Introduction
Glioblastoma multiforme (GBM), also known simply as
glioblastoma, is one of the most prevalent tumors of the central
nervous system in adults. It originates from glial brain cells and
is associated with an unfavorable prognosis (1,2).
Glioblastoma is the most prevalent and lethal primary brain tumor
in adults, with an estimated annual incidence of approximately
three cases per 100,000 individuals (1,3). These
tumors can be classified into two main categories based on the
presence or absence of specific genetic mutations: Primary
glioblastomas arise de novo, without a history of low-grade
gliomas, while secondary glioblastomas result from the progression
of low-grade gliomas to higher-grade tumors. In contrast to primary
GBMs, TP53 mutations associated with methylation of the MGMT
promoter are observed in most secondary GBMs, along with partial
loss of heterozygosity of 10q, 13q, 19q, and 22q (4).
Despite the administration of optimal treatments,
including radical surgical resection in conjunction with standard
radiotherapy and chemotherapy, patients with GBM have a median
survival duration of only ~16 months following diagnosis (5). In contrast to extracranial cancers,
GBM has the capacity to infiltrate deeply into the surrounding
brain tissue, with a low propensity for metastasis outside of the
brain (6). The recurrence patterns
and diffuse infiltration of GBM are in part attributable to the
tortuous blood vessels of these tumors, which provide migration
routes for tumor cells (7). At
present, surgical intervention remains the primary treatment for
GBM, with only a limited number of specific therapeutic agents
being employed. Consequently, there is a pressing need for the
identification of novel and efficacious targets.
GBM is highly heterogeneous, as evidenced by diverse
histological features and alterations in epigenetics, genetics and
transcriptomics (1). A number of
biomarkers and therapeutic targets have been identified for GBM,
including TP53 mutations, loss of heterozygosity 10q,
PTEN mutations, EGFR amplification and the aberrant
expression of E3 ubiquitin (Ub) ligases (E3s) (8–10). E3s
are proteins that regulate the turnover and activity of numerous
target proteins (10). Among them,
RING finger E3s are crucial in maintaining the equilibrium between
cell proliferation and apoptosis (11,12).
RING finger protein 135 (RNF135) is a member of the
E3 family, which is characterized by an N-terminal RING finger
domain and C-terminal SPRY and PRY motifs (13). A study of Schwann tumor cells from
malignant peripheral nerve sheath tumors revealed the
downregulation of RNF135, and postulated that RNF135 may contribute
to the heightened malignant risk observed in patients with
neurofibromatosis 1 gene microdeletion (14). In addition, another study identified
that RNF135 is upregulated in glioblastoma tissue and demonstrated
that RNF135 promotes the proliferation of human glioblastoma cells,
primarily via the ERK pathway (10). However, the specific substrate
targeted by RNF135 was not investigated in that study. In general,
E3 ligases exert their biological functions mainly through the
ubiquitination of substrates, so the underlying mechanism requires
further elucidation. In the present study, the substrate of RNF135
was identified and its functions in GBM were extensively
studied.
Materials and methods
Data analysis using public
databases
Gene Expression Profiling Interactive Analysis 2
(GEPIA2; http://gepia2.cancer-pku.cn) is a
tool for the analysis of gene expression data from The Cancer
Genome Atlas (TCGA) and the Tissue Genotype Expression database
(15). GEPIA2 was used to compare
the mRNA expression of RNF135 in human GBM with that in normal
tissue, and the P-value was calculated using an unpaired Student's
t-test. A significant difference was defined as an absolute log2
fold change of ≥1 and P<0.05. GEPIA2 was also used to perform a
prognostic value analysis of RNF135 by calculation of the overall
survival rate of patients with GBM. The survival graph was
generated directly by GEPIA2, using the log-rank test as the sole
option for analysis. Hazard ratios with 95% confidence intervals
and log-rank P-values were calculated.
Plasmid construction
Plasmids containing RNF135 or p21 with Myc or Flag
tag were inserted into empty vectors, including pCDH, pCADNA3.0,
pGEX4T-1, pET22b, pDEST32 or pDEST22 were purchased from Shanghai
Cell Researcher Biotech Co., Ltd. The pGEX4T-1-RNF135 and
pET22b-p21 vectors were employed to express recombinant proteins
for the purpose of conducting a GST pull-down and in vitro
ubiquitination assay. Mutations of p21 were generated using
site-directed mutagenesis, as previously described (16). Table
I presents the specific sequences of short hairpin RNAs
(shRNAs) targeting RNF135 in pLKO.1 vector and p21 in pGPU6/Hygro
vectors, which were also purchased from Shanghai Cell Researcher
Biotech Co., Ltd.
 | Table I.Sequences of shRNAs targeting RNF135
and cyclin-dependent kinase inhibitor 1A/p21. |
Table I.
Sequences of shRNAs targeting RNF135
and cyclin-dependent kinase inhibitor 1A/p21.
| shRNA | Target site sequence
(5′-3′) |
|---|
| Scramble |
GCGCGATAGCGCTAATAATTT |
| shRNF135-1 |
CATCCATCCAACCTTTAACTT |
| shRNF135-2 |
GCAGTAGAGAAGAGCATCACA |
| shRNF135-3 |
GTGGACAATCAGGAGAAGCTT |
| shp21-1 |
GCTGATCTTCTCCAAGAGGAA |
| shp21-2 |
CGCTCTACATCTTCTGCCTTA |
| shp21-3 |
GACAGATTTCTACCACTCCAA |
Cell culture, transfection and
reagents
The human U87 glioblastoma of unknown origin (cat.
no. TCHu138) and U251 GBM cell lines, as well as the human 293T
cell line, were purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. The authenticity of
all three cell lines was validated through the use of short tandem
repeat profiling, as detailed on the National Collection of
Authenticated Cell Cultures website (https://www.cellbank.org.cn/). The cell lines were
cultured in high-glucose Dulbecco's Modified Eagle Medium
supplemented with 10% fetal bovine serum and 100 µg/l
streptomycin/penicillin. The cells were transfected with the
overexpression vectors and shRNA using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The ratio of plasmid mass to
Lipofectamine® 2000 volume was 1:1 (2 µg:2 µl). The two
components were mixed together at room temperature for 20 min,
after which they were added to cells for continued culture at 37°C.
Stable cell lines were generated by selecting with puromycin (2
µg/ml; Beyotime Institute of Biotechnology) or hygromycin B (400
µg/ml; Beyotime Institute of Biotechnology) for ≥7 days.
Cycloheximide (CHX; Selleck Chemicals) was dissolved
in dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) at a
concentration of 100 mM and stored at −40°C. The U87 and U251 cells
were treated with 100 µM CHX for 3 or 6 h at 37°C in an incubator
before immunoblotting (IB) to assess protein degradation. For
analysis of the p21 degradation pathway, U87 and U251 cells were
treated with 1 µM proteasomal inhibitor bortezomib (BTZ; Selleck
Chemicals) or 20 nM autophagy inhibitor bafilomycin (BAF; Selleck
Chemicals) in combination with 100 µM CHX for 6 h at 37°C prior to
IB analysis.
Cell proliferation assay
Stably transfected U87 and U251 cells were seeded
into 96-well plates at a density of 5,000 cells per well. At the 0,
24, 48 and 72 h time points, where the 0 h time point was 6 h after
seeding, cells were incubated with Cell Counting Kit-8 (CCK-8)
solution (Beyotime Institute of Biotechnology) for 3 h. The
absorbance was then measured using a microplate reader (Bio-Rad
Laboratories, Inc.) at a wavelength of 450 nm. The experiments were
conducted with six replicates and repeated at least three
times.
Colony formation assay
The transfected U87 and U251 cells were seeded into
6-well plates at a density of 1,000 cells per well. After 7 days of
incubation at 37°C, the cells were fixed with 4% paraformaldehyde
(Sigma-Aldrich; Merck KGaA) for 15 min at room temperature, stained
with 0.2% crystal violet (Beyotime Institute of Biotechnology) for
10 min at room temperature. The number of colonies was manually
counted using a light microscope (CKX53; Olympus Corporation). A
colony was considered to comprise ≥50 cells.
Recombinant protein purification
Recombinant proteins tagged with glutathione
S-transferase (GST) or hexahistidine (His6) were purified from the
BL21 Escherichia coli (E. coli) system, following
previously described methods (16).
Following induction with isopropyl-β-D-mercapto-galactopyranoside
(Sangon Biotech Co., Ltd.) overnight at 4°C, the cells transfected
with protein-encoding plasmids were centrifuged and lysed in
phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM
Na2HPO4 and 2 mM
KH2PO4, pH 7.6). The samples were incubated
with glutathione or Ni2+ affinity gels (Sangon Biotech
Co., Ltd.), and eluted with 20 mM reduced L-glutathione solution
(pH 8.0) or 400 mM imidazole solution (pH 8.0). The eluate was
subsequently dialyzed in PBS buffer with 20% glycerol overnight at
4°C, after which aliquots were taken and stored at −80°C.
GST pull-down assay
GST-tagged protein (20 µg), His6-tagged protein (20
µg) and 50 µl Glutathione Sepharose™ 4B (Sangon Biotech
Co., Ltd.) were incubated in 600 µl GST pull-down buffer [20 mM
Tris-Cl, 1 mM EDTA, 5 mM MgCl2, 100 mM NaCl, 1% NP-40
and 1 mM dithiothreitol (DTT), pH 7. 6] overnight at 4°C, with the
addition of 10 mg/ml of fresh bovine serum albumin (Beyotime
Institute of Biotechnology). The samples were then centrifuged at
2,000 × g for 2 min at 4°C, and washed three times with GST
pull-down buffer. The immunoprecipitates were denatured in 40 µl 2X
SDS protein loading buffer at 100°C for 10 min before analysis by
IB.
Yeast two-hybrid (Y2H) screening
The Y2H assay was performed as described previously
(17). Briefly, empty vectors or
pDEST32-RNF135 were co-transformed with
pDEST22-p21/cyclin-dependent kinase inhibitor 1A (CDKN1A) into
yeast strain MaV203 (Thermo Fisher Scientific, Inc.). The positive
colonies were selected and tested for their ability to survive in
SD-2 medium (deficient in leucine and tryptophan) and SD-4 medium
(deficient in uracil, histidine, leucine and tryptophan) which were
purchased from Shanghai Cell Researcher Biotech Co., Ltd. This was
followed by staining with
5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-gal; Sangon
Biotech Co., Ltd.).
Co-immunoprecipitation (Co-IP),
immunoprecipitation (IP) and IB
For the Co-IP assay, transfected U87 and U251 cells
were lysed in 600 µl Co-IP buffer (50 mM Tris-HCl, 5 mM EDTA, 150
mM NaCl and 1% NP-40, pH 7.5) supplemented with a fresh protease
inhibitor cocktail (Roche Diagnostics GmbH). The cell lysates (500
µl) were then incubated overnight at 4°C with an anti-p21 antibody
(1:100 dilution; 10355-1-AP; Wuhan Sanying Biotechnology;
Proteintech Group, Inc.) and 25 µl Protein G magnetic beads
(L-1002; Biolinkedin). For the IP assay, transfected cells were
lysed in 600 µl RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 5 mM
EDTA, 0.1% SDS and 1% NP-40, pH 7.5) containing a fresh protease
inhibitor cocktail. Subsequently, the cell lysates (500 µl) were
incubated with either the anti-p21 antibody or anti-Flag antibody
(1:100 dilution; 66008-4-Ig; Proteintech Group, Inc.) and 25 µl
Protein G magnetic beads overnight at 4°C. The immunoprecipitates
were pelleted and washed three times with Co-IP or RIPA buffer as
aforementioned, followed by denaturation for 10 min at 100°C in 40
µl 2X SDS protein loading buffer. Subsequently, the
immunoprecipitates, along with inputs and other lysates, were
subjected to 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes (MilliporeSigma). After blocking with 5% non-fat
milk at room temperature for 1 h, the membranes were incubated with
the following primary antibodies overnight at 4°C: Anti-RNF135
(1:1,000 dilution; 25061-1-AP), anti-GAPDH (1:5,000 dilution;
60004-1-Ig), anti-p21 (1:1,000 dilution; 10355-1-AP), anti-p53
(1:1,000 dilution; 10442-1-AP), anti-Myc (1:5,000 dilution;
16286-1-AP), anti-His (1:5,000 dilution; 66005-1-Ig), anti-GST
(1:10,000 dilution; HRP-66001), anti-Flag (1:2,000 dilution;
20543-1-AP), anti-hemagglutinin (1:2,000 dilution; 81290-1-RR) or
anti-Ub (1:2,000, 10201-2-AP), all from Proteintech Group, Inc. The
membranes were then washed three times with Tris-buffered saline
with Tween-20 (50 mM Tris-HCl, 150 mM NaCl and 0.2% Tween-20, pH
8.0). The next day, the membranes were incubated with goat
anti-mouse IgG (1:5,000 dilution; SA00001-1; Proteintech Group,
Inc.) or goat anti-rabbit IgG (1:5,000 dilution; SA00001-2;
Proteintech Group, Inc.) at room temperature for 2 h. High-signal
ECL western blotting substrate (180–5001; Tanon Science &
Technology Co., Ltd.) was used to visualize the signals. Images
were captured using a Tanon 5200 imaging system (Tanon Science
& Technology Co., Ltd.).
In vitro ubiquitination assay
An in vitro ubiquitination assay was
conducted following the previously described method (16). Briefly, 20 ng His6-UBA1 (E1), 50 ng
His6-UBCH5A (E2), 100 ng GST-RNF135 (E3), 100 ng His6-p21 and 20 ng
His6-Ub were added to in vitro ubiquitination buffer (25 mM
Tris-Cl, 5 mM MgCl2 and 100 mM NaCl, pH 7.6,
supplemented with 0.5 mM DTT and 1 mM fresh ATP), brought to a
final volume of 25 µl and incubated for 1 h at 37°C. Then, 100 ng
Ub carboxyl-terminal hydrolase 2 catalytic core (Usp2cc; Shanghai
Cell Researcher Biotech Co., Ltd.) was added to the mixture, which
was then incubated at 37°C for 30 min. The ubiquitination of p21
was detected through IB analysis using an anti-p21 antibody.
Statistical analysis
Data were analyzed using GraphPad Prism 5 (GraphPad;
Dotmatics) and expressed as the mean ± SD. Statistical significance
was determined using unpaired Student's t-test or one-way ANOVA
with Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
RNF135 promotes the proliferation of
glioblastoma cells
To investigate the potential role of RNF135 in GBM,
TCGA data was analyzed using GEPIA2. At the mRNA level, the
expression of RNF135 was found to be significantly upregulated in
GBM tissues compared with relevant normal tissues (Fig. 1A). The association between RNF135
and the clinical outcome of GBM was also analyzed using GEPIA2. It
was found that patients with GBM who exhibited high mRNA expression
levels of RNF135 had poorer overall survival (Fig. 1B).
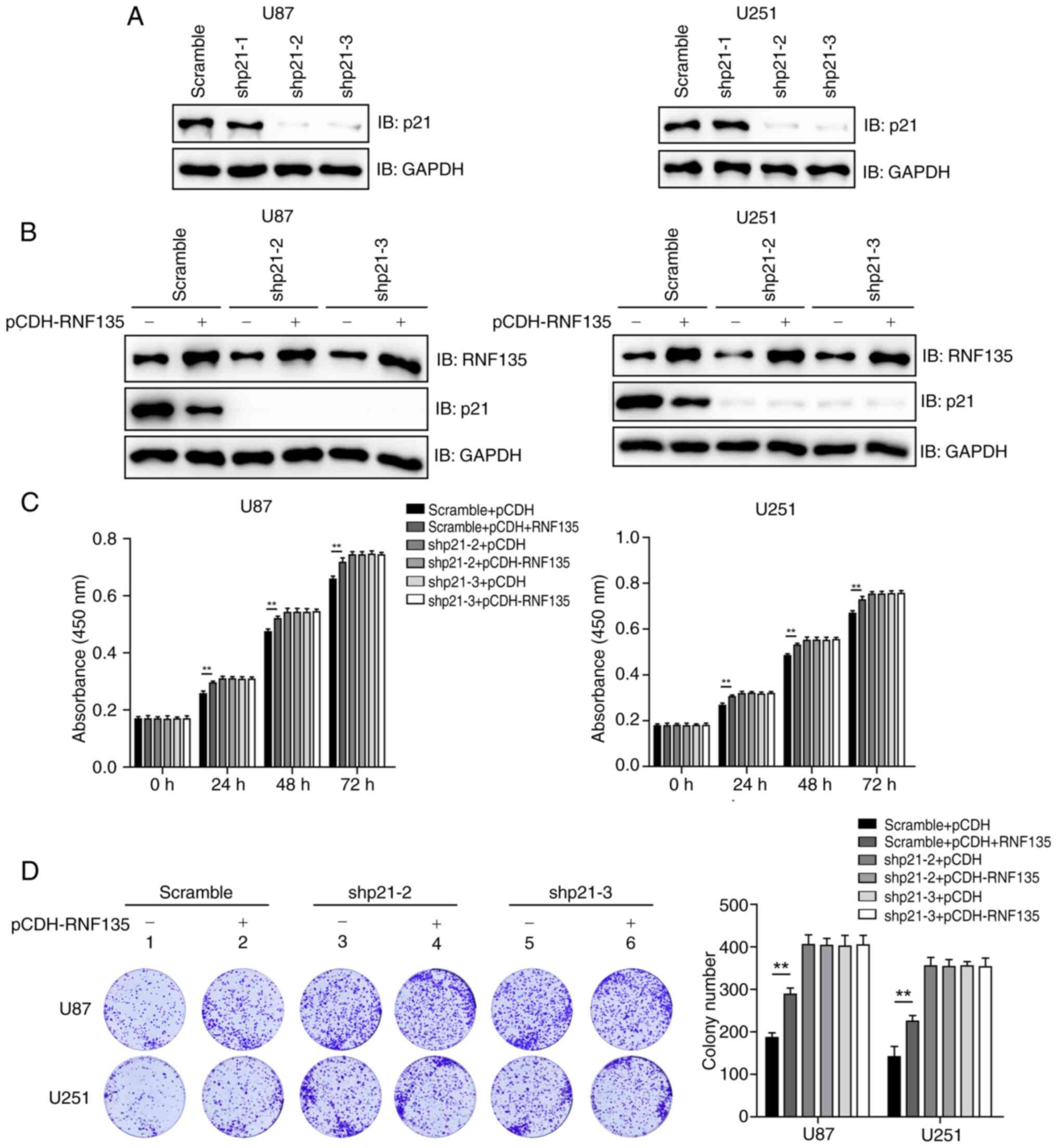
 | Figure 1.RNF135 promotes the proliferation of
glioblastoma cells. (A) Expression of RNF135 at the mRNA level was
higher in GBM than in corresponding normal tissues, according to
GEPIA2 database analysis. (B) Patients with GBM exhibiting high
RNF135 mRNA expression levels exhibited poor overall survival rates
according to Kaplan-Meier analysis performed using the GEPIA2
database. (C) Efficiency of an RNF135 overexpression vector in U87
and U251 cells was determined by IB. (D) RNF135 overexpression
promotes the proliferation of U87 and U251 cells as detected by
CCK-8 assay. The 0 h time point was defined as 6 h after cell
seeding. This experiment was repeated three times with six
replicates. (E) Overexpression of RNF135 promotes the colony
formation of GBM cells. Three samples per group were analyzed. (F)
Knockdown efficiency of three RNF135 shRNAs was determined by IB in
stably transfected U87 and U251 cells established using puromycin
selection. (G) RNF135 knockdown inhibits the proliferation of GBM
cells stably expressing RNF135 shRNAs as assessed using the CCK-8
assay. The experiment was repeated three times with six replicates.
(H) RNF135 knockdown inhibits the colony formation of GBM cells.
Three samples per group were analyzed. *P<0.05 and **P<0.01.
RNF135, RING finger protein 135; GBM, human glioblastoma; GEPIA2,
Gene Expression Profiling Interactive Analysis 2; T, tumors; N,
normal tissue; TPM, transcript per million; HR, hazard ratio;
p(HR), P-value for the HR; IB, immunoblotting; CCK-8, Cell Counting
Kit-8; shRNA, short hairpin RNA; shRNF135, shRNA targeting
RNF135. |
U87 and U251 cells were transfected with either an
empty vector (pCDH) or pCDH-RNF135. The expression of RNF135 was
detected using IB analysis, which confirmed that pCDH-RNF135
increased the expression of RNF135 (Fig. 1C). CCK-8 assays conducted at 24, 48
and 72 h revealed that cell growth was promoted in the U87 and U251
cells transfected with pCDH-RNF135 in comparison with that in the
pCDH groups (Fig. 1D). Colony
formation assays were also conducted, and the results were found to
be consistent with those of the CCK-8 assays, showing that the
overexpression of RNF135 increased colony formation (Fig. 1E).
Three shRNAs targeting RNF135 were designed and
tested in U87 and U251 cells. The knockdown efficiencies were
determined by IB analysis (Fig.
1F). shRNF135-1 and shRNF135-2 exhibited high knockdown
efficacy and were therefore selected for use in further
experiments. CCK-8 and colony formation assays demonstrated that
cell growth and colony formation was inhibited in the
RNF135-knockdown cells transfected with shRNF135-1 and shRNF135-2
in comparison with those in the negative control (scramble) groups
(Fig. 1G and H). The results
suggest that RNF135 plays a regulatory role in the proliferation of
GBM cells.
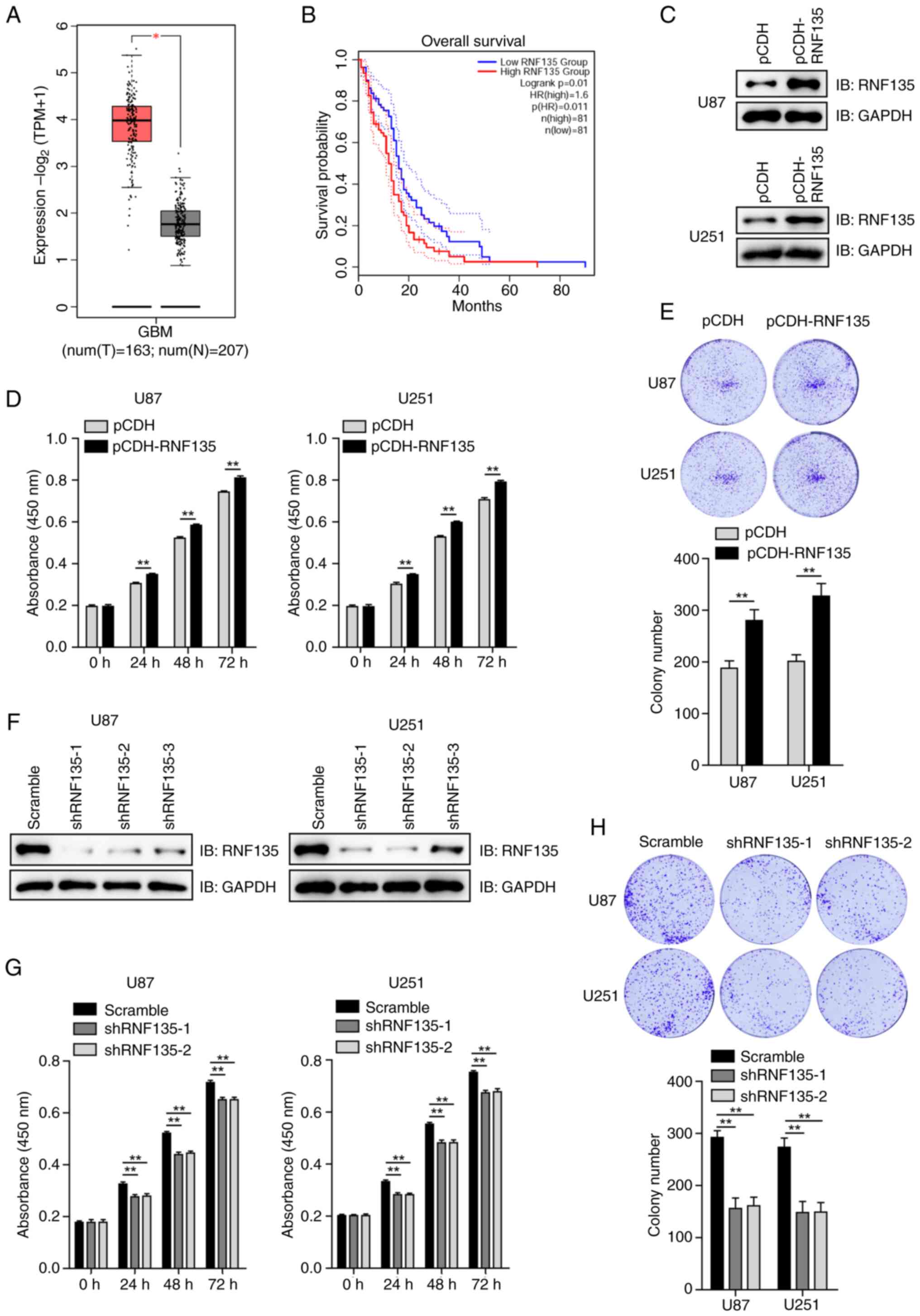
RNF135 interacts with and mediates the
degradation of p21
The mechanism by which RNF135 controls the
proliferation of GBM cells is unknown. As the p53-p21 signaling
pathway is the most classic cell cycle regulatory pathway (18), the effect of RNF135 on p53 and p21
was investigated. In the RNF135-knockdown U87 and U251 cells, the
protein levels of cell cycle-related p21 were markedly increased
compared with those in the scramble groups, whereas p53 protein
levels did not show any clear changes following RNF135 knockdown
(Fig. 2A). The impact of RNF135 on
the stability of p21 protein was then examined. A
pCDNA3.0-RNF135-Myc plasmid was constructed and transiently
transfected into U87 and U251 cells. The overexpression of RNF135
was then confirmed by IB analysis by comparison with cells
transfected with empty vector (Fig.
2B). Subsequently, U87 and U251 cells were transiently
transfected with varying amounts of the RNF135-Myc plasmid and
incubated with CHX. The results of IB analysis indicated that
RNF135 promoted the degradation of p21 in a concentration-dependent
manner (Fig. 2C). These findings
indicate that RNF135 facilitates the degradation of p21.
 | Figure 2.RNF135 interacts with and degrades
p21. (A) Knockdown of RNF135 in U87 and U251 cells using shRNF135
upregulates p21 as detected by IB. (B) RNF135 overexpression
efficiency was verified in U87 and U251 cells transiently
transfected with pCDNA3.0-RNF135-Myc vector by IB analysis. (C)
RNF135 promotes the degradation of p21 in a concentration-dependent
manner. This was evaluated in U87 and U251 cells transiently
transfected with different amounts of pCDNA3.0-RNF135-Myc vector,
treated with 100 µg/ml CHX for 6 h and analyzed by IB. (D) RNF135
mediates p21 degradation primarily via the proteasomal pathway. U87
and U251 cells transfected with empty or RNF135-encoding vectors
were treated with either proteasomal inhibitor BTZ or autophagy
inhibitor BAF plus CHX for 6 h and then subjected to IB analysis.
(E) RNF135 directly interacts with p21 as revealed by a GST
pull-down assay. (F) RNF135 is an interacting partner for p21 as
evidenced by a yeast two-hybrid assay. (G) Co-IP assays demonstrate
that endogenous p21 forms a complex with RNF135 in U87 and U251
cells. RNF135, RING finger protein 135; shRNF135, short hairpin RNA
targeting RNF135; IB, immunoblotting; CHX, cycloheximide; BTZ,
bortezomib; BAF, bafilomycin; GST, glutathione S-transferase; SD-2,
medium deficient in leucine and tryptophan; SD-4, medium deficient
in uracil, histidine, leucine and tryptophan; IP,
immunoprecipitation; PD, pull-down. |
RNF135-mediated degradation of p21
proceeds primarily through the proteasomal pathway
U87 and U251 cells transfected with plasmids
encoding RNF135 or with empty vector were treated with the CHX and
either proteasomal inhibitor BTZ or the autophagy inhibitor BAF for
6 h, and then subjected to IB analysis. The results in Fig. 2D demonstrate that RNF135-mediated
p21 degradation occurs primarily via the proteasomal pathway.
Recombinant RNF135 and p21 proteins were purified
from an E. coli system. The GST pull-down assay demonstrated
that GST-tagged RNF135 directly interacted with His6-tagged p21
in vitro (Fig. 2E). The Y2H
technology employs nutrient deficiency to monitor gene expression
and protein interactions. The RNF135 gene was cloned into pDEST32,
which has a fused activating domain (AD), while the p21/CDKN1A gene
was cloned into pDEST22, which has a fused DNA binding domain (BD).
It is only when the RNF135 and p21 genes are both expressed and AD
and BC interact that downstream gene expression is initiated,
allowing the yeast to grow on SD-4 plates and be stained by X-gal.
The results confirmed the interaction between RNF135 and p21
(Fig. 2F). Furthermore, Co-IP
assays utilizing anti-p21 antibodies demonstrated that endogenous
RNF135 formed complexes with p21 in both U87 and U251 cells
(Fig. 2G). These findings indicate
that RNF135 is an interacting partner for p21 and promotes its
degradation primarily through the proteasomal pathway.
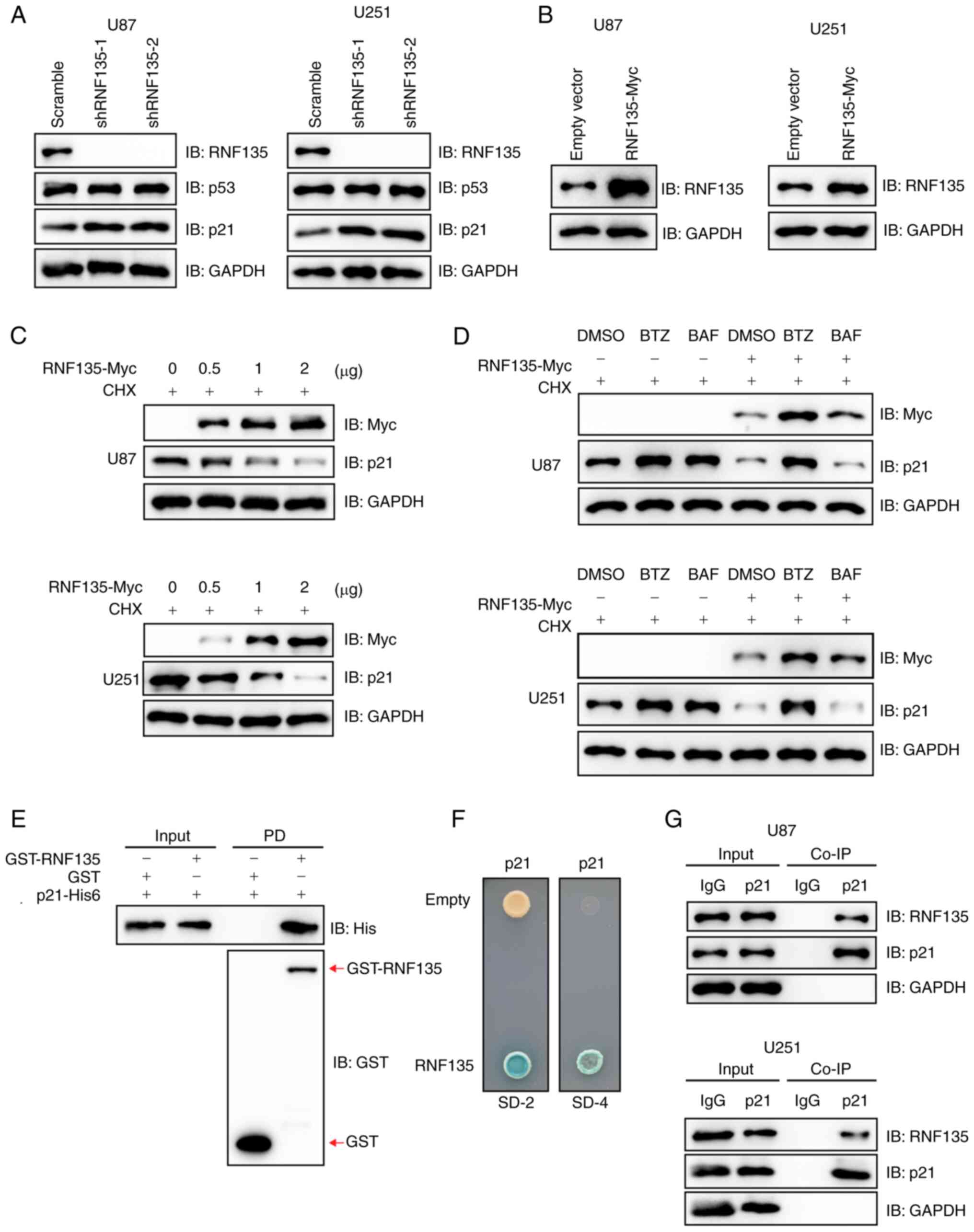
RNF135 mediates the ubiquitination of
p21
Several experiments were conducted to ascertain
whether RNF135 is an E3 ligase for p21. In U87 and U251 cells
stably expressing pCDH-RNF135, there was a clear increase in the
ubiquitination of p21 compared with that in control cells stably
transfected with the empty vector pCDH (Fig. 3A). Conversely, RNF135 knockdown
resulted in a reduction in the ubiquitination of p21 and an
increase in p21 protein levels in both U87 and U251 cells (Fig. 3B). An in vitro ubiquitination
assay was then performed to detect the ubiquitination of p21 in the
presence of E1 (UBA1), E2 (UBCH5A) and E3 (RNF135) by IB. The
modification induced by the combination of these three enzymes was
efficiently eliminated by Usp2cc, the catalytic core of the human
deubiquitinase enzyme USP2 (Fig.
3C). The p21 protein contains six lysine (K) residues (Fig. 3D). To map the RNF135-mediated
ubiquitination site on p21, pCDNA3.0-p21-Flag and several mutant
plasmids were constructed, transiently transfected into 293T cells,
and the p21 expression levels of the cells were detected by IB
analysis (Fig. 3D). K163 was
identified as the primary site for RNF135-mediated ubiquitination.
This was confirmed by the p21 mutant K163R, with a
lysine-to-arginine (K-to-R) substitution, which showed almost
complete resistance to RNF135-mediated ubiquitination on p21
(Fig. 3E). Further investigation
revealed that RNF135 facilitated the degradation of wild-type p21
protein, but had a minimal effect on K163R mutant p21 protein in
both U87 and U251 cells (Fig. 3F),
which corroborates the importance of this site. In conclusion, the
results demonstrate that p21 is a substrate of RNF135.
 | Figure 3.RNF135 mediates the ubiquitination of
p21. (A) RNF135 promotes the ubiquitination of endogenous p21 as
shown by the IP of RNF135 overexpressing cells with an anti-p21
antibody followed by IB. (B) Ablation of RNF135 reduces the
ubiquitination of endogenous p21. U87 and U251 cells stably
transfected with shRNF135 were subjected to IP with anti-p21
antibody followed by IB analysis. (C) RNF135 mediates the
ubiquitination of p21 in vitro, as detected by IB analysis.
(D) The overexpression of p21 induced by the transient transfection
of 293T cells with plasmids encoding p21 or its mutants in
comparison with an empty vector was detected by IB analysis. (E)
K163 was identified as the major site of RNF135-mediated p21
ubiquitination by submitting the lysates of 293T cells transiently
transfected with HA-Ub, RNF135-Myc and WT or mutant p21-Flag to IP
using anti-Flag affinity gels, followed by IB. (F) RNF135
facilitates the degradation of WT p21 protein but not the K163R
mutant as indicated by the IB of co-transfected U87 and U251 cell
lysates following treatment with 100 µg/ml CHX. RNF135, RING finger
protein 135; IP, immunoprecipitation; IB, immunoblotting; shRNF135,
short hairpin RNA targeting RNF135; HA, hemagglutinin; Ub,
ubiquitin; His6, hexahistidine; WT, wild-type; Usp2cc, Ub
carboxyl-terminal hydrolase 2 catalytic core; CHX,
cycloheximide. |
RNF135 promotes the proliferation of
GBM cells mainly though p21
Further experiments were performed to investigate
whether RNF135 promotes the proliferation of GBM cells in a
p21-dependent manner. Three shRNAs were designed to target p21 and
transfected into U87 and U251 cells. The knockdown efficiencies
were detected through IB analysis, as shown in Fig. 4A. Two of the shRNAs, shp21-2 and
shp21-3, exhibited high knockdown efficacy and were thus selected
for use in further experiments. U87 and U251 cells stably
expressing scramble shRNA, shp21-2 or shp21-3 were stably
transfected with either empty vector (pCDH) or pCDH-RNF135. The
protein levels of RNF135 and p21 were then detected by IB analysis
(Fig. 4B). The results of CCK-8
assays conducted at 24, 48 or 72 h demonstrated that RNF135
suppressed the proliferation of U87 and U251 cells with intact p21,
but not that of p21-knockdown cells (Fig. 4C). Colony formation assays were also
conducted, and the results were consistent with those of the CCK-8
assays, showing that RNF135 inhibited the colony formation only of
cells with intact p21 (Fig. 4D).
These data suggests that RNF135 promotes the proliferation of GBM
cells in a p21-dependent manner.
Discussion
In the present study, analyses performed using the
GEPIA2 public database revealed that RNF135 was upregulated in
human GBM and served as a prognostic marker for overall survival.
Furthermore, in vitro, the overexpression of RNF135 was
found to promote the proliferation of GBM cells, while the
knockdown of RNF135 inhibited it, which is consistent with a
previous study (10). These
findings provide evidence to suggest that RNF135 may act as an
oncogene in human GBM.
The Ub-proteasome pathway is a selective protein
degradation pathway that breaks down intracellular proteins. It
plays a crucial role in cell proliferation, differentiation,
apoptosis, autophagy and other cellular processes (19–21).
Dysfunctions in ubiquitination have been found to be associated
with a range of issues, including cancers and neurodegenerative
diseases (17,22,23).
RNF135 is a RING-type E3 ubiquitin ligase with an N-terminal RING
domain. It has been reported that RNF135 ubiquitinates the retinoic
acid-inducible gene-I protein, and increases its ability to
transmit signals, resulting in the production of antiviral IFN
(13). It has also been identified
that RNF135 is upregulated in glioblastoma tissue and promotes the
proliferation of human glioblastoma cells via ERK (10). However, the underlying mechanism
remains incompletely understood. In the present study, the cell
cycle inhibitor CDKN1A/p21, which is involved in the tumor
suppressor p53-associated signaling pathway, was identified as a
novel substrate for RNF135. The results of the present study also
suggest that RNF135 promotes the proliferation of GBM cells mainly
via p21.
The cyclin-dependent kinase inhibitor CDKN1A/p21 is
a pivotal downstream effector of p53, which acts as a cell cycle
inhibitor and anti-proliferative effector in normal cells (24). Dysregulation of this protein has
been found to be common in numerous types of cancer, and it has
been shown to affect several cellular processes including
apoptosis, DNA damage response and actin cytoskeleton remodeling
(25,26). In response to p53 transcription
factor activity, p21 induction may result in tumor growth arrest
through the inhibition of cyclin-dependent kinase complexes,
proliferation cell nuclear antigen, transcription factors and
coactivators (25). The
overexpression of p21 has been demonstrated to promote cell death
and induce senescence in human glioblastoma (27). Consistent with this, the present
study has confirmed that p21-knockdown promotes the proliferation
of GBM cells.
To date, >10 E3 ligases for p21 have been
reported, including MDM2, CHIP, makorin-1 and TRIM21 (28). The ubiquitination and degradation of
p21 play an important role in regulation of the cell cycle and
tumorigenesis (29). The findings
of the present study indicate that the RNF135-p21 axis contributes
to GBM cell proliferation, suggesting that RNF135 may serve as a
therapeutic target for the treatment of GBM. The present study
demonstrated that the knockdown of RNF135 significantly inhibited
the proliferation of GBM cells. Consequently, it is possible that
agents that inhibit RNF135 activity may have the potential to be
developed for the treatment of GBM.
It must be noted that there are certain limitations
to the present study. For example, the absence of data from animal
experiments and clinical data limits the translatability of the
research. In addition, the construction of RNF135 knockout cell
lines was unsuccessful using a CRISPR-based method; therefore, only
shRNAs for RNF135 were used and rescue experiments were omitted.
Furthermore, the RNF135 antibody used in the study is not
compatible with immunofluorescence assay, so it was not possible to
perform immunofluorescence experiments. Despite these limitations,
the present study elucidated the molecular mechanism by which
RNF135 regulates GBM cell proliferation and identified a potential
therapeutic target for the treatment of GBM.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant no. 81972339) and the Shanghai Municipal
Science and Technology Commission (grant no. 18XD1403400).
Data availability
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YC and ZW designed and supervised the project. WG
and MW performed experiments and data analysis. WG, YC and ZW wrote
the manuscript. YC and ZW confirm the authenticity of all the raw
data. All authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Isachesku E, Braicu C, Pirlog R,
Kocijancic A, Busuioc C, Pruteanu LL, Pandey DP and Berindan-Neagoe
I: The role of non-coding RNAs in epigenetic dysregulation in
glioblastoma development. Int J Mol Sci. 24:163202023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tu W, Zheng H, Li L, Zhou C, Feng M, Chen
L, Li D, Chen X, Hao B, Sun H, et al: Secreted phosphoprotein 1
promotes angiogenesis of glioblastoma through upregulating PSMA
expression via transcription factor HIF1α. Acta Biochim Biophys Sin
(Shanghai). 55:417–425. 2022.PubMed/NCBI
|
|
3
|
Chen B, Chen C, Zhang Y and Xu J: Recent
incidence trend of elderly patients with glioblastoma in the United
States, 2000–2017. BMC Cancer. 21:542021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Delgado-Martin B and Medina MÁ: Advances
in the knowledge of the molecular biology of glioblastoma and its
impact in patient diagnosis, stratification, and treatment. Adv Sci
(Weinh). 7:19029712020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khaddour K, Johanns TM and Ansstas G: The
landscape of novel therapeutics and challenges in glioblastoma
multiforme: Contemporary state and future directions.
Pharmaceuticals (Basel). 13:3892020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li C, Wang S, Yan JL, Torheim T, Boonzaier
NR, Sinha R, Matys T, Markowetz F and Price SJ: Characterizing
tumor invasiveness of glioblastoma using multiparametric magnetic
resonance imaging. J Neurosurg. 132:1465–1472. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cuddapah VA, Robel S, Watkins S and
Sontheimer H: A neurocentric perspective on glioma invasion. Nat
Rev Neurosci. 15:455–465. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y: E3 ubiquitin ligases as cancer
targets and biomarkers. Neoplasia. 8:645–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Wang F, Liu Y, Yao Y, Lv X, Dong B,
Li J, Ren S, Yao Y and Xu Y: RNF135, RING finger protein, promotes
the proliferation of human glioblastoma cells in vivo and in vitro
via the ERK pathway. Sci Rep. 6:206422016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng X, Song D, Liu X, Liang Y, Jiang P,
Wu S and Liu F: RNF125-mediated ubiquitination of MCM6 regulates
the proliferation of human liver hepatocellular carcinoma cells.
Oncol Lett. 27:1052024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia C, Tang H, Yang Y, Yuan S, Han T, Fang
M, Huang S, Hu R, Li C and Geng W: Ubiquitination of IGF2BP3 by E3
ligase MKRN2 regulates the proliferation and migration of human
neuroblastoma SHSY5Y cells. Biochem Biophys Res Commun. 529:43–50.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oshiumi H, Matsumoto M, Hatakeyama S and
Seya T: Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I
to promote interferon-beta induction during the early phase of
viral infection. J Biol Chem. 284:807–817. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pasmant E, Masliah-Planchon J, Lévy P,
Laurendeau I, Ortonne N, Parfait B, Valeyrie-Allanore L, Leroy K,
Wolkenstein P, Vidaud M, et al: Identification of genes potentially
involved in the increased risk of malignancy in NF1-microdeleted
patients. Mol Med. 17:79–87. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45
(W1):W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C, Han T, Li Q, Zhang M, Guo R, Yang Y,
Lu W, Li Z, Peng C, Wu P, et al: MKRN3-mediated ubiquitination of
Poly(A)-binding proteins modulates the stability and translation of
GNRH1 mRNA in mammalian puberty. Nucleic Acids Res. 49:3796–3813.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu X, Li C, Gao X, Xia K, Guo H, Li Y, Hao
Z, Zhang L, Gao D, Xu C, et al: Excessive UBE3A dosage impairs
retinoic acid signaling and synaptic plasticity in autism spectrum
disorders. Cell Res. 28:48–68. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Engeland K: Cell cycle regulation:
p53-p21-RB signaling. Cell Death Differ. 29:946–960. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mansour MA: Ubiquitination: Friend and foe
in cancer. Int J Biochem Cell Biol. 101:80–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang CC, Peng H, Wang Z, Yang J, Hu RG, Li
CY and Geng WJ: TRIM72-mediated degradation of the short form of
p62/SQSTM1 rheostatically controls selective autophagy in human
cells. Mil Med Res. 9:352022.PubMed/NCBI
|
|
21
|
Yang Y, Luo Y, Yang C, Hu R, Qin X and Li
C: TRIM25-mediated ubiquitination of G3BP1 regulates the
proliferation and migration of human neuroblastoma cells. Biochim
Biophys Acta Gene Regul Mech. 1866:1949542023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng
H, Xu X, Wang H, Yang M, Liu X, et al: Ubiquitylation of autophagy
receptor Optineurin by HACE1 activates selective autophagy for
tumor suppression. Cancer Cell. 26:106–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li C, Lu W, Yang L, Li Z, Zhou X, Guo R,
Wang J, Wu Z, Dong Z, Ning G, et al: MKRN3 regulates the epigenetic
switch of mammalian puberty via ubiquitination of MBD3. Natl Sci
Rev. 7:671–685. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shamloo B and Usluer S: p21 in cancer
research. Cancers (Basel). 11:11782019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu L, Yang Z, Dai G, Fan B, Yuan J, Liu Y,
Liu P and Ou Z: SOX5 promotes cell growth and migration through
modulating the DNMT1/p21 pathway in bladder cancer. Acta Biochim
Biophys Sin (Shanghai). 54:987–998. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mansour MA, Rahman M, Ayad AA, Warrington
AE and Burns TC: P21 overexpression promotes cell death and induces
senescence in human glioblastoma. Cancers (Basel). 15:12792023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang F, Wu Z, Li Q, Ni Z, Wang C and Lu J:
Ubiquitination of p21 by E3 ligase TRIM21 promotes the
proliferation of human neuroblastoma cells. Neuromolecular Med.
23:549–560. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dang F, Nie L and Wei W: Ubiquitin
signaling in cell cycle control and tumorigenesis. Cell Death
Differ. 28:427–438. 2021. View Article : Google Scholar : PubMed/NCBI
|