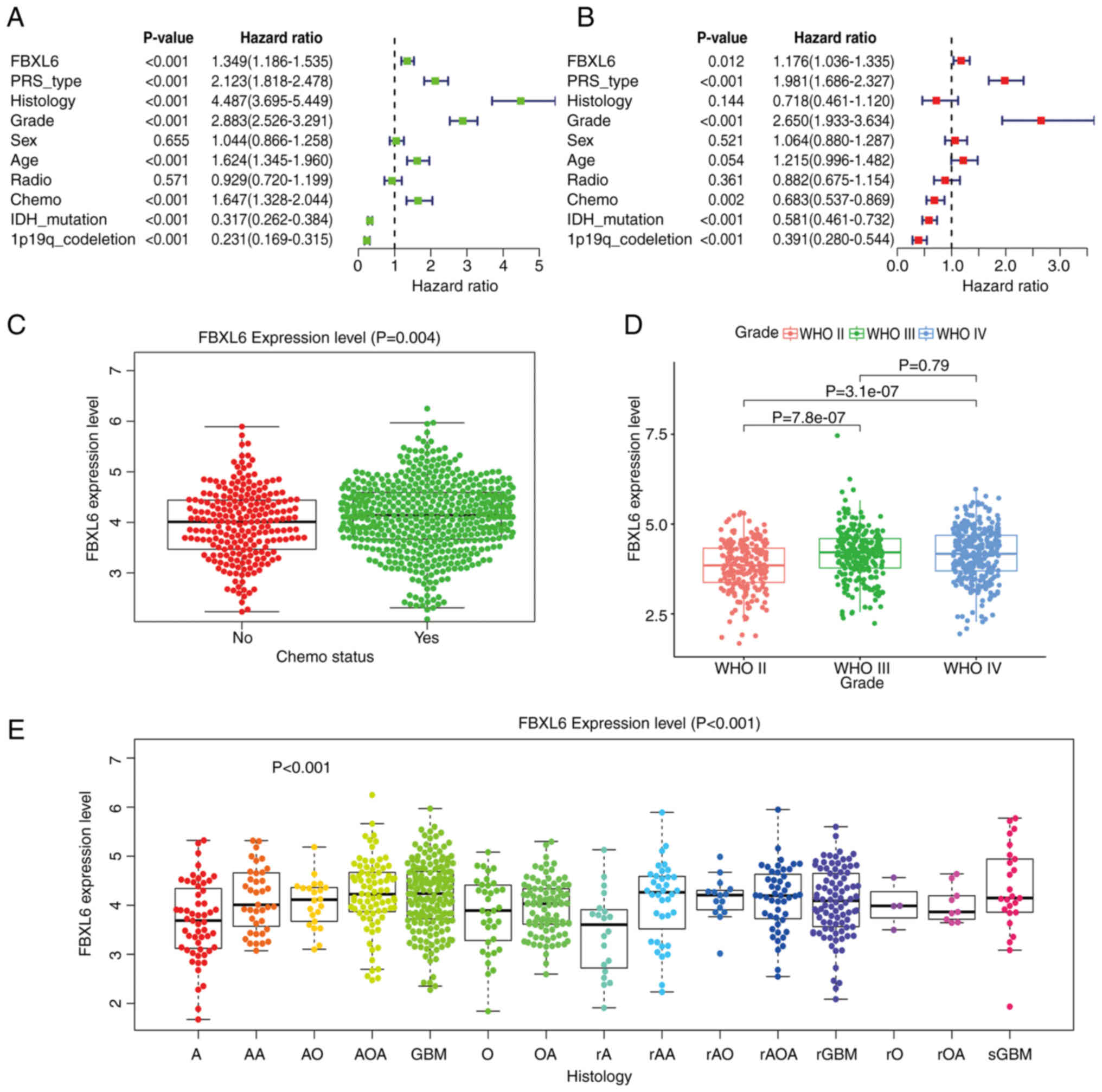 | Figure 4.Association between FBXL6 expression
levels and clinical traits in the Chinese Glioma Genome Atlas
database. (A) Univariate and (B) multivariate analysis
demonstrating a significant association between tumor grade,
recurrence, IDH mutation and 1p19q co-deletion. (C) Higher FBXL6
expression levels were demonstrated in patients who received
chemotherapy compared with patients who did not receive
chemotherapy. (D) Higher FBXL6 expression levels were demonstrated
in patients with WHO Grade IV compared with grades III and II. (E)
Higher FBXL6 expression levels were demonstrated in GBM compared
with low-grade gliomas. Data were presented as the median ±
interquartile range and analyzed using the Kruskal-Wallis test
followed by Dunn's post hoc test. GBM, glioblastoma; FBXL6, F-box
and leucine-rich repeat protein 6; WHO, World Health Organization;
IDH, isocitrate dehydrogenase; radio, radiotherapy; chemo,
chemotherapy; PRS, Polygenic Risk Score; GBM, glioblastoma. |
ROC curve prediction and nomogram
construction for glioma prognosis
Given the significant correlation between FBXL6
expression and clinical factors such as tumor grade and age
(Fig. 5A), the survival rates of
patients with glioblastoma over 1-, 3- and 5-year periods were
predicted using FBXL6 expression levels. The ROC curve demonstrated
that FBXL6 expression was strongly predictive of the survival rates
of patients with glioblastoma, with AUC values of 0.573, 0.639 and
0.631 for the 1-, 3- and 5-year survival rates, respectively
(Fig. 5B). A nomogram was
constructed to predict the correlation between FBXL6 expression and
age, sex and tumor grade, which demonstrated a significant clinical
predictive value for FBXL6 expression according to age and grade.
Confidence in the validity of the nomogram was further reinforced
by clinical calibration curve data (Fig. 5C,D).
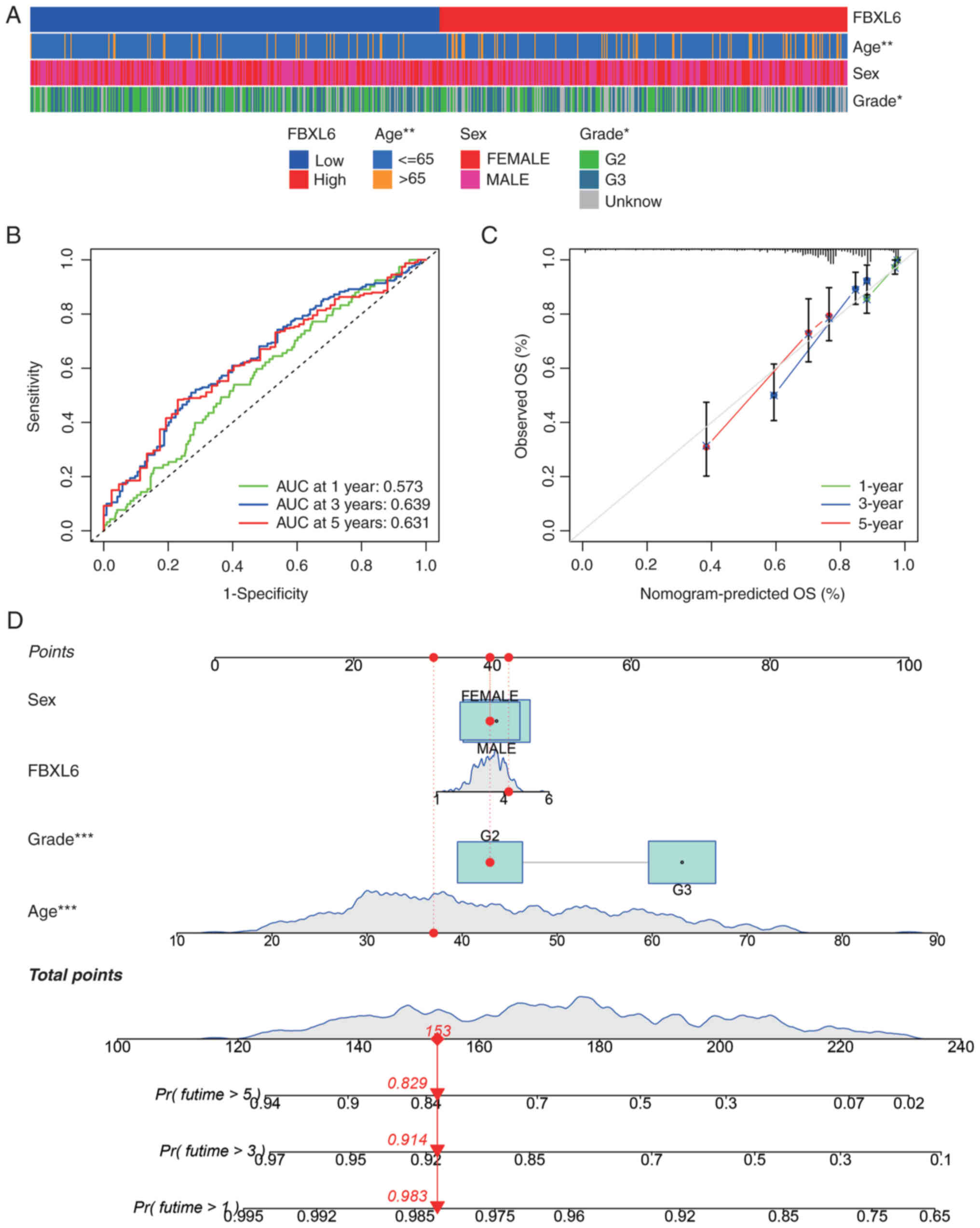 | Figure 5.Clinical phenotypes and nomogram
prediction based on FBXL6. (A) Heatmap of FBXL6 expression and
clinical feature correlation in The Cancer Genome Atlas glioma
data. (B) Receiver Operating Characteristic curve for 1, 3 and
5-year survival rate predictions. (C) Calibration curves for 1-, 3-
and 5-year survival rate predictions in patients with glioblastoma.
(D) Nomogram integrating sex, age and grading. *P<0.05,
**P<0.01, ***P<0.001. FBXL6, F-box and leucine-rich repeat
protein 6; OS, overall survival; AUC, area under the curve; G,
grade; Pr, Probability; Futime, follow-up time. |
Genetic correlations, biological
functions and GSEA enrichment analysis of FBXL6 in glioma
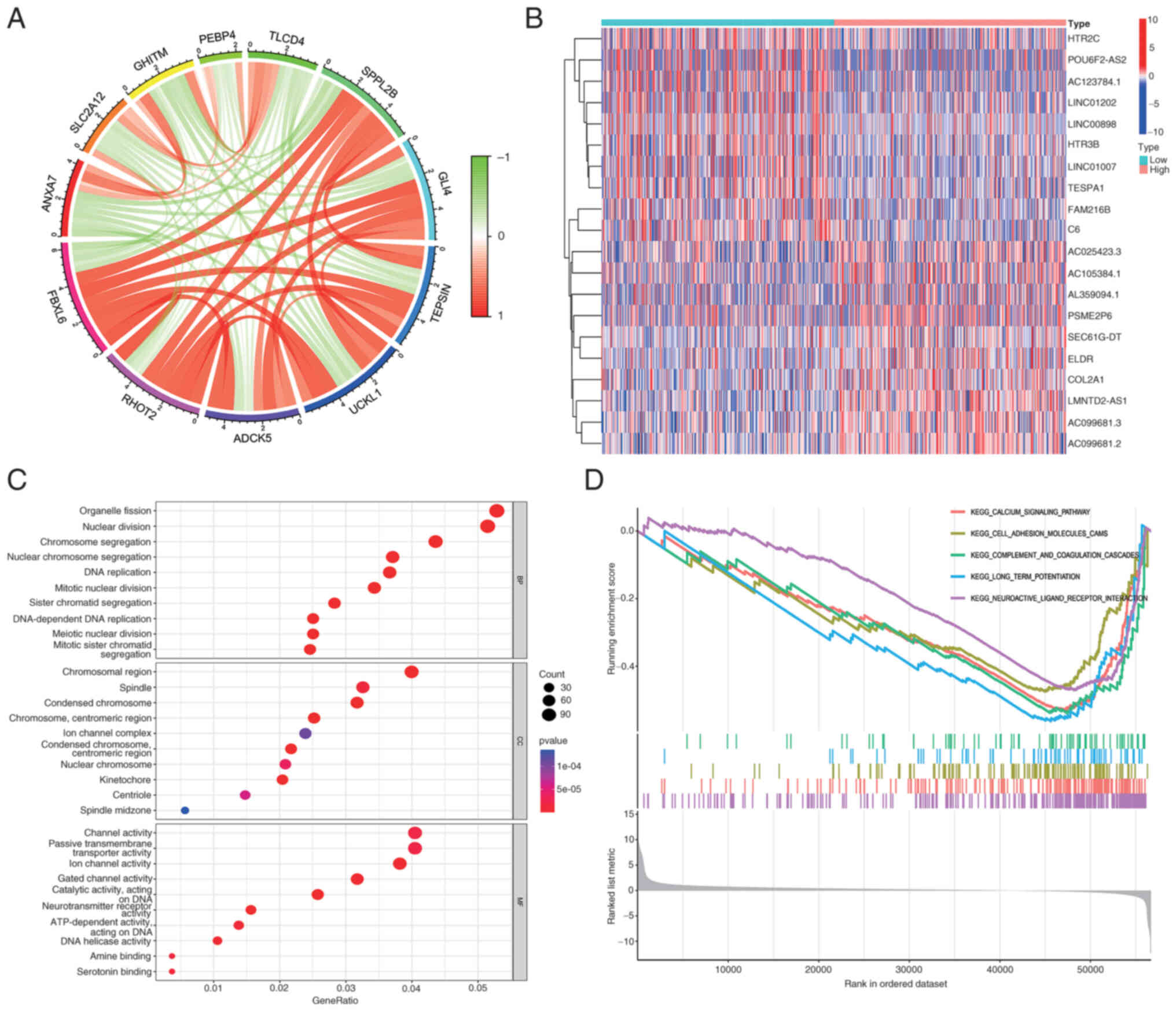
To explore the specific biological functions of
FBXL6 in glioblastoma, gene correlation analysis was performed with
a correlation threshold of 0.6 and P<0.001. The results
demonstrated that FBXL6 expression was positively correlated with
mitochondrial RHO GTPase 2, aarF domain-containing protein kinase
5, uridine-cytidine kinase-like 1, AP-4 complex accessory subunit
Tepsin, zinc finger protein GLI4 and signal peptide peptidase-like
2B and significantly negatively correlated with solute carrier
family 2, facilitated glucose transporter member 12, growth
hormone-inducible transmembrane protein,
phosphatidylethanolamine-binding protein 4 and TLC
domain-containing protein 4 (Fig.
6A). The general control of nucleotide synthesis 5 (GCN5) is an
enzyme that serves a critical role in the modification of histones,
influencing gene expression by adding acetyl groups to the histone
proteins, which impacts the structure and function of chromatin.
Gene correlation analysis demonstrated a significant correlation
between FBXL6 and GCN5, which was consistent with previous research
(Fig. S2). Furthermore, the
expression of FBXL6 was positively correlated with IDH1 and
negatively correlated with MGMT. However, the expression of GCN5
demonstrated no significant correlation with IDH1 and was
negatively correlated with MGMT (Fig.
S3). Gene Expression Profiling Interactive Analysis 2 database
(gepia2.cancer-pku.cn/#index) investigates the relationship between
the expression of FBXL6 and GCN5 and their correlation with
isocitrate dehydrogenase 1 (IDH1) and methylated
DNA-protein-cysteine methyltransferase (MGMT). These findings
indicated that FBXL6 and these correlated genes may influence the
progression of glioblastoma.
Gene correlation analysis identified FBXL6 as
positively associated with SPPL2B, GLI4, and ADCK5, and negatively
with TLCD4, PEBP4, and GHITM (Fig.
6A,B). The GO analysis revealed significant connections to
biological processes like ‘organelle image’, ‘nuclear division’,
and ‘chromosome aggregation’, underscoring FBXL6′s involvement in
key cellular activities and structures. In terms of molecular
function, FBXL6 expression was associated with ‘channel activity’,
‘passive transmembrane transport’ and ‘transporter activity’
(Fig. 6C). GSEA enrichment analysis
demonstrated that the calcium signaling pathway, cell adhesion
molecule cascades and long-term potentiation were differentially
enriched in patients with high FBXL6 expression (Fig. 6D).
FBXL6 expression and its impact on
immune cell infiltration in glioma
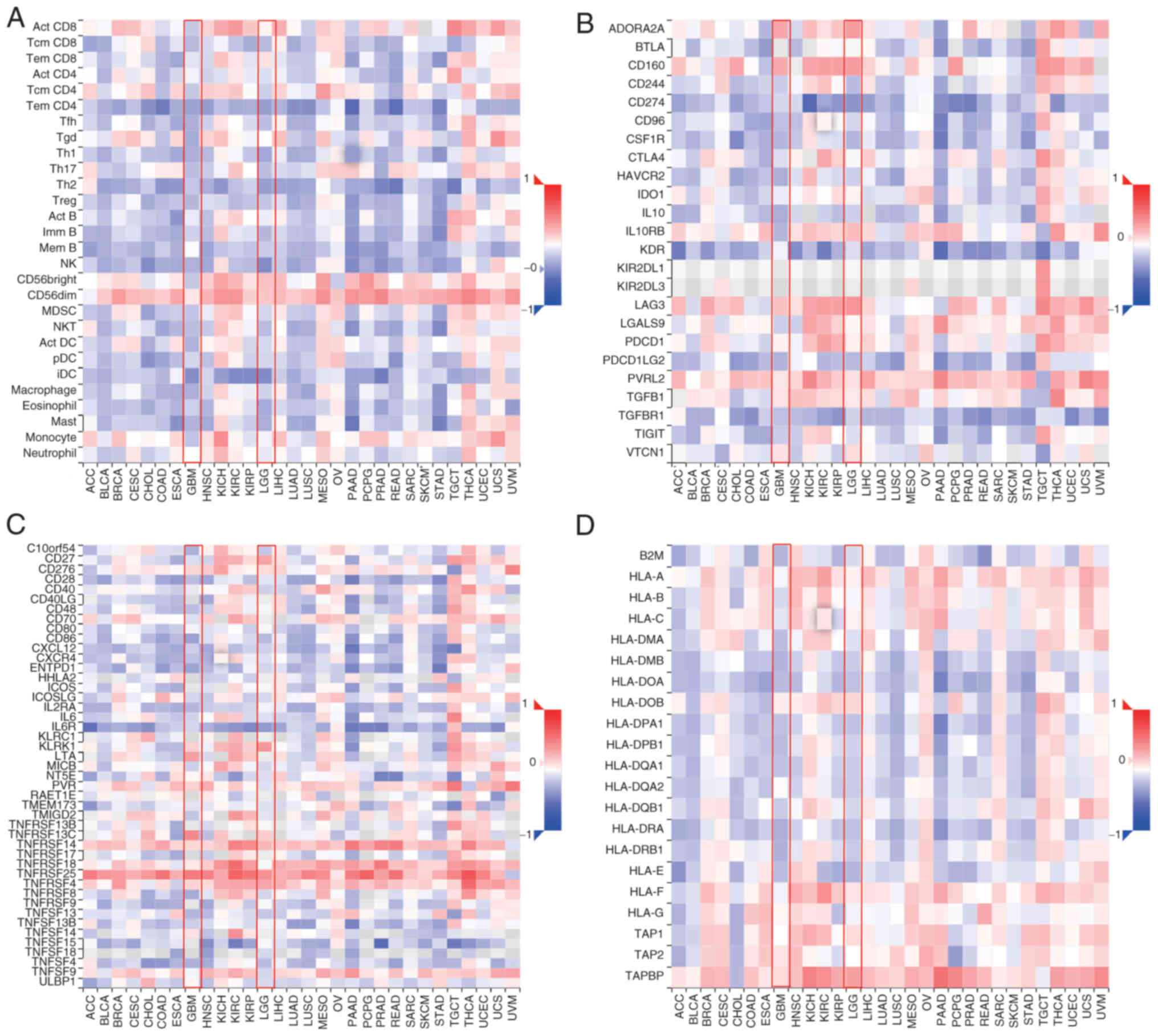
FBXL6 expression was correlated with
immunomodulators in gliomas including lymphocytes, immunoinhibitory
and immunostimulatory molecules and major histocompatibility
complex (MHC) molecules. The association between FBXL6 expression
and the expression of immunomodulatory genes in gliomas was also
examined. In lymphocytes, FBXL6 expression was positively
correlated with CD56dim, whereas it showed significant negative
correlations with effector memory CD4 T cells and interstitial
dendritic cells (Fig. 7A).
Furthermore, FBXL6 expression was significantly positively
correlated with the expression of immune-inhibiting genes,
adenosine receptor 2a and lymphocyte activation gene 3 protein,
whereas it was negatively correlated with TGF-β receptor 1
expression (Fig. 7B). Considering
immunostimulatory factors, FBXL6 expression in glioblastomas
demonstrated a positive correlation with tumor Necrosis Factor
Receptor Superfamily Member 25), whereas it showed a negative
correlation with IL-6 receptor expression (Fig. 7C). However, for MHC molecules, the
expression of FBXL6 demonstrates a significant positive correlation
with TAPBP (TAP Binding Protein; Fig.
7D). The analysis of the TIMER2 database indicated that in GBM
and low-grade glioma subtypes, the expression of FBXL6 was
negatively correlated with CD8 T cells and positively correlated
with B cells (Fig. S4). These
results suggested that FBXL6 may contribute to the malignant
progression of glioblastoma through its multifaceted involvement in
the tumor immune interaction system.
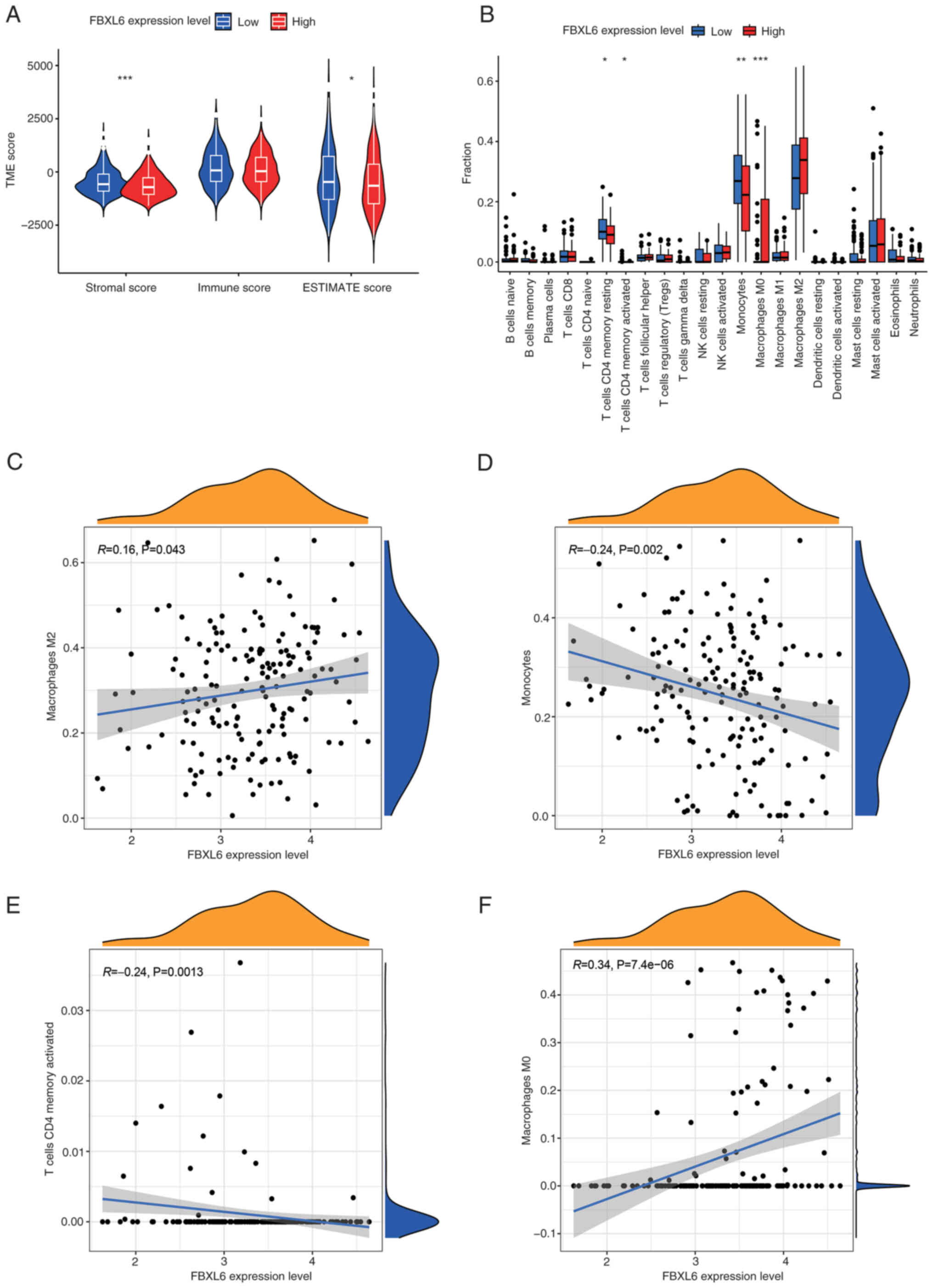
In addition, the relevance of FBXL6 expression in
the tumor immune microenvironment was assessed by estimating the
stromal and immune scores of the high- and low-expression groups.
Low expression of FBXL6 was significantly associated with lower
stromal and immune cores (Fig. 8A),
which may contribute to tumor immune escape and suppression,
thereby promoting glioblastoma progression (30). Similarly, in immune-infiltrating
cells, low expression of FBXL6 was significantly correlated with
CD4+ memory cells and monocytes, which was consistent with our
previous results and may contribute to immune evasion and promote
malignant progression in patients with glioblastoma (30). Additionally, a high expression level
of FBXL6 was significantly associated with M0 macrophages (Fig. 8B), whereas FBXL6 expression
predominantly correlated with memory resting CD4 T cells, memory
activated CD4 T cells and monocytes (Fig. 8B). The association between immune
cells and FBXL6 expression using CIBERSORT was further explored and
it was demonstrated that M2 and M0 macrophages were significantly
positively correlated with FBXL6 expression and significantly
negatively correlated with monocytes and memory resting CD4 T cells
(Fig. 8C-F).
Analyzing the relationship between
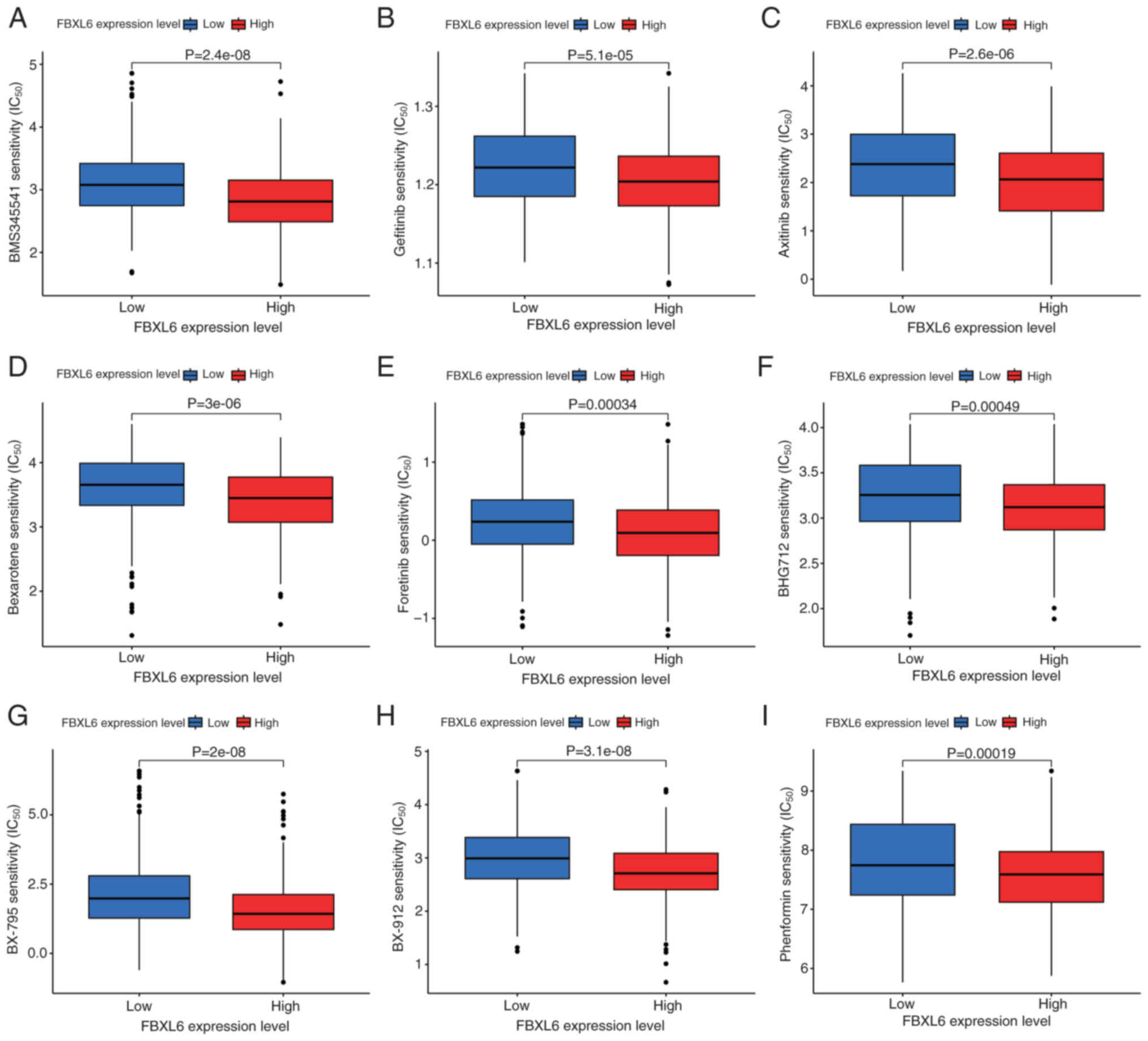
FBXL6 expression and drug sensitivity in glioma
With the continued advancement of precision medicine
and increasing demand for personalized treatment, the relationship
between FBXL6 expression and the IC50 of certain drugs
in glioma treatments was investigated to elucidate their possible
application in the individualized treatment of glioma. The R
package ‘oncoPredict’ was used to predict the relationship between
FBXL6 expression and drugs with a screening condition of
P<0.001. Statistically significant differences in the
sensitivity of eight anti-cancer drugs between the high and low
groups of FBXL6 expression were found. A total of nine of these
drugs (BMS345541, gefitinib, axitinib, bexarotene, foretinib,
BHG712, BX-795, BX-912 and phenformin) had a higher IC50 in the low
FBXL6 expression group compared with the high FBXL6 expression
group, which demonstrated that the patients with high levels of
FBXL6 expression may be more sensitive to treatment with these
particular anticancer drugs (Fig.
9). These results suggested that these drugs may serve a
potential role in the future treatment of gliomas with high FBXL6
expression.
Discussion
Glioblastoma is the most common type of brain tumor
with a relatively high mortality rate due to its high recurrence
rate and the difficulties associated with complete surgical
resection. Although the conventional treatment method for
glioblastoma involves a combination of surgery, radiotherapy and
chemotherapy, this approach has not resulted in substantial
improvements in patient survival rates (31). According to the WHO, the typical
survival period for Grade IV malignant glioma is <20 months
after diagnosis (32). The 5-year
survival rate for patients with glioblastoma is <5% and it is
lower for elderly patients aged over 65 years old. (33). Whilst Liu et al (34) previously reported progress with
bevacizumab in improving the survival of patients with glioma, its
effectiveness remains unclear. Early detection and treatment can
significantly improve the prognosis of patients with gliomas
(35). Therefore, it is crucial to
identify new biomarkers for the early diagnosis of gliomas.
Ubiquitination serves a crucial role as an important
component of various biological processes such as the cell cycle,
apoptosis and DNA damage repair. The impact of ubiquitination on
tumors has previously been reported (14). An enzyme that mediates
ubiquitination is the E3 ubiquitin ligase, which determines the
specificity of substrate ubiquitination and degradation (36). Dysregulation of F-box
protein-mediated protein degradation has been implicated in the
development of human malignancies. F-box proteins can be divided
into three subfamilies based on the presence of specific substrate
recognition domains (37). The Fbxw
subfamily consists of 10 proteins, including β-TrCP1, Fbxw7 (also
known as Fbw7 and Cdc4),and β-TrCP2 (Fbxw11) (18). Owing to the close association of
F-box family members with tumorigenesis, previous studies have
reported certain biological functions attributed to several
initially uncharacterized domains of FBXO proteins (38–41).
For example, FBXO6 binds to glycosylated degradation proteins on
the alpha chain of T cell receptors (42), whereas FBXO2 can ubiquitinate
proteins with N-linked high-mannose oligosaccharides, such as the
precursor form of β1 integrin (43). F-box proteins serve important roles
in tumorigenesis by regulating substrate turnover. Substrate
turnover can be dependent or independent of E3 ligase activity
(19,20). In particular, several F-box proteins
have emerged as potential therapeutic targets for cancer treatment
because their dysregulation is associated with
tumorigenesis(18). Emerging
experimental and clinical data suggest aberrations in cell cycle
regulatory factors, many of which have tumor-suppressive or
oncogenic functions (44). Based on
the crucial role of F-box proteins in cell cycle regulation, the
key role of F-box proteins in tumorigenesis has been reported
(45). In a study involving FBXL7
knockout mice, severe and progressive hematopoietic failure was
observed by 12 weeks of age, with the development of T-cell acute
lymphoblastic leukemia within 16 weeks (46). In tumor multi-omics research, a
large number of genes associated with tumor prognosis have been
discovered through bioinformatics (47). However, the effect of FBXL6 on
gliomas remains unclear and requires further investigation. The
present study demonstrated that a high expression level of FBXL6
was indicative of poor prognosis in patients with glioma and was
associated with decreased survival rates in these patients.
The expression of GCN5 has a considerable influence
on glioma proliferation and invasion and is significantly
associated with tumor grade (48,49). A
previous study reported that GCN5 negatively regulated autophagy by
inhibiting the biogenesis of autophagosomes and lysosomes,
primarily through the targeting of transcription factor EB, a key
autophagy and lysosome-related gene expression regulator (48). While the present study did not
elucidate how the correlation between FBXL6 and GCN5 influenced
glioma progression, future investigations are expected to provide a
more comprehensive understanding of this relationship. A previous
study reported that GCN5 can influence the progression of gliomas
via the STAT3 and AKT pathways (48). As a histone acetyltransferase, GCN5
may work in conjunction with HMGA2 to facilitate the invasion and
metastasis of glioma cells (50).
The present study did not investigate the impact of epigenetics,
thereby it is currently unclear whether FBXL6 impacts the
epigenetic landscape in glioma cells. Nevertheless, future research
should focus on epigenetic studies to investigate this further.
Nonetheless, these results collectively indicated
the clinical value of FBXL6 as a potential prognostic biomarker for
gliomas. The enrichment analysis conducted using GSEA demonstrated
that FBXL6 was involved in various signaling pathways such as
‘BUTANOATE_METABOLISM’, ‘CALCIUM_SIGNALING_PATHWAY’,
‘LONG_TERM_POTENTIATION’, ‘NOTCH_SIGNALING_PATHWAY’ and
‘PRIMARY_BILE_ACID_BIOSYNTHESIS’. These findings suggested that
FBXL6 may affect the progression of glioma in patients through
these pathways.
The role of FBXL6 in cell cycle regulation and
tumorigenesis is important. However, current understanding of its
prognostic value, underlying molecular mechanisms and drug
sensitivity in gliomas remains incomplete. The present study
confirmed that FBXL6 expression was significantly higher in gliomas
compared with normal tissues, which suggested its potential impact
on glioma malignancy. These findings highlighted FBXL6 as a
candidate for a prognostic biomarker in glioma and as a potential
target for neuroglioma treatment. Moreover, bioinformatics analysis
demonstrated that FBXL6 may represent a viable future therapeutic
target in glioma. Furthermore, IC50 values indicated
that drugs such as BMS345541, gefitinib, axitinib, bexarotene,
foretinib, BHG712, BX-795, BX-912 and phenformin exhibited enhanced
efficacy in treating patients with glioma characterized by low
FBXL6 expression levels.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by research grants from the
National Natural Science Foundation of China (grant no. 81830052)
and the Shanghai Key Laboratory of Molecular Imaging (grant no.
18DZ2260400).
Availability of data and materials
The datasets generated in the present study may be
requested from the corresponding author.
Authors' contributions
QL and SN guided the conception and design of the
study. QL, JZ, WZ, HZ and ML collected clinical data and
constructed figures. QL, JZ, HZ, SJ and JZ performed statistical
analysis. JZ, SJ and SN revised the manuscript. QL and JZ conducted
the second round of image acquisition and modifications. QL, JZ,
WZ, HZ, ML, JZ, SJ and SN confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the Ninth People's Hospital, Shanghai Jiaotong
University School of Medicine (approval no. H9H-2023-T489-1,
Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molinaro AM, Taylor JW, Wiencke JK and
Wrensch MR: Genetic and molecular epidemiology of adult diffuse
glioma. Nat Rev Neurol. 15:405–417. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Research Network, .
Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA,
Rheinbay E, Miller CR, Vitucci M, et al: Comprehensive, integrative
genomic analysis of diffuse lower-grade gliomas. N Engl J Med.
372:2481–2498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan B, Wang G, Tang X, Tong A and Zhou L:
Immunotherapy of glioblastoma: Recent advances and future
prospects. Hum Vaccin Immunother. 18:20554172022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roukens MG, Alloul-Ramdhani M, Moghadasi
S, Op den Brouw M and Baker DA: Downregulation of vertebrate Tel
(ETV6) and Drosophila Yan is facilitated by an evolutionarily
conserved mechanism of F-box-mediated ubiquitination. Mol Cell
Biol. 28:4394–4406. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonacci T and Emanuele MJ: Dissenting
degradation: Deubiquitinases in cell cycle and cancer. Semin Cancer
Biol. 67:145–158. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abbas R and Larisch S: Killing by
degradation: Regulation of apoptosis by the
ubiquitin-proteasome-system. Cells. 10:34652021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Daulny A and Tansey WP: Damage control:
DNA repair, transcription, and the ubiquitin-proteasome system. DNA
Repair (Amst). 8:444–448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bendotti C, Marino M, Cheroni C, Fontana
E, Crippa V, Poletti A and De Biasi S: Dysfunction of constitutive
and inducible ubiquitin-proteasome system in amyotrophic lateral
sclerosis: Implication for protein aggregation and immune response.
Prog Neurobiol. 97:101–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han D, Wang L, Jiang S and Yang Q: The
ubiquitin-proteasome system in breast cancer. Trends Mol Med.
29:599–621. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reinstein E and Ciechanover A: Narrative
review: Protein degradation and human diseases: The ubiquitin
connection. Ann Intern Med. 145:676–684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang F, Nie L and Wei W: Ubiquitin
signaling in cell cycle control and tumorigenesis. Cell Death
Differ. 28:427–438. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ciechanover A: The unravelling of the
ubiquitin system. Nat Rev Mol Cell Biol. 16:322–324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chan CH, Li CF, Yang WL, Gao Y, Lee SW,
Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al: The Skp2-SCF
E3 ligase regulates Akt ubiquitination, glycolysis, herceptin
sensitivity, and tumorigenesis. Cell. 149:1098–1111. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng X, Liu X, Guo X, Jiang S, Chen T, Hu
Z, Liu H, Bai Y, Xue M, Hu R, et al: FBXO38 mediates PD-1
ubiquitination and regulates anti-tumour immunity of T cells.
Nature. 564:130–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Liu P, Inuzuka H and Wei W: Roles
of F-box proteins in cancer. Nat Rev Cancer. 14:233–247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu J, Zhang X, Zhang L, Wu CY, Rezaeian
AH, Chan CH, Li JM, Wang J, Gao Y, Han F, et al: Skp2 E3 ligase
integrates ATM activation and homologous recombination repair by
ubiquitinating NBS1. Mol Cell. 46:351–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nelson DE, Randle SJ and Laman H: Beyond
ubiquitination: The atypical functions of Fbxo7 and other F-box
proteins. Open Biol. 3:1301312013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen D, Liu X, Xia T, Tekcham DS, Wang W,
Chen H, Li T, Lu C, Ning Z, Liu X, et al: A multidimensional
characterization of E3 ubiquitin ligase and substrate interaction
network. iScience. 16:177–191. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Cui K, Zhang Q, Li X, Lin X, Tang Y,
Prochownik EV and Li Y: FBXL6 degrades phosphorylated p53 to
promote tumor growth. Cell Death Differ. 28:2112–2125. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu Y, Yao W, Wang T, Xue W, Meng Y, Cai L,
Jian W, Yu Y and Zhang C: FBXL6 depletion restrains clear cell
renal cell carcinoma progression. Transl Oncol. 26:1015502022.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi W, Feng L, Dong S, Ning Z, Hua Y, Liu
L, Chen Z and Meng Z: FBXL6 governs c-MYC to promote hepatocellular
carcinoma through ubiquitination and stabilization of HSP90AA1.
Cell Commun Signal. 18:1002020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schiff D: Low-grade gliomas. Continuum
(Minneap Minn). 23:1564–1579. 2017.PubMed/NCBI
|
|
26
|
Russell B, Collins A, Dally M, Dowling A,
Gold M, Murphy M and Philip J: Living longer with adult high-grade
glioma: Setting a research agenda for patients and their
caregivers. J Neurooncol. 120:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding H, Zhao J, Zhang Y, Yu J, Liu M, Li
X, Xu L, Lin M, Liu C, He Z, et al: Systematic analysis of drug
vulnerabilities conferred by tumor suppressor loss. Cell Rep.
27:3331–3344.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dai Z, Wu J, Chen F, Cheng Q, Zhang M,
Wang Y, Guo Y and Song T: CXCL5 promotes the proliferation and
migration of glioma cells in autocrine- and paracrine-dependent
manners. Oncol Rep. 36:3303–3310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Lin XT, Yu HQ, Fang L, Wu D, Luo
YD, Zhang YJ and Xie CM: Elevated FBXL6 expression in hepatocytes
activates VRK2-transketolase-ROS-mTOR-mediated immune evasion and
liver cancer metastasis in mice. Exp Mol Med. 55:2162–2176. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu W, Klockow JL, Zhang M, Lafortune F,
Chang E, Jin L, Wu Y and Daldrup-Link HE: Glioblastoma multiforme
(GBM): An overview of current therapies and mechanisms of
resistance. Pharmacol Res. 171:1057802021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chinot OL, Wick W, Mason W, Henriksson R,
Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea
D, et al: Bevacizumab plus radiotherapy-temozolomide for newly
diagnosed glioblastoma. N Engl J Med. 370:709–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen J, McKay RM and Parada LF: Malignant
glioma: Lessons from genomics, mouse models, and stem cells. Cell.
149:36–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu LY, Ji MS, Nguyen NT, Chow FE, Molaie
DM, Pianka ST, Green RM, Liau LM, Ellingson BM, Nghiemphu PL, et
al: Patterns of long-term survivorship following bevacizumab
treatment for recurrent glioma: A case series. CNS Oncol.
8:CNS352019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghorbani A, Avery LM, Sohaei D,
Soosaipillai A, Richer M, Horbinski C, McCortney K, Xu W, Diamandis
EP and Prassas I: Discovery of novel glioma serum biomarkers by
proximity extension assay. Clin Proteomics. 20:122023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng J, Guo J, Wang Z, North BJ, Tao K,
Dai X and Wei W: Functional analysis of Cullin 3 E3 ligases in
tumorigenesis. Biochim Biophys Acta Rev Cancer. 1869:11–28. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Naseem Y, Zhang C, Zhou X, Dong J, Xie J,
Zhang H, Agboyibor C, Bi Y and Liu H: Inhibitors targeting the
F-BOX proteins. Cell Biochem Biophys. 81:577–597. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
D'Angiolella V, Donato V, Vijayakumar S,
Saraf A, Florens L, Washburn MP, Dynlacht B and Pagano M:
SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity
through CP110 degradation. Nature. 466:138–142. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rye MS, Wiertsema SP, Scaman ES, Oommen J,
Sun W, Francis RW, Ang W, Pennell CE, Burgner D, Richmond P, et al:
FBXO11, a regulator of the TGFβ pathway, is associated with severe
otitis media in Western Australian children. Genes Immun.
12:352–359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Duan S, Cermak L, Pagan JK, Rossi M,
Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R and Pagano
M: FBXO11 targets BCL6 for degradation and is inactivated in
diffuse large B-cell lymphomas. Nature. 481:90–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Santra MK, Wajapeyee N and Green MR: F-box
protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest
after DNA damage. Nature. 459:722–725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida Y, Tokunaga F, Chiba T, Iwai K,
Tanaka K and Tai T: Fbs2 is a new member of the E3 ubiquitin ligase
family that recognizes sugar chains. J Biol Chem. 278:43877–43884.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoshida Y, Chiba T, Tokunaga F, Kawasaki
H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K and Tai
T: E3 ubiquitin ligase that recognizes sugar chains. Nature.
418:438–442. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Frescas D and Pagano M: Deregulated
proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the
scales of cancer. Nat Rev Cancer. 8:438–449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nakayama KI and Nakayama K: Regulation of
the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol.
16:323–333. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Maser RS, Choudhury B, Campbell PJ, Feng
B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna
I, et al: Chromosomally unstable mouse tumours have genomic
alterations similar to diverse human cancers. Nature. 447:966–971.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Menyhárt O and Győrffy B: Multi-omics
approaches in cancer research with applications in tumor subtyping,
prognosis, and diagnosis. Comput Struct Biotechnol J. 19:949–960.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu K, Zhang Q, Lan H, Wang L, Mou P, Shao
W, Liu D, Yang W, Lin Z, Lin Q and Ji T: GCN5 Potentiates glioma
proliferation and invasion via STAT3 and AKT signaling pathways.
Int J Mol Sci. 16:21897–21910. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ouyang C, Mu J, Lu Q, Li J, Zhu H, Wang Q,
Zou MH and Xie Z: Autophagic degradation of KAT2A/GCN5 promotes
directional migration of vascular smooth muscle cells by reducing
TUBA/α-tubulin acetylation. Autophagy. 16:1753–1770. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang S, Zhang H and Yu L: HMGA2 promotes
glioma invasion and poor prognosis via a long-range chromatin
interaction. Cancer Med. 7:3226–3239. 2018. View Article : Google Scholar : PubMed/NCBI
|