Introduction
Primary hepatocellular carcinoma (HCC) is currently
the most common malignancy and the leading cause of cancer-related
mortality worldwide. According to the International Agency for
Research on Cancer, liver cancer became the 6th most prevalent
disease (4.7%) and 3rd most common cause of cancer deaths (8.3%)
globally in 2020 (1). However,
certain drugs for HCC may not be fully effective due to the unique
immune microenvironment of the liver (2–4).
Therefore, further research is urgently needed to identify new
targets for the treatment of HCC.
The mucin (MUC) family is a group of highly
glycosylated macromolecules that are expressed at high levels in
mammalian epithelial cells. They contribute to the formation of
mucus barriers that prevent infection (5). Notably, certain MUCs are abnormally
expressed in cancer cells and involved in tumor development,
including cell proliferation, apoptosis suppression, chemical
resistance, metabolic reprogramming and immune evasion (6–8). MUC1,
also known as tumor-associated epithelial membrane antigen or
CD227, was the first MUC to be discovered (9). MUC1 expression is primarily detected
in malignant tumors that originate from epithelial cells and is
characterized by increased expression with loss of polarity and
structural changes (10). MUC1 can
promote the evasion of stress-induced apoptosis by cancer cells by
binding directly to the p53 regulatory domain and p53 response
elements (11). Meanwhile, MUC1 can
downregulate the expression of E-calcium mucus protein
(E-cadherin), which is one of the manifestations of increased tumor
cell invasion (12). According to a
study of primary liver cancer, patients with a high MUC1 expression
accounted for ≤68% of all patients, with the highest rate of
recurrence after surgery and a positive association with MUC1
expression intensity (13).
Multiple studies have reported that the increased expression of
MUC1 in HCC tissue is associated with HCC development (14–16).
Therefore, downregulating MUC1 expression in HCC may have potential
in the clinical treatment of HCC. Furthermore, it is crucial to
have a comprehensive understanding of the regulatory mechanism of
MUC1 for the development of molecularly targeted therapies against
HCC.
Circular (circ)RNAs have a loop structure, unlike
traditional linear RNAs. This unique structure makes them resistant
to RNA exonucleases, which increases their expression stability and
decreases their susceptibility to degradation (17). circRNAs contain numerous micro
(mi)RNA (miR)-binding sites that can act as ‘sponges’ of miRNA in
cells, interfering with the expression of miRNA-regulated target
genes (18). This ability of
circRNAs is known as the competitive endogenous RNA mechanism
(19,20). circRNAs serve a crucial regulatory
role in disease development by interacting with disease-associated
miRNAs. In the present study, circRNAs and miRNAs that may regulate
MUC1 expression were assessed, providing new insights for the
future clinical treatment of HCC.
Materials and methods
Patients and tissues
A total of five HCC tissues and their corresponding
adjacent normal tissues were collected from patients who underwent
surgery between March and May 2023 at the Department of General
Surgery, Third Hospital of Shanxi Medical University, Shanxi
Bethune Hospital (Taiyuan, China). The tissue samples were promptly
preserved at −80°C. The current study was approved by the Ethics
Committee of the Third Hospital of Shanxi Medical University,
Shanxi Bethune Hospital (approval no. YXLL-2023-226). All
participants provided written informed consent to participate in
the study.
Experimental cell lines
The human MIHA normal liver cell line and the
MHCC97L, MHCC97H, Huh7, Hep3B and HCCLM3 liver cancer cell lines
were purchased from Hunan Fenghui Biotechnology Co., Ltd. All cell
lines were subjected to mycoplasma testing. The cells were cultured
in DMEM (Boster Biological Technology) supplemented with 15% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Boster Biological Technology). The cells
were maintained at 37°C and 5% CO2.
Bioinformatics analysis
The best cut-off point of MUC1 expression value in
all samples of HCC was taken as a threshold, and the samples were
divided into MUC1 high and low expression groups. The difference in
overall survival (OS) and progression-free survival (PFS) between
the two groups was assessed using the log-rank test. Cox
multifactorial regression analysis of MUC1 expression and clinical
characteristics was used to assess whether MUC1 could serve as an
independent HCC prognostic factor. Correlations were assessed using
Pearson's correlation coefficient. The Cancer Genome Atlas
(https://portal.gdc.cancer.gov) database
was used to identify miRNAs that are downregulated in HCC
[log2(fold change) <-1; Padj<0.05] and
have the ability to regulate MUC1 expression (correlation <-0.2;
P<0.05). The Gene Expression Omnibus dataset GSE97332
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97332)
was used to identify circRNAs expressed at high levels
[log2(fold change) >1; Padj<0.05] in
HCC. RNA22 version 2 (https://cm.jefferson.edu/rna22/Interactive/;
P<0.05) and miRanda version 3.3a (http://www.bioinformatics.com.cn/local_miranda_miRNA_target_prediction_120;
MaxScore >140; MaxEnergy <-15) were used to predict the
relationships between miRNAs and circRNAs.
Total RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from the MHCC97L and MIHA
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and quantified using Qubit 4 Fluorometer
(Invitrogen; Thermo Fisher Scientific, Inc.). The M5 Super plus
qPCR RT Kit with genomic DNA remover (cat. no. MF166-plus-01; Mei5
Biotechnology, Co., Ltd.) was used; 1 µl of 10X gDNA plus remover
mix and 1 µg of RNA template, made up to 10 µl using (RNase)-free
double-distilled H2O and incubated at 42°C for 2 min,
then 4 µl 5X M5 RT Super plus Mix and as 6 µl (RNase)-free
double-distilled H2O, was added and incubated at 37°C
for 15 min and then 85°C for 5 sec. The riboSCRIPT
mRNA/lncRNA qRT-PCR Starter Kit (cat. no. C11030-2; Guangzhou
RiboBio Co., Ltd. was also employed; 1 µg of RNA template, 1 µl
Random Primer, 1 µl Oligo (dT)18, 2 µl 5X reverse
transcription buffer and 2 µl RTase Mix, made up to 10 µl using
(RNase)-free double-distilled H2O and incubated at 42°C
for 60 min and then 70°C for 10 min. These were used to
reverse-transcribe miRNA, mRNA and circRNA into cDNA on the C1000
Touch Thermal Cycler (Bio-Rad Laboratories, Inc.). Subsequent
RT-qPCR was performed using 2X M5 HiPer SYBR Premix EsTaq (Mei5
Biotechnology, Co., Ltd.) which included 2 µl cDNA, 12.5 µl 2X M5
Hiper SYBR Premix EsTaq, 0.5 µl forward and reverse primers and 9.5
µl ribonuclease (RNase)-free double-distilled H2O added
to the CFX96 Touch Real-Time PCR Detection System (Bio-Rad
Laboratories). The thermocycling conditions were as follows: 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30
sec. The relative expression of the target mRNA and miRNA were
quantified using the 2−ΔΔCq method (21). The primers sequences were as
follows: MUC1, 5′-CTGCTGGTGCTGGTCTGTGTTC-3′ forward and
5′-GGGTACTCGCTCATAGGATGGTAGG-3′ reverse; GAPDH,
5′-CAGGAGGCATTGCTGATGAT-3′ forward and 5′-GAAGGCTGGGGCTCATTT-3′
reverse; U6, 5′-CTCGCTTCGGCAGCACA-3′ forward and
5′-AACGCTTCACGAATTTGCGT-3′ reverse; miR-122-5p,
5′-CCTGGAGTGTGACAATGGTGTTTG-3′ forward; hsa_circ_0055054,
5′-TGATGTTGCAGCAGTAGTGGATGG-3′ forward and
5′-CCACACGAGAGAGATTGCAGC-3′ reverse; linear_0055054,
5′-GCTGCAATCTCTCTCGTGTGG-3′ forward and
5′-TCCATCCACTACTGCTGCAACATC-3′ reverse. The aforementioned
sequences were purchased from Sangon Biotech Co., Ltd. GAPDH or U6
were used as controls for mRNA, circRNA or miRNA. The reverse
miR-122-5p sequence (cat. no. B661601; Sangon Biotech Co., Ltd.)
could not be disclosed due to confidentiality agreements with the
reagent company.
RNase R assay
To evaluate the stabilization of hsa_circ_0055054,
the following reaction system was prepared in sterile
microcentrifuge tubes: 17.9 µl diethyl pyrocarbonate-treated water,
1 µg template total RNA, 2 µl 10X RNase R Reaction Buffer and 2 U
RNase R (20 U/µl; Shanghai Yeasen Biotechnology Co., Ltd.).
Subsequently, the mixture was incubated for 15 min at 37°C. RT-qPCR
was used to detect the relative abundance of circRNA and linear
RNA.
Western blotting
Protein were extracted from MHCC97L cells and MIHA
cells were using radioimmunoprecipitation assay lysis buffer
(Boster Biological Technology) and 1% protease/phosphatase
inhibitor cocktail (MedChemExpress). Protein concentration was
determined using the bicinchoninic acid protein assay kit (Boster
Biological Technology). The proteins were diluted to the desired
concentration by adding 5X SDS-PAGE loading buffer (Boster
Biological Technology) and PBS (Boster Biological Technology). The
proteins were subsequently denatured through incubation for 5 min
at 95°C in a metal bath. The proteins (20 µg per lane for each
sample) were separated by 10% SDS-PAGE and subsequently transferred
onto a PVDF membrane that had been previously treated with
anhydrous ethanol for 1 min. The membrane was blocked with
protein-free rapid sealing solution (Boster Biological Technology)
for 20 min at room temperature, after which anti-MUC1 rabbit
polyclonal antibodies (1:1,500; cat. no. A0333; ABclonal Biotech
Co., Ltd.) and anti-β-actin polyclonal antibodies (1:10,000; cat.
no. BA2305; Boster Biological Technology) were added and incubated
overnight at 4°C. The membrane was washed for 30 min with TBS
containing 0.05% Tween-20. Subsequently, secondary antibodies
(HRP-conjugated AffiniPure goat anti-rabbit IgG; 1:20,000; cat. no.
BA1039; Boster Biological Technology) was added, and the samples
were incubated at room temperature for 2 h and washed three times
for 10 min each. Eventually, protein visualization was performed
using the ChemiDoc Imaging System (Bio-Rad Laboratories, Inc.) and
the FG supersensitive ECL luminescence reagent (Dalian Meilum
Biotechnology Co., Ltd.).
Cell transfection
The following specific small interfering (si)RNA
sequences were purchased from Sangon Biotech Co., Ltd.: si-MUC1,
sense: 5′-CUCUCGAUAUAACCUGACGAUTT-3′ and antisense:
5′-AUCGUCAGGUUAUAUCGAGAGTT-3′; si-negative control (NC), sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense:
5′-ACGUGACACGUUCGGAGAATT-3′; miR-122-5p mimic, sense:
5′-UGGAGUGUGACAAUGGUGUUUG-3′ and antisense:
5′-AACACCAUUGUCACACUCCAUU-3′; miR-122-5p inhibitor, sense:
5′-CAAACACCAUUGUCACACUCCA-3′; miR-122-5p mimic-NC, sense:
5′-UUGUACUACACAAAAGUACUG-3′ and antisense:
5′-GUACUUUUGUGUAGUACAAUU-3′; miR-122-5p inhibitor-NC, sense:
5′-CAGUACUUUUGUGUAGUACAA-3′; and si-hsa_circ_0055054, sense:
5′-GCGUCAUCUUUGCAAAGACAATT-3′ and antisense:
5′-UUGUCUUUGCAAAGAUGACGCTT-3′. For si-MUC1 and si-hsa_circ_0055054,
a universal negative control was used, namely a common negative
control with no homology to the target gene sequence and no
significant homology to other mRNAs of the same species. MHCC97L
cells were cultured in 24-well plates (3×105 cells/well)
or 96-well plates (2×104 cells/well) and incubated for
24 h. Subsequently, 1.25 µl 20 µM miRNA was diluted with 30 µl 1X
riboFECT™ CP buffer (Guangzhou RiboBio Co., Ltd.). A total
of 3 µl riboFECT CP reagent was then added, the solution was mixed
gently and incubated at room temperature for 15 min, followed by
incubation with DMEM supplemented with 15% FBS at 37°C and 5%
CO2 for 48 h. After 48 h, RNAs and proteins were
extracted as in the western blotting and RT-qPCR section from the
cells and subsequently validated by western blotting and
RT-qPCR.
Dual-luciferase reporter assay
The sequences containing miR-122-5p-binding sites in
hsa_circ_0055054 and MUC1 3′-UTR, as well as site-directed mutation
of the binding sites produced by Sangon Biotech Co., Ltd.
(sequences as in Cell transfection section), were subcloned into
pmirGLO reporting luciferase vectors (Promega Corporation). The
wild-type (WT) or mutant (MT) luciferase reporter vectors were
constructed. Subsequently, miR-122-5p mimic or miR-122-5p mimic-NC
vectors were co-transfected into MHCC97L cells with the reporter
plasmids by Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). After 48 h, luciferase activity was
measured using the Dual-Luciferase Reporter Assay System (Promega
Corporation)and expressed as relative activity. In the two groups
transfected with the same Luciferase plasmid, the relative
expression of luciferase in the miRNA-NC group was normalized to 1,
and the value of the target miRNA group was divided by the value of
the miRNA-NC group to obtain the value of its relative expression
of luciferase.
Cell proliferation assay
MHCC97L cells in the logarithmic growth phase with a
cell fusion rate of ~85% were made into a cell suspension at a
concentration of 1×105 cells/ml. Subsequently, 100 µl
cell suspension was added to each well in a 96-well plate.
Following overnight incubation at 37°C and 5% CO2, the
cells were transfected as aforementioned. Cell proliferation was
assessed using the Cell Counting Kit-8 (Boster Biological
Technology). The plate was incubated for 1 h in a cell incubator.
Subsequently, optical density values were assessed using a
microplate reader (Thermo Fisher Scientific, Inc.) at 450 nm.
Wound healing assay
The MHCC97L cells were cultured in a 24-well plate
at a density of 2×105 cells/well. When the cell fusion
rate reached 100% confluence, the culture medium was removed and
wounds were made by a 200 µl pipette tip. Then serum-free medium
was added. Cellular wound healing was observed under an inverted
light microscope (magnification, ×100; Nikon Corporation) at both
the 0 and 48 h timepoints at the same location. The experiment was
repeated three times, and the relative scratch width was used to
quantify the migratory ability of the HCC cells. The relative
scratch width was calculated by dividing the distance of the
scratch zone at 48 h by the distance of the scratch zone at 0 h,
subtracting this result from 1, and then multiplying it by
100%.
Cell invasion assays
Experiments were performed using transfected MHCC97L
cells in a Transwell chamber containing 60 µl Matrigel (BD
Biosciences) diluted at 1:8 with serum-free medium (Boster
Biological Technology) and incubated 2 h at 37°C and 5%
CO2. Then the excess liquid of the upper chamber in the
completed incubation chambers was aspirated and 100 µl of
serum-free medium was added to each well and incubated for 30 min
at 37°C and 5% CO2. The upper compartment of the 24-well
Transwell chamber (8-mm pore size; Guangzhou Jet Bio-Filtration
Co., Ltd.) contained 3×105 cells and 200 µl DMEM
(without serum), whilst the lower compartment contained 600 µl
complete medium (20% FBS). After 24 h of continuous incubation at
37°C and 5% CO2, the cells in the lower layer were fixed
with 4% paraformaldehyde for 20 min and stained with 0.1% crystal
violet for 10 min at room temperature. Subsequently, the cells were
observed under an inverted light microscope (magnification, ×100;
Nikon Corporation).
In vivo study
Subcutaneous transplantation models of tumors were
constructed using 4-week-old female BALB/c nude mice provided by
SPF Biotechnology Co, Ltd. To construct short hairpin (sh)RNA,
si-NC and si-hsa_circ_0055054 were selected and assessed in
cellular experiments and lentivirally packaged. These cells were
then turned into stable transfected MHCC97L cells by Suzhou Haixing
biotechnology Co., Ltd. The mice were acclimatized to the specific
pathogen-free animal laboratory for 1 week under a 12-h light/dark
cycle. Following acclimatization, 10 mice were randomly assigned to
two groups of five mice using a random number table. Stably
transfected MHCC97L cells harboring sh-NC or sh-hsa_circ_0055054
were thawed and the expression levels of hsa_circ_0055054 were
determined by RT-qPCR as aforementioned. A total of
~5×106 MHCC97L cells (100 µl) transfected with sh-NC or
sh-hsa_circ_0055054 were subcutaneously injected into the
mid-posterior area of the right axilla of both groups of mice. The
mice were provided unrestricted access to food and water at a
temperature of 25°C throughout the experimentation period. The mice
with tumors were examined three times a week, and the maximum
allowable tumor volume (length × width2 × 0.5) was 2
cm3. In addition, if the nude mice lost >20% of their
body weight, showed obvious pain or self-injury, developed ulcers
or infections at the site of subcutaneous tumors, or the tumors
metastasized to other parts of the body during the course of the
experiments, the experiments were terminated, and the animals were
euthanized. After 4 weeks, the mice were euthanized by
CO2 overdose: CO2 was infused into the
euthanasia chamber at a rate of 30% of its volume per min, and the
tumors were removed. Subsequently, the volume and weight of each
tumor was calculated, and the tumors were fixed with 4%
paraformaldehyde (Boster Biological Technology) for further
experiments. During euthanasia, if an animal had no respiration or
pulse, no heartbeat for >5 min determined by stethoscope or by
touching the heart in the chest cavity, and if the corneal reflexes
of the animal were absent, pupils were dilated, and neural reflexes
had disappeared, the animal was considered to be dead. The animal
experimental procedures were approved and performed following the
guidelines of the Animal Ethics Committee of Shanxi Provincial
People's Hospital (approval no. 2023-451).
Statistical analysis
Data are expressed as the mean ± standard deviation,
and all experiments were repeated three times. SPSS (version 26.0;
IBM Corp.) was used for statistical analysis. The graphs were
produced using GraphPad Prism (version 9.5.1; Dotmatics) and ImageJ
(version 1.5.4, National Institutes of Health). Data from
proliferation, migration, invasion, tumor volume and weight,
western blotting and RT-qPCR were analyzed according to the type of
data and methods of comparison, depending on whether the data
conformed to a normal distribution. A paired Student's t-test was
used to assess the mean values of the HCC tissues and the
corresponding paraneoplastic normal tissues. The mean values
between the other two groups were compared using an unpaired
Student's t-test, and differences between multiple groups were
compared using one-way ANOVA with Dunnett's post-hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
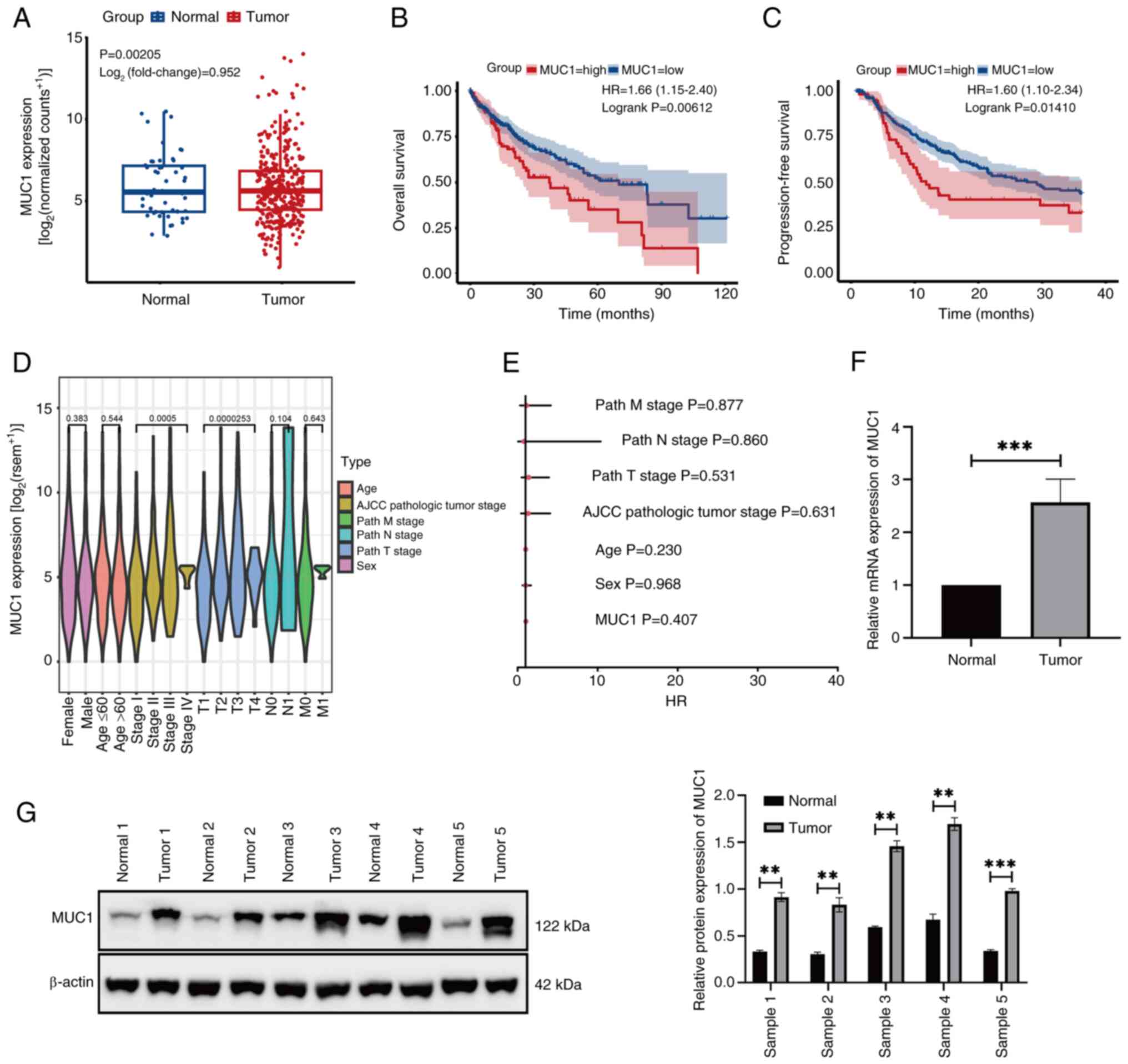
High MUC1 expression in HCC is
associated with a poor survival rate
The bioinformatics data demonstrated that MUC1 was
expressed at significantly higher levels in HCC tissues compared
with normal tissues (Fig. 1A).
Furthermore, Kaplan-Meier survival curve analysis indicated that
patients with a high MUC1 expression had significantly shorter OS
compared with those with a low MUC1 expression (Fig. 1B). Similarly, patients with high
MUC1 expression had a significantly shorter PFS than those with low
MUC1 expression (Fig. 1C). However,
further data analysis revealed that MUC1 expression in HCC was not
significantly associated with the clinical characteristics of the
patients (Fig. 1D and E). RT-qPCR
(Fig. 1F) and western blotting
(Fig. 1G) demonstrated that MUC1
was expressed at significantly higher levels in HCC tissues than in
normal tissues. These findings suggest that MUC1 overexpression is
associated with HCC development.
MUC1 downregulation impedes the
function of HCC
MUC1 expression was evaluated in the five liver
cancer lines MHCC97L, MHCC97H, Huh7, Hep3B and HCCLM3 using western
blotting and RT-qPCR. MIHA human normal liver cells were used as a
control. The results revealed that the levels of MUC1 in liver
cancer cells, particularly in MHCC97L cells, were significantly
higher than those in control cells (Fig. 2A). Therefore, the MHCC97L cell line
was selected for subsequent experiments. The experimental group was
co-cultured with si-MUC1, whilst the control group received either
only the transfection reagent or the transfection reagent and
si-NC. The successful transfection of si-MUC1 and a notable
reduction in MUC1 expression were confirmed by both western
blotting (Fig. 2B) and RT-qPCR
(Fig. 2C). Furthermore, reducing
the expression of MUC1 in MHCC97L cells led to a significant
reduction in migration (Fig. 2D),
invasion (Fig. 2E) and
proliferation (Fig. 2F) compared
with the Blank group.
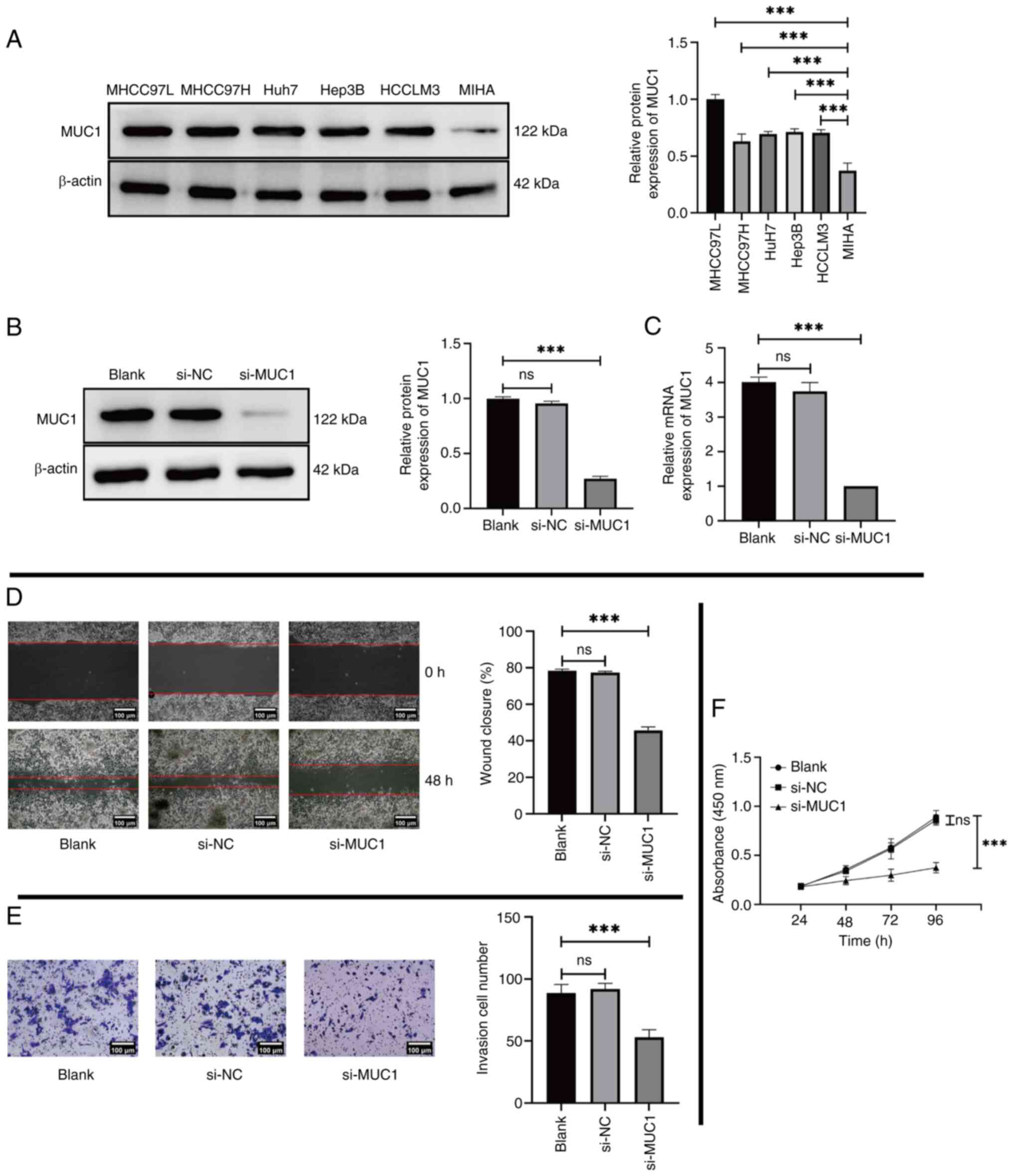 | Figure 2.Expression of MUC1 in several HCC
cell lines and the impact of MUC1 knockdown on the phenotypic
function of HCC cells. (A) Expression level of MUC1 in MHCC97L,
MHCC97H, Huh7, Hep3B and HCCLM3 cells was assessed using western
blotting, with MIHA cells used as control. MUC1 expression in
MHCC97L cells following transfection with only transfection
reagents (Blank) compared with si-NC and si-MUC1, assessed using
(B) western blotting and (C) reverse transcription-quantitative
PCR. (D) MHCC97L cell migration evaluated using a wound healing
assay following transfection with only transfection reagents
(Blank) and compared with si-NC and si-MUC1 (scale bar, 100 µm).
(E) Cell invasion assay assessing the invasion of MHCC97L cells in
the Blank group compared with si-NC and si-MUC1 groups (scale bar,
100 µm). (F) MHCC97L cell proliferation following transfection with
only transfection reagents (Blank) compared with si-NC and si-MUC1
was assessed using the Cell Counting Kit-8 assay. ***P<0.001.
ns, not significant; MUC1, mucin 1; HCC, hepatocellular carcinoma;
si, small interfering; NC, negative control. |
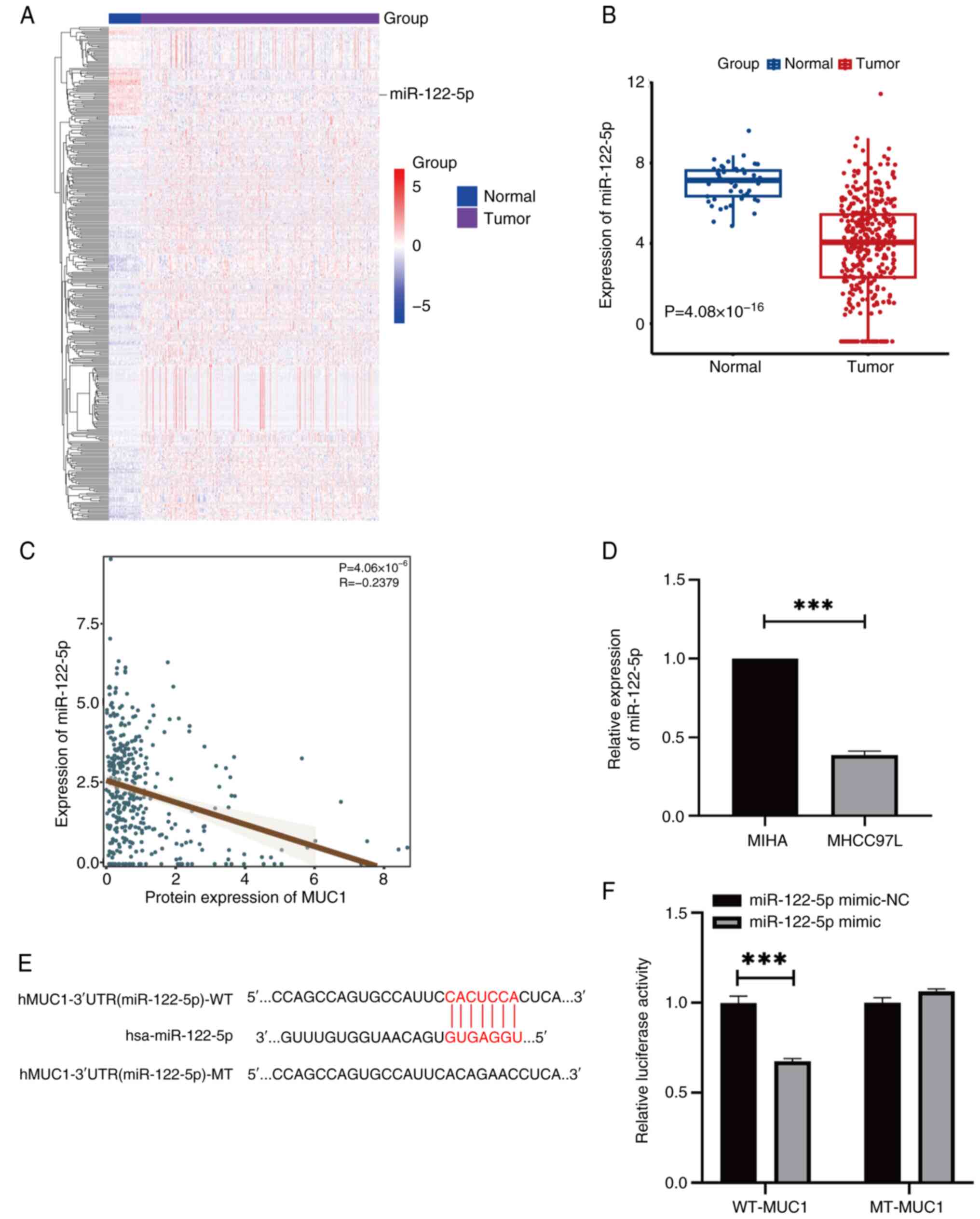
MUC1 is regulated by miR-122-5p
A total of 39 downregulated miRNAs were identified
in HCC, five of which were significantly negatively associated with
MUC1 expression. Considering its expression level and negative
associated with MUC1, miR-122-5p was selected for further
experiments (Fig. 3A). Pearson's
correlation coefficient and RT-qPCR revealed a significant negative
correlation between MUC1 and miR-122-5p (Fig. 3B-D) and the binding regions between
MUC1 and miR-122-5p were identified (Fig. 3E). In MHCC97L cells, the co-culture
of the miR-122-5p mimic with WT-MUC1 resulted in a significant
decrease in dual-luciferase reporter activity, whilst no
significant change was observed when the cells were co-transfected
with MT-MUC1 (Fig. 3F). These
findings suggest that miR-122-5p can bind to MUC1.
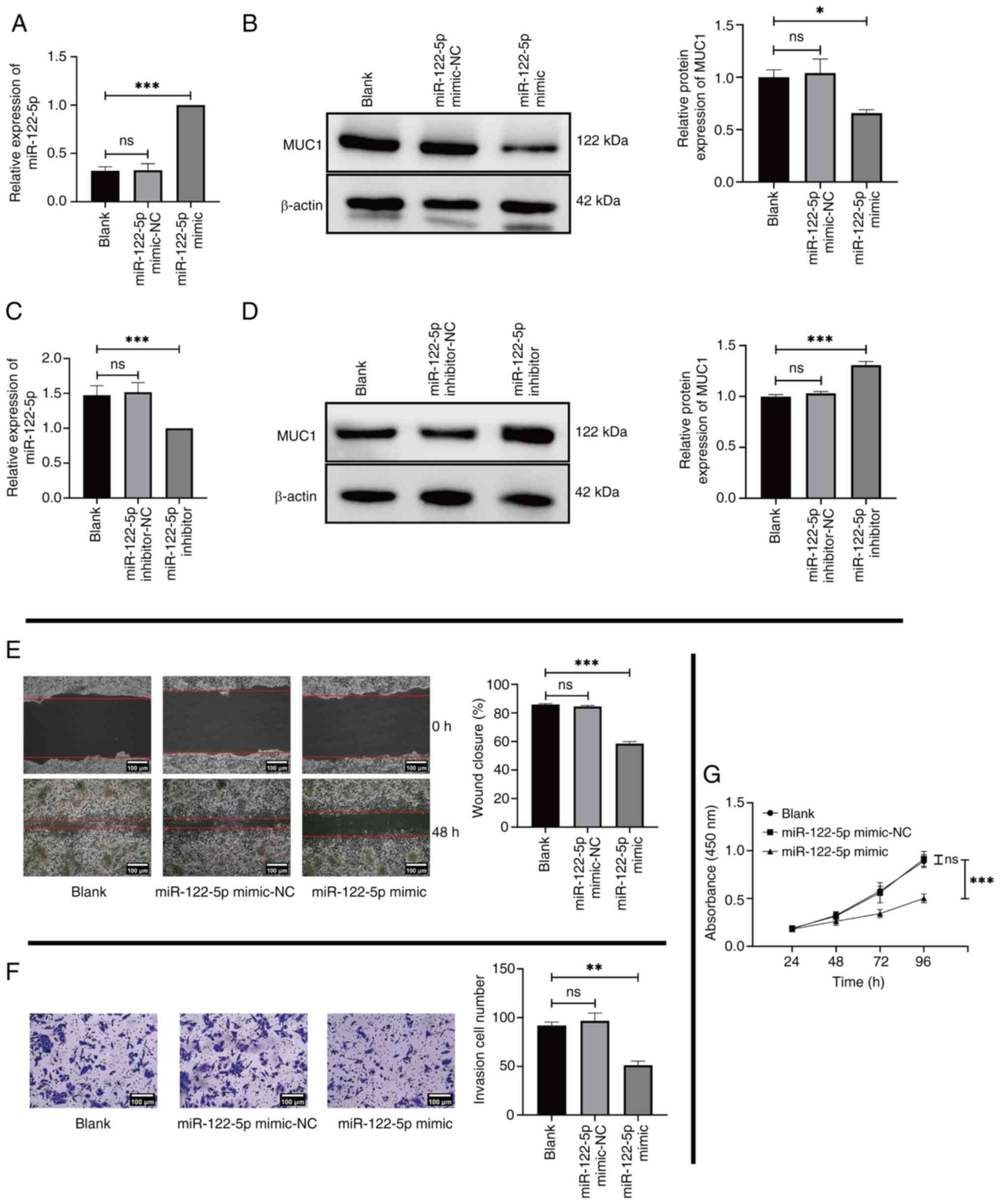
Effects of miR-122-5p on MHCC97L cells
through the regulation of MUC1 expression
To assess the role of miR-122-5p in HCC, MHCC97L
cells were transfected with miR-122-5p mimic or miR-122-5p
inhibitor. RT-qPCR and western blotting were used to determine the
expression of the miR-122-5p mimic/inhibitor and MUC1 in the
transfected cells. The results demonstrated that miR-122-5p mimic
was successfully transfected into MHCC97L cells (Fig. 4A). A significant reduction in MUC1
expression was observed in MHCC97L cells transfected with the
miR-122-5p mimic compared with the Blank group (Fig. 4B). Subsequently, miR-122-5p
inhibitor was used to evaluate the results of the previous
experiments by transfecting MHCC97L cells. The results revealed
that miR-122-5p inhibitor was successfully transfected into MHCC97L
cells (Fig. 4C) and MUC1 expression
was significantly elevated in the experimental group compared with
the Blank group (Fig. 4D). These
findings were consistent with aforementioned bioinformatics
analysis, which demonstrate that miR-122-5p negatively regulates
MUC1 expression. At the cellular level, MHCC97L cells transfected
with the miR-122-5p mimic were used to perform cell phenotype
experiments. The results revealed significant decreases in the
migration (Fig. 4E), invasion
(Fig. 4F) and proliferation
(Fig. 4G) of the miR-122-5p mimic
group compared with the Blank group.
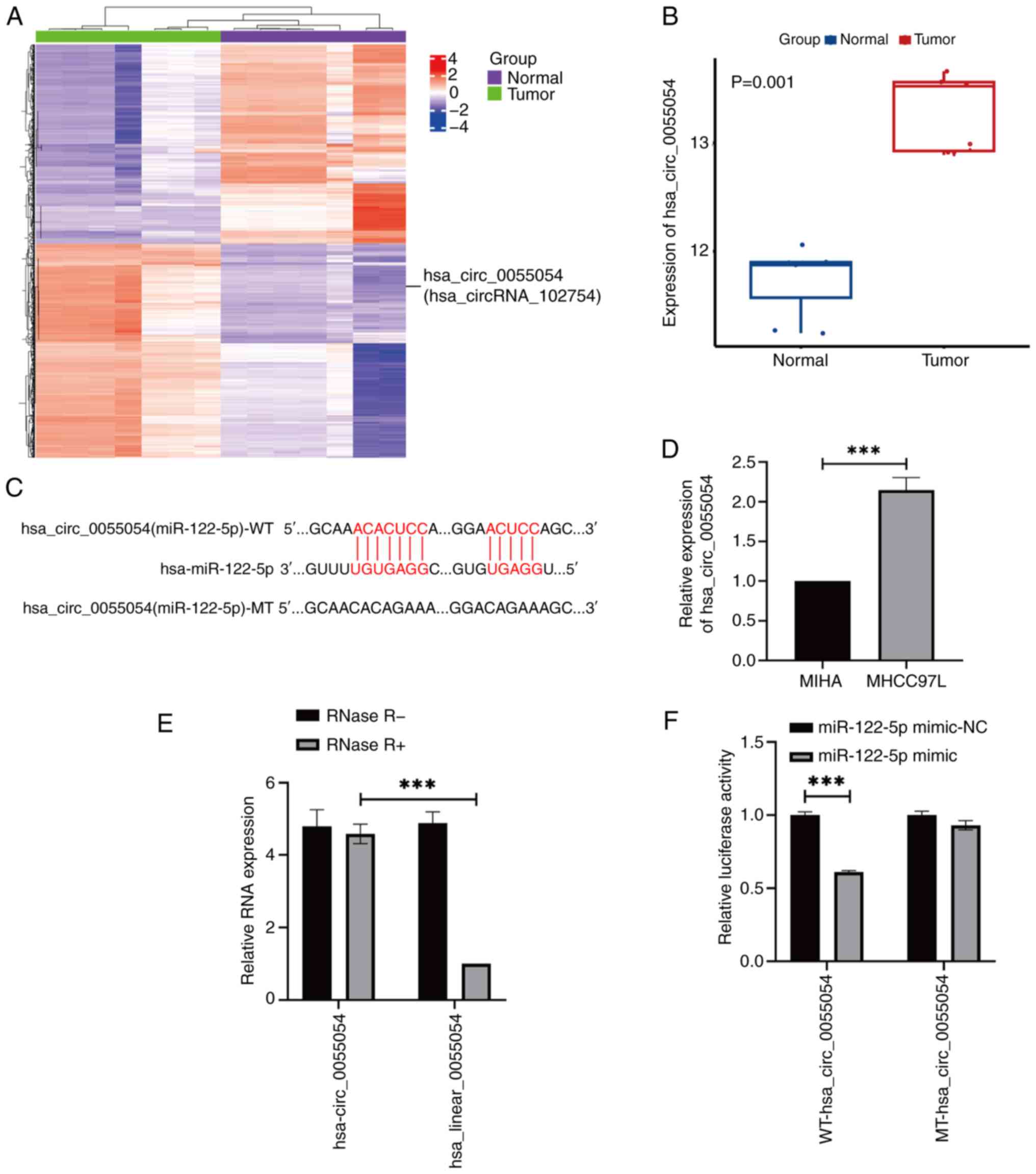
hsa_circ_0055054 can bind to
miR-122-5p
By analyzing the dataset GSE97332, it was found that
415 circRNAs were specifically expressed at high levels in HCC.
RNA22 and miRanda were then used to predict the binding sites of
these circRNAs to miR-122-5p. It was determined that
hsa_circ_0055054 satisfied the study criteria (Fig. 5A and B) and contained a binding site
for miR-122-5p (Fig. 5C).
Subsequently, RT-qPCR revealed that hsa_circ_0055054 was
significantly expressed at high levels in MHCC97L cells compared
with in MIHA cells (Fig. 5D). The
RNase R assay was then used to ascertain the stability of
hsa_circ_0055054 in MHCC97L cells. After RNase R treatment the
linear RNA was almost completely digested and the circRNA was
enriched. The findings indicated that hsa_circ_0055054 exhibited
resistance to RNase R treatments, whereas linear_0055054 was
significantly degraded (Fig. 5E).
Furthermore, dual-luciferase activity in MHCC97L cells
significantly decreased following transfection with the miR-122-5p
mimic containing WT-hsa_circ_0055054. However, no significant
change in dual-luciferase reporter activity was observed when the
miR-122-5p mimic was co-transfected with MT-hsa_circ_0055054
(Fig. 5F). This finding suggests an
interaction between miR-122-5p and hsa_circ_0055054.
hsa_circ_0055054 promotes MHCC97L cell
proliferation, migration and invasion by binding to miR-122-5p
To assess whether hsa_circ_0055054 regulates the
expression of MUC1 and its potential regulatory effects, MHCC97L
cells were co-cultured with si-NC or si-hsa_circ_0055054. RT-qPCR
and western blotting revealed significantly lower expression levels
of hsa_circ_0055054 (Fig. 6A) and
MUC1 (Fig. 6B) in the
si-hsa_circ_0055054 group compared with that in the Blank group.
Following transfection, cell phenotype experiments were performed,
which revealed that transfection with si-hsa_circ_0055054
significantly inhibited the migration (Fig. 6C), invasion (Fig. 6D) and proliferation (Fig. 6E) of MHCC97L cells to varying
degrees compared with the Blank group.
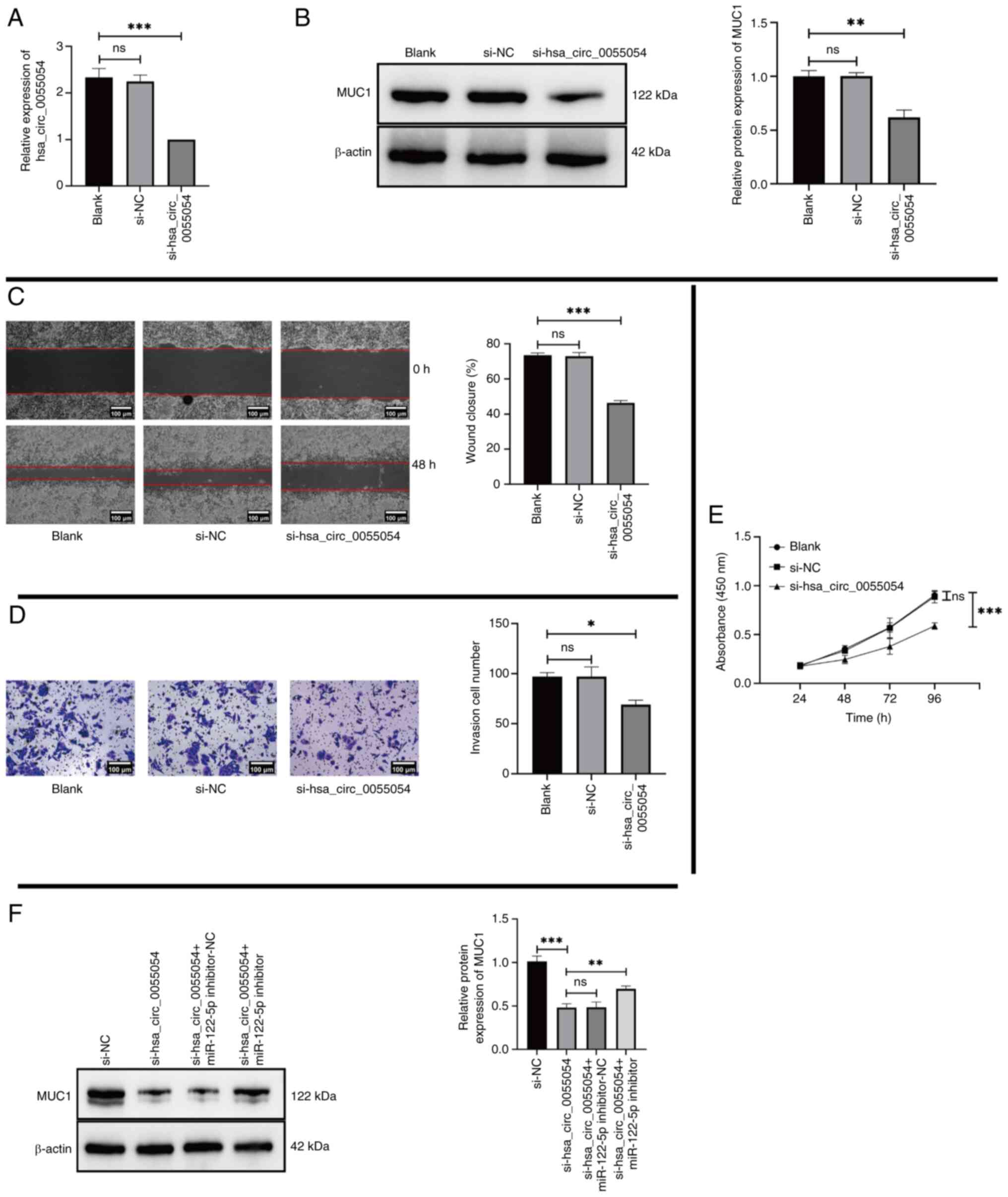 | Figure 6.hsa_circ_0055054 promotes MHCC97L
cell proliferation, migration and invasion by binding to
miR-122-5p. (A) Reverse transcription-quantitative PCR for the
detection of hsa_circ_0055054 expression and (B) western blotting
for the detection of MUC1 expression in MHCC97L cells following
transfection with only transfection reagents (Blank) compared with
si-NC and si-hsa_circ_0055054. (C) Migration of MHCC97L cells
evaluated using a wound healing assay following transfection with
only transfection reagents (Blank) compared with si-NC and
si-hsa_circ_0055054 (scale bar, 100 µm). (D) Cell invasion assay
performed to assess the invasion of MHCC97L cells in the Blank
group compared with si-NC and si-hsa_circ_0055054 groups (scale
bar, 100 µm). (E) MHCC97L cell proliferation following transfection
with only transfection reagents (Blank) compared with si-NC and
si-hsa_circ_0055054, assessed using the Cell Counting Kit-8 assay.
(F) Western blotting for the detection of MUC1 expression in
MHCC97L cells following transfection with si-hsa_circ_0055054
compared with si-NC, si-hsa_circ_0055054 with miR-122-5p
inhibitor-NC, and si-hsa_circ_0055054 with miR-122-5p inhibitor.
*P<0.05; **P<0.01; ***P<0.001. ns, not significant; miR,
microRNA; MUC1, mucin 1; si, small interfering RNA; NC, negative
control; circ, circular RNA. |
To confirm whether hsa_circ_0055054 regulates MUC1
expression by binding to miR-122-5p, subsequent rescue experiments
were performed. Si-NC, si-hsa_circ_0055054 co-transfected with
miR-122-5p inhibitor-NC, and si-hsa_circ_0055054 co-transfected
with miR-122-5p inhibitor were compared with si-hsa_circ_0055054,
respectively. The results indicated that the introduction of an
miR-122-5p inhibitor could partially restore the decreased
expression of MUC1 due to the suppression of hsa_circ_0055054
(Fig. 6F) and that hsa_circ_0055054
promoted MHCC97L cell proliferation, migration and invasion by
binding to miR-122-5p.
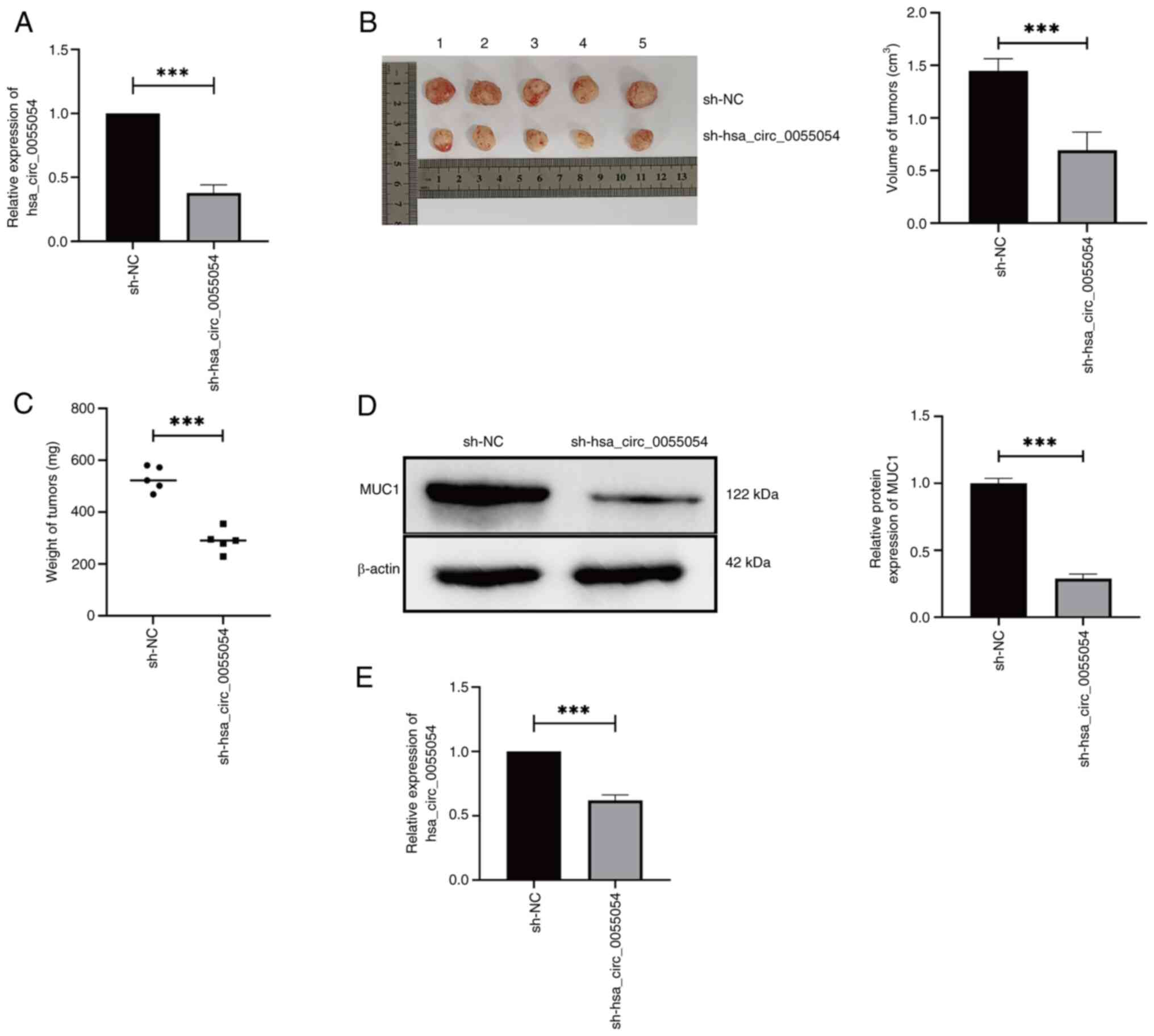
hsa_circ_0055054 knockdown inhibits
HCC growth in vivo
The RT-qPCR results indicated that the expression of
hsa_circ_0055054 was significantly lower in the experimental group
sh-hsa_circ_0055054 compared with that in the control group sh-NC.
These findings confirmed the successful generation of MHCC97L cells
stably transfected with sh-hsa_circ_0055054 (Fig. 7A). The results showed that MHCC97L
cells transfected with sh-hsa_circ_0055054 formed subcutaneous
tumors of significantly lower volume (Fig. 7B) and weight (Fig. 7C) compared with cells transfected
with sh-NC. Subsequently, a significant reduction in MUC1 (Fig. 7D) and hsa_circ_0055054 (Fig. 7E) expression were demonstrated in
the subcutaneous tumors formed by MHCC97L cells transfected with
sh-hsa_circ_0055054 compared with those transfected with sh-NC. The
aforementioned animal experiments provided evidence that
hsa_circ_0055054 knockdown successfully inhibited HCC
progression.
Discussion
Previous studies have reported that MUC1
overexpression in several tumor settings, such as thyroid cancer
(22), clear cell renal cell
carcinoma (23) and breast
carcinoma (24), promotes tumor
progression. With the development of bioinformatics and molecular
biology, the mechanism through which MUC1 may promote HCC
progression has been studied extensively. MUC1 may diminish the
impact of mitochondrial apoptotic factors and shield HCC cells from
anticancer genotoxic agents (25).
In established tumors, high levels of MUC1 may hinder the
proliferation of cytotoxic T lymphocyte (CTL) cells or cause CTL
death, thereby weakening the destructive effect of immune cells
(26). In addition, Wang et
al (27) and Bozkaya et
al (28) identified a
cooperative interaction of MUC1 with the JNK/TGF-β or HGF/c-Met
signaling pathway during hepatocarcinogenesis. In combination,
these findings indicated that MUC1 has potential as a tumor
biomarker for biological treatment (29–31).
Furthermore, the search for the upstream regulatory mechanisms of
MUC1 has become a new area of research interest in the field of
tumor therapy (8). The present
study demonstrated that MUC1 expression was significantly higher in
HCC tissues and cell lines compared with corresponding adjacent
normal tissues and normal liver cells. Furthermore, bioinformatics
analysis revealed that patients with a high MUC1 expression in HCC
had lower survival rates; however, bioinformatics analysis also
demonstrated that MUC1 was not significantly associated with the
clinical characteristics of patients with HCC. The results that
appeared to be contradictory lead to the investigation of the
potential ability of MUC1 inhibition in HCC to prevent HCC
progression. The present study demonstrated that the inhibition of
MUC1 expression in MHCC97L cells suppressed the proliferation,
migration and invasion of MHCC97L cells.
Epigenetics, a heritable change in the function of a
gene without a change in the DNA sequence of the gene, ultimately
leads to a change in phenotype. The regulation of gene
transcription by non-coding RNAs through certain mechanisms is one
of the forms of epigenetics (32).
For example, several diseases are treated by altering the
expression of certain circRNAs and miRNAs in the organism in such a
way as to affect their downstream targets in different signaling
pathways. Several RNA therapies are currently in clinical use
(33). Therefore, targeted therapy
of non-coding RNA (such as miRNA and circRNA) holds promise for the
treatment of several diseases.
miRNA is a post-transcriptional regulatory factor
that decreases the expression of target genes by acting on the mRNA
transcribed from genes (34).
Previous studies have reported that miR-122-5p is an abundant and
conserved liver-specific miRNA that regulates liver metabolism and
functions as a tumor suppressor (35–38);
however, the interpretation of the miR-122-5p target network
remains incomplete. The present study demonstrated that miR-122-5p
expression was significantly lower in HCC tissues compared with
corresponding adjacent normal tissues, which was consistent with
the results of Luna et al (39). Notably, the bioinformatics results
of the current study suggested that miR-122-5p can bind with MUC1
and regulate its expression. MHCC97L cells were transfected with
miR-122-5p mimic and miR-122-5p inhibitor, respectively, with the
aim of detecting MUC1 protein expression to demonstrate that
miR-122-5p negatively regulates MUC1 expression. Furthermore,
inhibition of miR-122-5p promoted MUC1 expression in MHCC97L cells,
which complemented the subsequent rescue experiments. These
findings were confirmed by dual-luciferase reporter assays and cell
function experiments, and indicated that the decreased expression
of miR-122-5p in MHCC97L cells promotes MUC1 overexpression, which
accelerates MHCC97L cell proliferation, migration and invasion.
There is increasing evidence that circRNAs serve a
significant regulatory role in disease through interactions with
disease-associated miRNAs (40–42).
In HCC, circ-GPR173B (14),
circ-ZEB1 (15) and circ-cSMARCA5
(16) have been reported to
regulate the growth and metastasis of HCC by acting as sponges for
different miRNAs. The hsa_circ_0055054 is formed through the
cyclization of exons 10–11 of the glucosamine-fructose-6-phosphate
aminotransferase isomerizing 1 (GFPT1) gene. Previous studies have
reported that GFPT1 may contribute to the progression of cervical
(43), pancreatic (44) and esophageal cancer (45), but the function of hsa_circ_0055054
remains unknown. The present study demonstrated that the expression
of hsa_circ_0055054 was significantly higher in HCC tissues
compared with the corresponding adjacent normal tissues, and that
it binds to miR-122-5p. Furthermore, the present study demonstrated
that hsa_circ_0055054 knockdown in MHCC97L cells led to a reduction
in MUC1 expression and inhibited the proliferation, migration and
invasion of MHCC97L cells. Furthermore, rescue experiments revealed
that hsa_circ_0055054 inhibited the phenotypic function of HCC
cells through a mechanism dependent on MUC1 by upregulating
miR-122-5p.
To determine whether hsa_circ_0055054 knockdown
could inhibit the proliferation, migration and invasion of MHCC97L
cells in vivo, animal experiments were performed. Lentiviral
packaging was used to construct cells that could stably proliferate
for a long period of time in animals using si-hsa_circ_0055054
sequences previously validated in cellular experiments. The results
were consistent with aforementioned cellular experiments,
suggesting that the hsa_circ_0055054/miR-122-5p/MUC1 regulatory
axis may serve a crucial role in HCC progression.
The present study had certain limitations; it
investigated whether hsa_circ_0055054 could regulate MUC1
expression through miR-122-5p, but did not investigate its pathway
and mechanism of action; meanwhile, other upstream regulatory
mechanisms of MUC1 need to be explored. In addition, it is
necessary to assess whether hsa_circ_0055054 and miR-122-5p can
influence the development of HCC by affecting other miRNAs and
mRNAs, and this is a topic that requires further investigation in
the future. Finally, since MUC1 can also serve an important role in
several tumors, further investigation is required to determine the
role of the hsa_circ_0055054/miR-122-5p/MUC1 regulatory axis in
other tumors.
In conclusion, the findings of the current study
indicate that hsa_circ_0055054 knockdown in MHCC97L cells can lead
to the increased expression of miR-122-5p and decreased expression
of MUC1. This can result in the inhibition of proliferation,
migration and invasion of HCC cells. Therefore, both
hsa_circ_0055054 and miR-122-5p show promise as future targets for
the clinical treatment of HCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PH and HZ conceived the project. PH and QL performed
the experiments. PH collected the data and drafted the manuscript.
HZ made contributions to the conception and design of the study, as
well as to the revision and correction of the manuscript. All
authors have read and approved the final manuscript. PH, QL and HZ
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Third Hospital of Shanxi Medical University, Shanxi
Bethune Hospital (approval no. YXLL-2023-226) and the Animal Ethics
Committee of Shanxi Provincial People's Hospital (approval no.
2023-451).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Junttila MR and de Sauvage FJ: Influence
of tumour micro- environment heterogeneity on therapeutic response.
Nature. 501:346–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ringel J and Löhr M: The MUC gene family:
Their role in diagnosis and early detection of pancreatic cancer.
Mol Cancer. 2:92003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhatia R, Gautam SK, Cannon A, Thompson C,
Hall BR, Aithal A, Banerjee K, Jain M, Solheim JC, Kumar S and
Batra SK: Cancer-associated mucins: Role in immune modulation and
metastasis. Cancer Metastasis Rev. 38:223–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Han Y, Sun C, Li X, Zheng J, Che J,
Yao X and Kufe D: Novel insights into the roles and therapeutic
implications of MUC1 oncoprotein via regulating proteins and
non-coding RNAs in cancer. Theranostics. 12:999–1011. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nabavinia MS, Gholoobi A, Charbgoo F,
Nabavinia M, Ramezani M and Abnous K: Anti-MUC1 aptamer: A
potential opportunity for cancer treatment. Med Res Rev.
37:1518–1539. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren J, Agata N, Chen D, Li Y, Yu WH, Huang
L, Raina D, Chen W, Kharbanda S and Kufe D: Human MUC1
carcinoma-associated protein confers resistance to genotoxic
anticancer agents. Cancer Cell. 5:163–175. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nath S and Mukherjee P: MUC1: A
multifaceted oncoprotein with a key role in cancer progression.
Trends Mol Med. 20:332–342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ye J, Wei X, Shang Y, Pan Q, Yang M, Tian
Y, He Y, Peng Z, Chen L, Chen W and Wang R: Core 3 mucin-type
O-glycan restoration in colorectal cancer cells promotes
MUC1/p53/miR-200c-dependent epithelial identity. Oncogene.
36:6391–6407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roy LD, Sahraei M, Subramani DB, Besmer D,
Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI, et al:
MUC1 enhances invasiveness of pancreatic cancer cells by inducing
epithelial to mesenchymal transition. Oncogene. 30:1449–1459. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan SF, Li KZ, Wang L, Dou KF, Yan Z, Han
W and Zhang YQ: Expression of MUC1 and its significance in
hepatocellular and cholangiocarcinoma tissue. World J
Gastroenterol. 11:4661–4666. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Gu M, Ma J, Wang Y, Li M, Wang H,
Yin X and Li X: CircGPR137B/miR-4739/FTO feedback loop suppresses
tumorigenesis and metastasis of hepatocellular carcinoma. Mol
Cancer. 21:1492022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu W, Zheng L, Zhang R, Hou P, Wang J, Wu
L and Li J: Circ-ZEB1 promotes PIK3CA expression by silencing
miR-199a-3p and affects the proliferation and apoptosis of
hepatocellular carcinoma. Mol Cancer. 21:722022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma
JZ, Sun SH, Yang F and Zhou WP: Circular RNA cSMARCA5 inhibits
growth and metastasis in hepatocellular carcinoma. J Hepatol.
68:1214–1227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19:1722020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ni J, Bucci J, Chang L, Malouf D, Graham P
and Li Y: Targeting MicroRNAs in prostate cancer radiotherapy.
Theranostics. 7:3243–3259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patel KN, Maghami E, Wreesmann VB, Shaha
AR, Shah JP, Ghossein R and Singh B: MUC1 plays a role in tumor
maintenance in aggressive thyroid carcinomas. Surgery.
138:994–1002. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lucarelli G, Netti GS, Rutigliano M,
Lasorsa F, Loizzo D, Milella M, Schirinzi A, Fontana A, Di Serio F,
Tamma R, et al: MUC1 expression affects the immunoflogosis in renal
cell carcinoma microenvironment through complement system
activation and immune infiltrate modulation. Int J Mol Sci.
24:48142023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schroeder JA, Adriance MC, Thompson MC,
Camenisch TD and Gendler SJ: MUC1 alters beta-catenin-dependent
tumor formation and promotes cellular invasion. Oncogene.
22:1324–1332. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan H, Wang J, Wang F, Zhang N, Li Q, Xie
F, Chen T, Zhai R, Wang F, Guo Y, et al: Mucin 1 gene silencing
inhibits the growth of SMMC-7721 human hepatoma cells through
Bax-mediated mitochondrial and caspase-8-mediated death receptor
apoptotic pathways. Mol Med Rep. 12:6782–6788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bose M and Mukherjee P: Microbe-MUC1
crosstalk in cancer-associated infections. Trends Mol Med.
26:324–336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Ni WH, Hu KB, Zhai XY, Xie F, Jie
J, Zhang NN, Jiang LN, Yuan HY and Tai GX: Targeting MUC1 and JNK
by RNA interference and inhibitor inhibit the development of
hepatocellular carcinoma. Cancer Sci. 108:504–511. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bozkaya G, Korhan P, Cokaklı M, Erdal E,
Sağol O, Karademir S, Korch C and Atabey N: Cooperative interaction
of MUC1 with the HGF/c-Met pathway during hepatocarcinogenesis. Mol
Cancer. 11:642012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Apostolopoulos V, Stojanovska L and
Gargosky SE: MUC1 (CD227): A multi-tasked molecule. Cell Mol Life
Sci. 72:4475–4500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carson DD: The cytoplasmic tail of MUC1: A
very busy place. Sci Signal. 1:pe352008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Supruniuk K and Radziejewska I: MUC1 is an
oncoprotein with a significant role in apoptosis (review). Int J
Oncol. 59:682021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Villanueva L, Álvarez-Errico D and
Esteller M: The contribution of epigenetics to cancer
immunotherapy. Trends Immunol. 41:676–691. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Winkle M, El-Daly SM, Fabbri M and Calin
GA: Noncoding RNA therapeutics-challenges and potential solutions.
Nat Rev Drug Discov. 20:629–651. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang N, Zheng J, Chen Z, Liu Y, Dura B,
Kwak M, Xavier-Ferrucio J, Lu YC, Zhang M, Roden C, et al:
Single-cell microRNA-mRNA co-sequencing reveals non-genetic
heterogeneity and mechanisms of microRNA regulation. Nat Commun.
10:952019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Zhang Z and Wang FS: The efficacy
of miRNA122, a novel therapeutic target, for predicting the
progression of hepatocellular carcinoma (HCC). Cell Mol Immunol.
9:103–104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: miR-122-a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakao K, Miyaaki H and Ichikawa T:
Antitumor function of microRNA-122 against hepatocellular
carcinoma. J Gastroenterol. 49:589–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z,
Liu J, Cui Y, Bian X, Bie P and Qian C: MicroRNA-122 sensitizes HCC
cancer cells to adriamycin and vincristine through modulating
expression of MDR and inducing cell cycle arrest. Cancer Lett.
310:160–169. 2011.PubMed/NCBI
|
|
39
|
Luna JM, Barajas JM, Teng KY, Sun HL,
Moore MJ, Rice CM, Darnell RB and Ghoshal K: Argonaute CLIP defines
a deregulated miR-122-bound transcriptome that correlates with
patient survival in human liver cancer. Mol Cell. 67:400–410.e7.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen S, Zhang Y, Ding X and Li W:
Identification of lncRNA/circRNA-miRNA-mRNA ceRNA network as
biomarkers for hepatocellular carcinoma. Front Genet.
13:8388692022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singh D, Kesharwani P, Alhakamy NA and
Siddique HR: Accentuating CircRNA-miRNA-transcription factors axis:
A conundrum in cancer research. Front Pharmacol. 12:7848012022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li D, Guan M, Cao X, Zha ZQ, Zhang P,
Xiang H, Zhou Y, Peng Q, Xu Z, Lu L and Liu G: GFPT1 promotes the
proliferation of cervical cancer via regulating the ubiquitination
and degradation of PTEN. Carcinogenesis. 43:969–979. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gong Y, Qian Y, Luo G, Liu Y, Wang R, Deng
S, Cheng H, Jin K, Ni Q, Yu X, et al: High GFPT1 expression
predicts unfavorable outcomes in patients with resectable
pancreatic ductal adenocarcinoma. World J Surg Oncol. 19:352021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang C, Lian H, Xie L, Yin N and Cui Y:
LncRNA ELFN1-AS1 promotes esophageal cancer progression by
up-regulating GFPT1 via sponging miR-183-3p. Biol Chem.
401:1053–1061. 2020. View Article : Google Scholar : PubMed/NCBI
|