Introduction
The global prevalence of intrahepatic
cholangiocarcinoma (ICC) is 0.01–0.46%, accounting for 15–20% of
all primary liver cancer cases, and its incidence has increased in
recent years (1,2). ICC is characterized by no specific
symptoms in the early stage of disease, a high degree of
malignancy, and high recurrence and metastasis rates. In total, 65%
of patients with ICC have reached an advanced stage of disease when
symptoms appear at first diagnosis, and only chemotherapy agents
are suitable for treatment (3).
Research on targeted therapy for ICC is scarce, and the sensitivity
to chemical agents is low, which directly leads to susceptibility
to late-stage ICC and metastasis, with a poor 5-year survival rate
of only 5–20% (1,2). Therefore, the early diagnosis of
recurrent ICC is an important scientific problem that urgently
requires further study.
Machine learning (ML) can process patient clinical
data and imaging reports, as well as analyze these data to discover
potential patterns and associations (4). Furthermore, ML can detect patterns and
features that are difficult to detect, thus providing more accurate
predicted diagnostic outcomes (5,6).
Therefore, ML can provide decision support to physicians, improving
the understanding and interpretation of clinical data. ML can also
provide physicians with probability and risk assessments regarding
tumor recurrence, leading to more optimal treatment planning
(7,8). The ML model is a reliable tool for
predicting early recurrence in patients with cancer following
curative resection due to exhibiting superior performance compared
with other models, including clinical models (9,10).
However, it is still unknown which ML-based model is more suitable
for identifying patients with early recurrence of ICC.
Therefore, the objective of the present study was to
determine the effectiveness of ML for early recurrent ICC
diagnosis, particularly in comparison with clinical models.
Furthermore, determining which ML model has the best diagnostic
performance for patients with recurrent ICC is novel and clinically
significant. To the best of our knowledge, although some related
studies have focused on topics similar to those of the present
study (11,12), no previous publications have
examined this topic using a networks meta-analysis.
Materials and methods
Preferred reporting items for
systematic reviews and meta-analyses (PRISMA)
The present study was conducted in accordance with
the PRISMA 2020 guidelines (13,14).
The study protocol is registered in the International Prospective
Register of Systematic Reviews (no. CRD42024487932; http://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=487932)
(15).
Search strategies and study
selection
A systematic search of the PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase
(www.embase.com) and Cochrane Library databases
(https://www.cochranelibrary.com/) from
their inception until November 2023 was performed using the
following search terms: ‘Machine learning’, ‘recurrent intrahepatic
cholangiocarcinoma’ and their Medical Subject Headings (MeSH)
terms, with no language restrictions. In the present study, early
recurrence was defined as recurrence within 1 year after surgery,
and a liver imaging report was the standard for determining whether
a patient had experienced recurrence. In total, two reviewers
independently screened the original studies that met the following
predefined selection criteria: Studies that reported the diagnostic
accuracy of ML models and studies that reported the diagnostic area
under the curve (AUC) of early recurrence in ICC. All superiority,
non-inferiority, retrospective and prospective studies were
included, and the clinical Tumor-Nodes-Metastasis (TNM) stage was
considered the control group in comparison with the ML-based group.
The recorded outcomes included the diagnostic AUC, accuracy,
sensitivity, specificity, negative predictive value and positive
predictive value. Studies that met any of the following criteria
were excluded from the present study: No diagnostic data can be
extracted for meta-analysis.
Data extraction, risk of bias
assessment and quality of evidence
In total, two independent reviewers extracted data
from the original studies using a standardized Excel table, and
disagreements were discussed and double-checked with an additional
experienced reviewer. The extracted data included the basic
characteristics of the studies, such as imaging, artificial
intelligence (AI) model, sample size (including % of male
patients), diagnosis of cirrhosis (yes/no), presence of viral
hepatitis (yes/no), extent of resection, major vascular invasion,
microvascular invasion, perineural invasion, histological grade,
adjuvant therapy and the outcomes reported. In addition, the
sources of bias were assessed using the Newcastle-Ottawa Scale
(NOS) score (16), with a score
>4 considered acceptable. In addition, the Grading of
Recommendations Assessment, Development and Evaluation framework
was used to assess the quality of evidence contributing to each
estimated network outcome (17).
Data and statistical analyses
Pairwise and frequent network meta-analyses were
used to determine the diagnostic efficacy of different AI models.
Subgroup outcomes were grouped into the training and validation
cohorts. The hazard ratios (HRs) with 95% confidence intervals for
all outcomes are summarized. P<0.05 or I2>50% in
the forest plots indicated heterogeneity in the outcomes, and a
random effects model was used. Furthermore, Begg's and Egger's
tests were performed to assess publication bias for the available
comparisons, and P<0.05 was considered to indicate the existence
of publication bias.
The surface under the cumulative ranking curve
(SUCRA) is presented as a percentage and was used to determine the
probability of a treatment being the most effective by treatment
hierarchy. A higher SUCRA score (close to 100%) indicated that the
ML model was most efficient for the early diagnosis of recurrent
ICC. A greater HR indicated that the ML model was more effective.
Global and local methods were used to avoid inconsistency (18,19).
In addition, the certainty of evidence was determined using the
Confidence in Network MetaAnalysis framework (20). Moreover, a comparison-adjusted
funnel plot was used to detect potential publication biases among
the outcomes. All analyses were performed using StataSE version
15.1 (StataCorp LP) and R version 4.2.3 (R Foundation for
Statistical Computing).
Results
Study characteristics and quality
In total, 5 eligible studies published between
September 2018 and May 2023 involving 1,247 patients were selected
to confirm the diagnostic accuracy of different AI models for
recurrent ICC (21–25). The literature search process is
shown in Fig. 1. Table I (further details in Table SI) shows that the characteristics
of the included studies and the baseline data were relatively
balanced, and all included studies were of acceptable quality
according to the NOS (Table SII).
The included studies used three different AI models: Random forest
(21,23), Light Gradient Boosting Machine
(LightGBM) (22) and nomograms
(24,25).
 | Table I.Baseline characteristics of the
included studies. |
Table I.
Baseline characteristics of the
included studies.
| First author,
year | Imaging | AI model | Grouping | Sample size, n
[females, n (%)] | Diagnosis of
cirrhosis, Yes/No | Clinical model | (Refs.) |
|---|
| Alaimo et al,
2023 | Not | Random | i) Training
cohort; | i) 376 [146
(38.8)]; | i) 56/320; | None | (21) |
|
| mentioned | forest | ii) Testing
cohort | ii) 160 [59
(36.9)] | ii) 26/134 |
|
|
| Sony et al,
2023 | CT scan | LightGBM | i) Derivation
cohort; | i) 140 [53
(37.9)]; | i) 34/106; | AJCC 8th | (22) |
|
|
|
| ii) Internal
validation cohort; | ii) 36 [14
(38.9)]; | ii) 9/27; | TNM model |
|
|
|
|
| iii) External
validation cohort-1; | iii) 74 [37
(54.4)]; | iii) 24/50; |
|
|
|
|
|
| iv) External
validation cohort-2 | iv) 61 [32
(52.5)] | iv) 9/52 |
|
|
| Jolissaint et
al, | CT scan | Random | i) Training
cohort; | i) 97 [55
(56.70)]; | - | Tumor size | (23) |
| 2022 |
| forest | ii) Validation
cohort | ii) 41 [20,
(47.78)] |
|
|
|
| Guo et al,
2022 | Ultrasound | CEUS- | i) Training
group; | i) 30 [7,
(23.33)]; | i) 15/15; | TNM model | (24) |
|
|
| clinical | ii) Validation
group | ii) 23 [4,
(17.39)] | ii) 17/6 |
|
|
|
|
| nomogram |
|
|
|
|
|
| Liang et al,
2018 | 3.0-T MRI | Clinical | i) Training
cohort; | i) 139 [54,
(38.85)]; | - | None | (25) |
|
| scanner | nomogram | ii) Independent
validation cohort | ii) 70 [24,
(34.29)] |
|
|
|
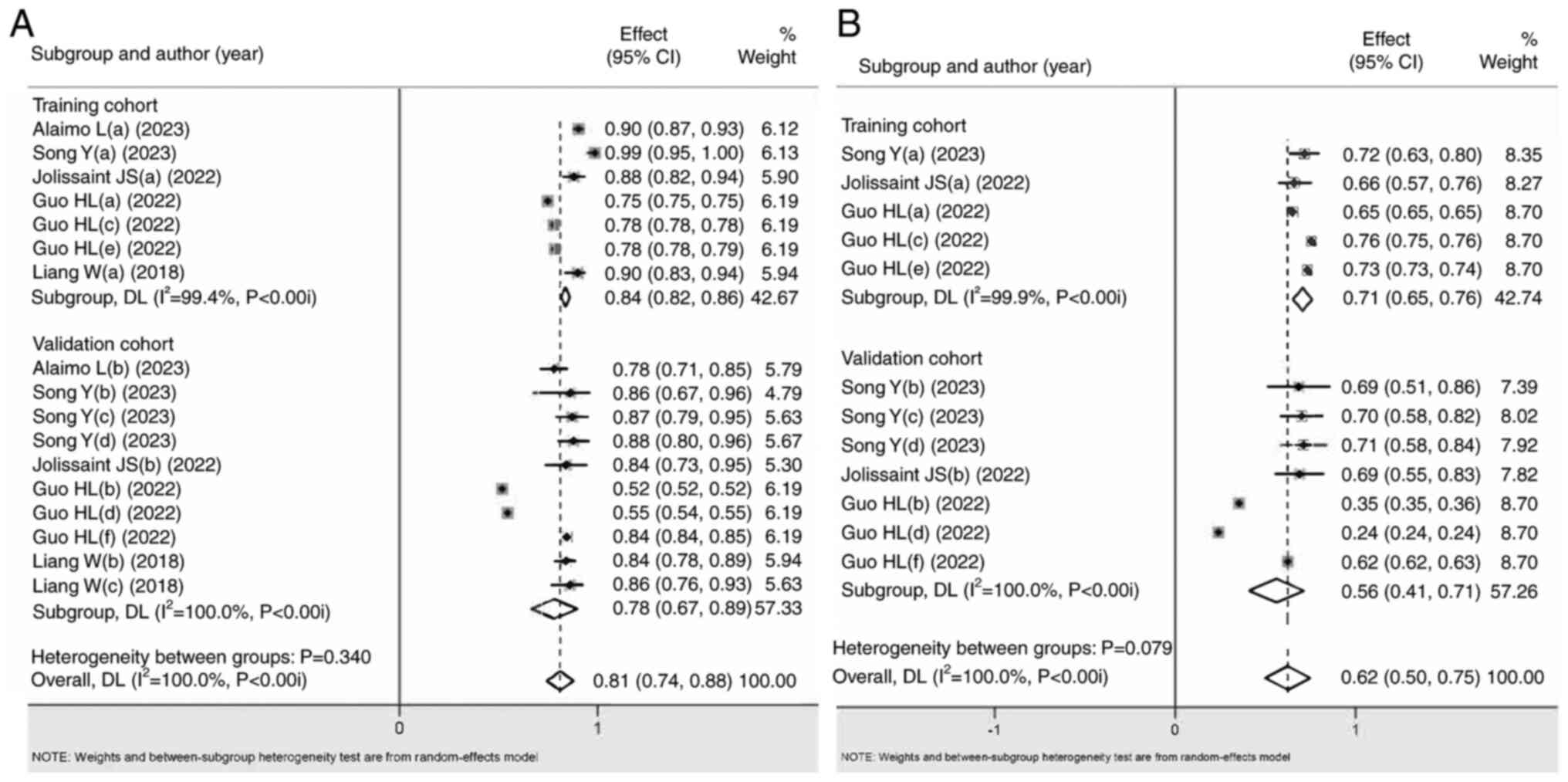
Pairwise meta-analysis outcomes
The AUC, accuracy, sensitivity, specificity,
negative predictive value and positive predictive value of the HR
data were used to determine the diagnostic accuracy of ML for ICC.
Significant differences were detected in almost all overall and
subgroup outcomes (Fig. 2; Table II) for the AUC, accuracy,
sensitivity, specificity, negative predictive value and positive
predictive value. Low to substantial heterogeneity was found in the
aforementioned subgroups, with no publication bias, and low to high
grade due to the influence of included study types. For the
aforementioned outcomes, it was found that the ML-based diagnostic
accuracy was greater than that of the clinical models (SUCRA score
closer to 1, with significant differences), which initially proved
that the ML-based diagnostic power was better than that of clinical
models.
 | Table II.Subgroup analysis of early diagnostic
accuracy of ML to predict early recurrence of intrahepatic
cholangiocarcinoma. |
Table II.
Subgroup analysis of early diagnostic
accuracy of ML to predict early recurrence of intrahepatic
cholangiocarcinoma.
| A, AUC |
|---|
|
|---|
| ML model | Clinical model |
|---|
|
|
|---|
| Subgroup | Combined result, ES
(95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade (17) | Sub-group | Combined result, ES
(95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|---|
| Overall | 0.810 | <0.001, | - | 0.275, | Low | Overall | 0.620 | 0.000, | - | 0.689, | Low |
|
|
(0.740–0.877)a | 100.°b |
| 0.202 |
|
|
(0.500–0.750)a | 100.00b |
| 0.988 |
|
| Training | 0.840 | <0.001, | 0.973 | 0.362, | Low | Training | 0.710 | 0.000, | 0.994 | 0.405 | Low |
| cohort (n=7) |
(0.820–0.86)a | 99.4b |
| 0.259 |
| cohort | (0.650–0.760) | 99.9b |
| 0.988 |
|
|
|
|
|
|
|
| (n=5) |
|
|
|
|
|
| Validation | 0.780 | <0.001, | - | 0.468, | Low | Validation | 0.560 | 0.000, | - | 1.000, | Low |
| cohort (n=10) |
(0.670–0.890)a | 100.0b |
| 0.887 |
| cohort |
(0.410–0.710)a | 100.00b |
| 0.949 |
|
|
|
|
|
|
|
| (n=7) |
|
|
|
|
|
|
| B,
Accuracy |
|
| ML
model | Clinical
model |
|
|
|
Subgroup | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade
(17) |
Sub-group | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|
| Overall | 0.870 | <0.001, | - | 1.000, | Low | Overall | 0.695 | 0.976, 0.0 | 0.859 | 0.089, | Moderate |
|
|
(0.790–0.950)a | 88.9b |
| 0.444 |
|
|
(0.646–0.743)a |
|
| 0.207 |
|
| Training | 0.890 | <0.001, | - | 0.317, - | Very | Training | 0.700 | - | - | - | - |
| cohort (n=2) | (0.750–1.03) | 96.7b |
|
| low | cohxort |
(0.629–0.771)a |
|
|
|
|
|
|
|
|
|
|
| (n=1) |
|
|
|
|
|
| Validation | 0.850 | 0.567, |
| 0.602, | Low | Validation | 0.690 | 0.919, 0.0 | - | 0.117, | Low |
| cohort (n=3) |
(0.800–0.900)a | 0.0 | - | 0.584 |
| cohort |
(0.624–0.757)a |
|
| 0.026c |
|
|
|
|
|
|
|
| (n=3) |
|
|
|
|
|
|
| C,
Sensitivity |
|
| ML
model | Clinical
model |
|
|
|
Subgroup | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade
(17) |
Sub-group | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|
| Overall | 0.920 | 0.017, | 0.955 | 0.355, | Low | Overall | 0.738 | 0.982, | 0.539 | 0.452, | Moderate |
|
|
(0.870–0.980)a | 61.0b |
| 0.194 |
|
|
(0.670–0.806)a | 0.0 |
| 0.000c |
|
| Training | 0.910 | 0.001, | - | 0.317, - | Very | Training | 0.762 | 0.680, | - | 0.317, - | Low |
| cohort (n=3) | (0.820–1.01) | 86.3b |
|
| low | cohort |
(0.665–0.859)a | 0.0 |
|
|
|
|
|
|
|
|
|
| (n=2) |
|
|
|
|
|
| Validation | 0.940 | 0.871, | - | 0.602, | Low | Validation | 0.715 | 0.992,0.0 | 0.593 | 0.497, | Low |
| cohort (n=4) | (0.860–1.01) | 0.0 |
| 0.418 |
| cohort |
(0.620–0.810)a |
|
| 0.563 |
|
|
|
|
|
|
|
| (n=4) |
|
|
|
|
|
|
| D,
Specificity |
|
| ML
model | Clinical
model |
|
|
|
Subgroup | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade
(17) |
Sub-group | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|
| Overall | 0.793 | <0.001, | 0.836 | 0.155, | Low | Overall | 0.617 | 0.739, | 0.686 | 1.000, | Moderate |
|
|
(0.686–0.899)a | 87.1b |
| 0.993 |
|
|
(0.551–0.682)a | 0.0 |
| 0.669 |
|
| Training | 0.804 | <0.001, | - | 0.117, | Very | Training | 0.604 | 0.336, | - | 0.317, | Low |
| cohort (n=3) |
(0.645–0.963)a | 94.1b |
| 0.279 | low | cohort |
(0.516–0.691)a | 0.0 |
|
|
|
|
|
|
|
|
|
| (n=2) |
|
|
|
|
|
| Validation | 0.778 | 0.011, | - | 0.497, | Very | Validation | 0.633 | 0.653, | - | 0.497, | Moderate |
| cohort (n=4) |
(0.614–0.942)a | 73.2b |
| 0.281 | low | cohort |
(0.535–0.732)a | 0.0 |
| 0.970 |
|
|
|
|
|
|
|
| (n=4) |
|
|
|
|
|
|
| E, Negative
predictive value |
|
| ML
model | Clinical
model |
|
|
|
Subgroup | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade
(17) |
Sub-group | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|
| Overall | 0.909 | 0.002,
71.3b | 0.697 | 0.393, | Low | Overall | 0.743 | 0.017, | 0.473 | 0.335, | Low |
|
|
(0.849–0.968)a |
|
| 0.995 |
|
|
(0.660–0.827)a | 63.7b |
| 0.000c |
|
| Training | 0.919 | <0.001, | - | 0.602, | Very | Training | 0.782 | 0.008, | - | 0.317, - | Very low |
| cohort (n=3) |
(0.825–1.013)a | 89.7b |
| 0.904 | low | cohort |
(0.603–0.960)a | 85.7b |
|
|
|
|
|
|
|
|
|
| (n=2) |
|
|
|
|
|
| Validation | 0.896 | 0.753, | - | 0.497, | Low | Validation | 0.716 | 0.249, | - | 0.497, | Moderate |
| cohort (n=4) |
(0.826–0.967)a | 0.0 |
| 0.120 |
| cohort |
(0.633–0.800)a | 27.1 |
| 0.864 |
|
|
|
|
|
|
|
| (n=4) |
|
|
|
|
|
|
| F, Positive
predictive value |
|
| ML
model | Clinical
model |
|
|
|
Subgroup | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade
(17) |
Sub-group | Combined result,
ES (95% CI) | Hetero-geneity,
P-value, I2 (%) |
Meta-regression | Publication
bias | Grade |
|
| Overall | 0.763 | <0.001, | 0.992 | 0.393, | Low | Overall | 0.576 | 0.000, | 0.799 | 1.000, | Low |
|
|
(0.613–0.912)a | 96.0b |
| 0.551 |
|
|
(0.448–0.704)a | 85.8b |
| 0.269 |
|
| Training | 0.765 | <0.001, | - | 0.117, | Very | Training | 0.552 | 0.000, | - | 0.317, - | Very low |
| cohort (n=3) |
(0.547–0.982)a | 98.0b |
| 0.300 | low | cohort |
(0.289–0.814)a | 93.7b |
|
|
|
|
|
|
|
|
|
| (n=2) |
|
|
|
|
|
| Validation | 0.763 | <0.001, | - | 1.000, | Very | Validation | 0.590 | 0.000, | - | 1.000, | Low |
| cohort (n=4) | (0.517–1.008) | 92.5b |
| 0.244 | low | cohort |
(0.422–0.759)a | 83.4b |
| 0.513 |
|
|
|
|
|
|
|
| (n=4) |
|
|
|
|
|
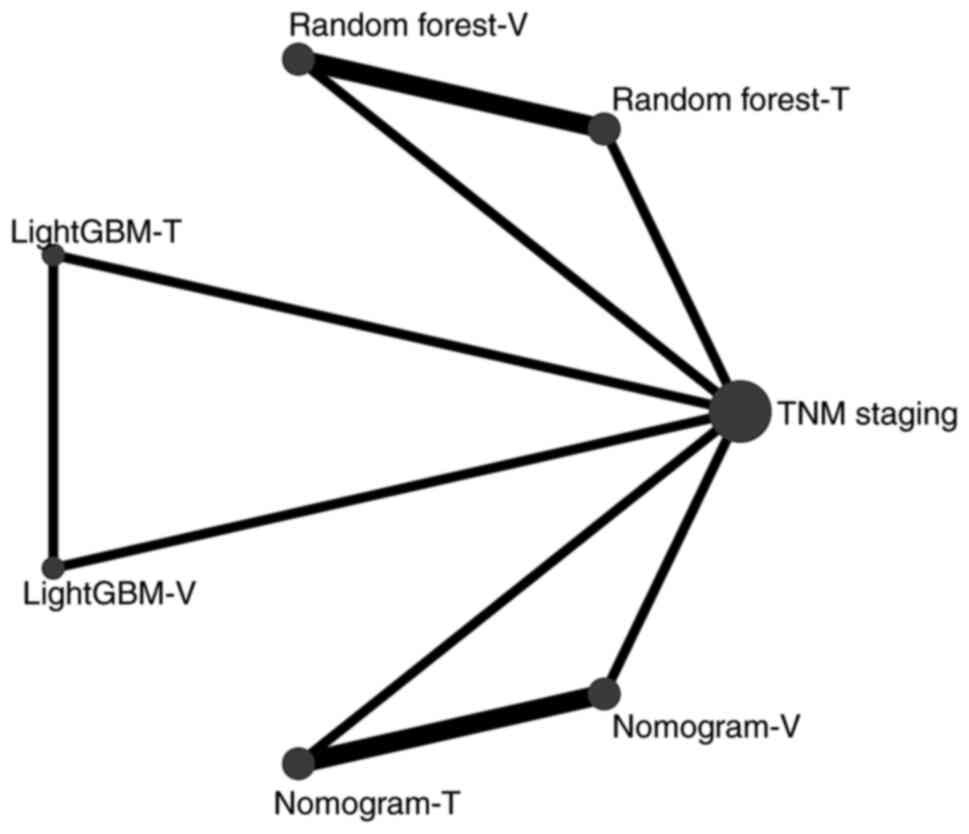
Network meta-analysis outcomes
It remained unclear which ML model had the best
diagnostic accuracy. Therefore, due to the small number of included
studies, a network meta-analysis was performed to determine the
diagnostic accuracy ranking of the ML and clinical models based on
the AUC. Fig. 3 shows the network
diagram of the AI models with AUC. No intersection between the
ML-based models was found, and thus, clinical staging was used as a
reference parameter in comparison with the ML-based group in the
networks meta-analysis. A league table was generated according to
the SUCRA score, and it was found that Nomogram-T (training cohort)
ranked first, followed by LightGBM-T, Random forest-T, LightGBM-V
(validation cohort), Random forest-V, Nomogram-V and TNM stage. In
addition, significant differences between most models were found
(Fig. 4). Of note, the training
cohorts ranked higher than the validation cohorts, possibly due to
the larger sample size of the training cohort, resulting in an
improved training performance. The nomogram ranked first, meaning
that this model had the best diagnostic accuracy for patients with
recurrent ICC.
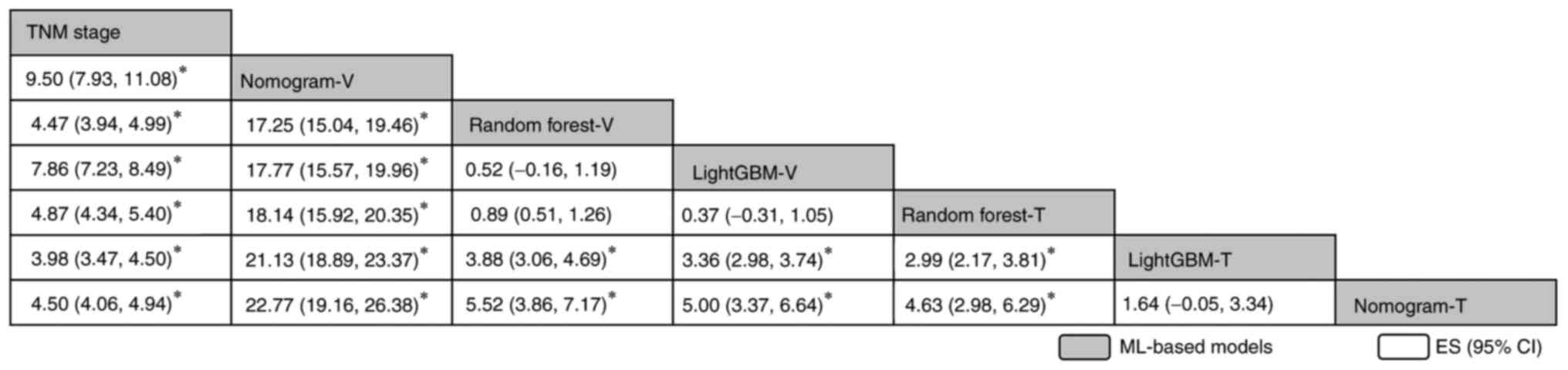 | Figure 4.Network comparisons of the diagnostic
accuracy when using area under the curve in all ML models included
in the present study, reported in order of the diagnosis effect
according to the surface under the cumulative ranking curve score.
The data in each grid represent the HRs and 95% CIs, which is used
to compare the row vs. the column. An HR of >0 is favorable for
the models stated in the rows. *P<0.05. CI, confidence interval;
ES, effect size; GBM, Gradient Boosting Machine; HR, hazard ratio;
ML, machine learning; TNM, Tumor-Nodes-Metastasis; T, training
cohort; V, validation cohort. |
Discussion
The present study focused on the diagnostic value of
ML-based models for recurrent ICC via pairwise and network
meta-analyses. First, 5 studies that included 1,247 patients with
ICC were selected and it was determined that the quality of the
studies was acceptable. Second, from the pairwise meta-analysis, it
was found that the ML-based diagnostic accuracy was greater than
that of clinical models (closer to 1, with significant
differences), which initially proved that the ML-based diagnostic
power was more optimal than that of the clinical models. Third,
according to the network meta-analysis, the nomogram achieved the
best diagnostic accuracy for patients with recurrent ICC.
Patients with ICC may have jaundice, abdominal pain,
weight loss, loss of appetite and other symptoms. The presence of
these symptoms may indicate abnormalities in the intrahepatic bile
ducts that require further examination. Abnormal liver function is
typically an early indicator of ICC, which can reflect the
functional status of the hepatobiliary system of the patient and
whether there is liver function damage. For patients with ICC,
ultrasound, CT, MRI and other imaging examinations can help
physicians find abnormalities in the hepatobiliary system, judge
whether there is bile duct dilatation, stones and other conditions,
and preliminarily assess the possible presence of
cholangiocarcinoma. A space-occupying lesion of >2 cm may
indicate the presence of ICC. Abnormal α-fetoprotein (AFP) blood
concentration is also a diagnostic marker for ICC. When the AFP
blood concentration is ≥400 ng/ml and imaging examination shows the
presence of space-occupying lesions, the possibility of ICC should
be highly suspected. Histological examination is the gold standard
for diagnosis of ICC. Pathological examination of resected tissues
can also clarify important information such as tumor type, grade
and invasion degree, which guides the choice of treatment plan
(26,27). ML algorithms can effectively process
and analyze large-scale, high-dimensional data. In the medical
field, this means that valuable information can be extracted from
large amounts of clinical data, imaging data and biomarkers,
providing support for diagnosis. Traditional methods typically rely
on observational tumor features (large and irregular in shape,
unclear in margin and high in density) for diagnosis, while ML
algorithms can automatically learn and extract useful features from
raw data. This makes the diagnostic model more flexible and
accurate, and able to adapt to the complexities of different
diseases and individuals.
The comprehensive nomogram constructed in the study
by Bu et al (28) is a
promising and convenient tool for evaluating the risk of frailty in
patients with diabetes and can aid clinicians in screening
high-risk populations. In addition, it was concluded that the
nomogram constructed by Lin et al (29) was highly predictive for gastric
cancer. It was also concluded by Chong et al (30) that a preoperative radiomics-based
random forest nomogram is a potential biomarker of microvascular
invasion and recurrence-free survival prediction for patients with
a solitary hepatocellular carcinoma ≤5 cm. These findings indicate
that nomograms can have important roles in improving diagnostic
efficiency.
Random forest is an algorithm based on ensemble
learning that constructs multiple decision trees and integrates
their outputs to obtain more stable and accurate prediction
results. The advantages of Random forest include high parallelism
of training, ability to process high-dimensional features and
insensitivity to partial feature loss. However, this approach also
has several shortcomings; for instance, features with more values
easily have a greater impact on the decision, thus affecting the
stability of the Random forest model. LightGBM is a ML model based
on the gradient boosting decision tree (GBDT). The main goal of
LightGBM is to address the efficiency and scalability issues of
GBDT during the training process. LightGBM has a wide range of
applications in multiple fields, particularly in handling
high-dimensional and large-scale datasets. A nomogram is a
visualization tool for predicting outcomes from multiple factor
conditions. A nomogram network uses a series of parallel lines to
represent the range and influence of different input factors. Users
can obtain the predicted values of the output factors by
intersecting different lines. This visualization allows users to
intuitively understand the relationships between factors and
quickly make predictions. Nomograms have important application
value in data analysis and decision-making processes (31,32).
Therefore, similar to our previous conclusions, nomogram models are
recommended for recurrent ICC.
In the present study, it was also found that the
training cohort ranked ahead of the validation cohort, which was a
notable result due to the frequently higher values of the combined
results. ML algorithms rely on a large amount of data in the
training set to predict recurrent tumor diagnosis, but due to the
particularity and difficulty of data acquisition, there may be data
bias and sample imbalance problems. This can compromise model
performance and generalization and requires appropriate processing
and optimization (33,34). For further research, the training
cohort used in the present study should be optimized for model
stability and reliability, and then research should be conducted
using the same ML-based model or a group of models that could
increase the stability of the model, and the ML-based diagnostic
tumor model was a unique innovation of the present study. There
were also some limitations to the present study. First, only 5
studies were included in the analysis, and the quality of these
studies was deemed acceptable. Follow-up studies could expand the
assessed disease types, such as from ICC to all liver cancer or
digestive tract tumors, and incorporate high-quality controlled
trials published in the future. Second, the heterogeneity from the
pairwise meta-analysis was large due to clinical factors. Third,
only one outcome could be included in the network meta-analysis due
to the small sample size.
In summary, the present meta-analysis concluded that
the application of an ML-based diagnostic model for patients with
recurrent ICC was more optimal than that of a clinical model, and
the nomogram model, which was ranked first, is recommended for
patients with recurrent ICC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The Scientific Research Fund of
Liaoning Provincial Education Department (grant no.
LJKQZ20222420).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
CY, YZ and CP conceived and designed the study; CY,
YZ and CP confirm the authenticity of all the raw data; CY, JX, SW,
YW, YZ and CP searched, retrieved and selected the studies; CY, YZ,
YW, SW and CP extracted the data; CY, JX, SW and YW analyzed and
interpretated the data; CY, YZ and CP performed the meta-analysis
and interpreted the results. YC, SW, YZ and CP wrote and edited the
draft of this manuscript. JX revised critically for important
intellectual content. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen X, Du J, Huang J, Zeng Y and Yuan K:
Neoadjuvant and adjuvant therapy in intrahepatic
cholangiocarcinoma. J Clin Transl Hepatol. 10:553–563. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Halder R, Amaraneni A and Shroff RT:
Cholangiocarcinoma: A review of the literature and future
directions in therapy. Hepatobiliary Surg Nutr. 11:555–566. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kubo S, Shinkawa H, Asaoka Y, Ioka T,
Igaki H, Izumi N, Itoi T, Unno M, Ohtsuka M, Okusaka T, et al:
Liver cancer study group of Japan clinical practice guidelines for
intrahepatic cholangiocarcinoma. Liver Cancer. 11:290–314. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ahmed Z, Mohamed K, Zeeshan S and Dong X:
Artificial intelligence with multi-functional machine learning
platform development for better healthcare and precision medicine.
Database (Oxford). 2020:baaa0102020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ibrahim I and Abdulazeez A: The role of
machine learning algorithms for diagnosing diseases. J Appl Sci
Technol Trend. 2:10–19. 2021. View Article : Google Scholar
|
|
6
|
Richens JG, Lee CM and Johri S: Improving
the accuracy of medical diagnosis with causal machine learning. Nat
Commun. 11:39232020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stenzinger A, Moltzen EK, Winkler E,
Molnar-Gabor F, Malek N, Costescu A, Jensen BN, Nowak F, Pinto C,
Ottersen OP, et al: Implementation of precision medicine in
healthcare-A European perspective. J Intern Med. 294:437–454. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee J, Yoo SK, Kim K, Lee BM, Park VY, Kim
JS and Kim YB: Machine learning-based radiomics models for
prediction of locoregional recurrence in patients with breast
cancer. Oncol Lett. 26:4222023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng J, Zeng J, Lin K, Lin H, Wu Q, Guo P,
Zhou W and Liu J: Development of a machine learning model to
predict early recurrence for hepatocellular carcinoma after
curative resection. Hepatobiliary Surg Nutr. 11:176–187. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin L, Zhao Q, Fu S, Cao F, Hou B and Ma
J: Development and validation of machine learning models to predict
survival of patients with resected stage-III NSCLC. Front Oncol.
13:10924782023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Likhitrattanapisal S, Tipanee J and
Janvilisri T: Meta-analysis of gene expression profiles identifies
differential biomarkers for hepatocellular carcinoma and
cholangiocarcinoma. Tumour Biol. 37:12755–12766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu HY, Xia S, Liu AG, Wei MD, Chen ZB, Li
YX, He Y, Liao MJ, Hu QP and Pan SL: Upregulation of miR-132-3p in
cholangiocarcinoma tissues: A study based on RT-qPCR, the cancer
genome atlas miRNA sequencing, gene expression omnibus microarray
data and bioinformatics analyses. Mol Med Rep. 20:5002–5020.
2019.PubMed/NCBI
|
|
13
|
Page MJ, McKenzie JE, Bossuyt PM, Boutron
I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan
SE, et al: The PRISMA 2020 statement: An updated guideline for
reporting systematic reviews. BMJ. 372:n712021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Tulder M, Furlan A, Bombardier C and
Bouter L; Editorial Board of the Cochrane Collaboration Back Review
Group, : Updated method guidelines for systematic reviews in the
cochrane collaboration back review group. Spine (Phila Pa 1976).
28:1290–1299. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
University of York Centre for Reviews and
Dissemination, . Jinatongthai P, Kongwatcharapong J, Phrommintikul
A, Nathisuwan S and Chaiyakunapruk N: Comparative efficacy and
safety of reperfusion therapy with fibrinolytic agents in patient
with ST-segment elevation myocardial infarction: A systematic
review and network meta-analysis. Prospero 2019: CRD42019161406.
2022.Available from:. https://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=487932(date
of access, 18/01/2024).
|
|
16
|
Lo CK, Mertz D and Loeb M:
Newcastle-Ottawa Scale: comparing reviewers' to authors'
assessments. BMC Med Res Methodol. 14(45):
doi:10.1186/1471-2288-14-45. 2014.PubMed/NCBI
|
|
17
|
Salanti G, Del Giovane C, Chaimani A,
Caldwell DM and Higgins JPT: Evaluating the quality of evidence
from a network meta-analysis. PLoS One. 9:e996822014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang C, Xu C, Li X and Zhang Y, Zhang S,
Zhang T and Zhang Y: Could camrelizumab plus chemotherapy improve
clinical outcomes in advanced malignancy? A systematic review and
network meta-analysis. Front Oncol. 11:7001652021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang C, Han X, Jin M, Xu J, Wang Y, Zhang
Y, Xu C, Zhang Y, Jin E and Piao C: The effect of video game-based
interventions on performance and cognitive function in older
adults: Bayesian network meta-analysis. JMIR Serious Games.
9:e270582021. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nikolakopoulou A, Higgins JPT,
Papakonstantinou T, Chaimani A, Del Giovane C, Egger M and Salanti
G: CINeMA: An approach for assessing confidence in the results of a
network meta-analysis. PLoS Med. 17:e10030822020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alaimo L, Lima HA, Moazzam Z, Endo Y, Yang
J, Ruzzenente A, Guglielmi A, Aldrighetti L, Weiss M, Bauer TW, et
al: Development and validation of a machine-learning model to
predict early recurrence of intrahepatic cholangiocarcinoma. Ann
Surg Oncol. 30:5406–5415. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song Y, Zhou G, Zhou Y, Xu Y, Zhang J,
Zhang K, He P, Chen M, Liu Y, Sun J, et al: Artificial intelligence
CT radiomics to predict early recurrence of intrahepatic
cholangiocarcinoma: A multicenter study. Hepatol Int. 17:1016–1027.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jolissaint JS, Wang T, Soares KC, Chou JF,
Gönen M, Pak LM, Boerner T, Do RKG, Balachandran VP, D'Angelica MI,
et al: Machine learning radiomics can predict early liver
recurrence after resection of intrahepatic cholangiocarcinoma. HPB
(Oxford). 24:1341–1350. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo Q, Sun C, Chang Q, Wang Y, Chen Y,
Wang Q, Li Z and Niu L: Contrast-enhanced ultrasound-based nomogram
for predicting malignant involvements among sonographically
indeterminate/suspicious cervical lymph nodes in patients with
differentiated thyroid carcinoma. Ultrasound Med Biol.
48:1579–1589. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang W, Xu L, Yang P, Zhang L, Wan D,
Huang Q, Niu T and Chen F: Novel nomogram for preoperative
prediction of early recurrence in intrahepatic cholangiocarcinoma.
Front Oncol. 8:3602018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moris D, Palta M, Kim C, Allen PJ, Morse
MA and Lidsky ME: Advances in the treatment of intrahepatic
cholangiocarcinoma: An overview of the current and future
therapeutic landscape for clinicians. CA Cancer J Clin. 73:198–222.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, Yang T, Wu M and Shen F:
Intrahepatic cholangiocarcinoma: Epidemiology, risk factors,
diagnosis and surgical management. Cancer Lett. 379:198–205. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bu F, Deng XH, Zhan NN, Cheng H, Wang ZL,
Tang L, Zhao Y and Lyu QY: Development and validation of a risk
prediction model for frailty in patients with diabetes. BMC
Geriatr. 23:1722023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin J, Su H, Zhou Q, Pan J and Zhou L:
Predictive value of nomogram based on Kyoto classification of
gastritis to diagnosis of gastric cancer. Scand J Gastroenterol.
57:574–580. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chong HH, Yang L, Sheng RF, Yu YL, Wu DJ,
Rao SX, Yang C and Zeng MS: Multi-scale and multi-parametric
radiomics of gadoxetate disodium-enhanced MRI predicts
microvascular invasion and outcome in patients with solitary
hepatocellular carcinoma ≤5 cm. Eur Radiol. 31:4824–4838. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Zhang Z, Wei L and Wei S:
Construction and validation of nomograms combined with novel
machine learning algorithms to predict early death of patients with
metastatic colorectal. Front Public Health. 10:10081372022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lei H, Li X, Ma W, Hong N, Liu C, Zhou W,
Zhou H, Gong M, Wang Y, Wang G and Wu Y: Comparison of nomogram and
machine-learning methods for predicting the survival of non-small
cell lung cancer patients. Cancer Innov. 1:135–145. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saleh H, Abd-El Ghany SF, Alyami H and
Alosaimi W: Predicting breast cancer based on optimized deep
learning approach. Comput Intell Neurosci. 2022:18207772022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ghoniem RM, Algarni AD, Refky B and Ewees
AA: Multi-modal evolutionary deep learning model for ovarian cancer
diagnosis. Symmetry. 13:6432021. View Article : Google Scholar
|