Introduction
Identifying efficacious targets and therapies for
hepatocellular carcinoma (HCC) remains a paramount focus in
oncological research. The advent of novel biomarkers and targeted
therapeutics has expanded the therapeutic landscape, rendering the
quest for effective targets less daunting (1). The emergence of targeted drugs has
revolutionized treatment strategies, ushering in an era of
heightened therapeutic efficacy.
Calnexin (CANX), a pivotal molecular chaperone
protein, is instrumental in facilitating accurate folding of
glycoproteins within the endoplasmic reticulum (ER) environment.
CANX is a highly reliable marker in serum that can differentiate
between patients with lung cancer and healthy individuals, aiding
in the early detection of lung cancer (2). N-acetylgalactosaminyltransferase
activation has been shown to stimulate the glycosylation process of
ER-resident CANX in both breast and liver cancer (3). CANX and ERp57 interact to facilitate
the degradation of the extracellular matrix. Anti-CANX antibodies
has demonstrated efficacy in inhibiting liver tumor growth and lung
metastasis of breast and liver cancer cells (3). It has long been believed that calcium
homeostatic regulatory signals contribute to the onset, progression
and metastasis of cancer (4). In
the context of calcium homeostasis, CANX serves a role in the
regulation of apoptosis in dendritic cells (5). MicroRNA (miR)-148a-3p can function as
a tumor promotor in colorectal cancer by targeting the CANX/major
histocompatibility complex I axis, suggesting the crucial
involvement of CANX in tumor immunological responses (6). CANX serves as a quality control factor
for protein folding in the ER. It promotes antitumor immunotherapy
and regulates several processes, including cancer cell adhesion,
migration, proliferation and energy metabolism (7).
Vacuolar (V)-ATPase, a component of the energy
metabolism system, influences tumor invasion and migration.
V-ATPase is comprised of two components, the transmembrane V0 part
and the cytoplasmic V1 part (8).
ATP6V1F, a protein belonging to the ATP6V1 domain protein,
constitutes a part of the V-type ATPase complex. Central to protein
folding mechanisms, it is instrumental in in upholding cellular
homeostasis and ensuring environmental stability within cells
(9). Overexpression of ATP6V1F in
HCC tissues can accelerate the development of HCC, suppress
apoptosis, encourage the migration and invasion of HCC cells, and
is strongly associated with a poor prognosis for patients (10). Furthermore, bioinformatics has
revealed a notable upregulation of ATP6V0B in renal cell carcinoma
(11). However, the interaction
between CANX and the ATP6V1 domain and V0 domain proteins is still
unclear.
Dihydroartemisinin (DHA) is a derivative of
artemisinin and a precursor to other active components of
artemisinin. Research has reported that DHA can suppress tumor cell
proliferation, induce apoptosis, reverse drug resistance and
influence tumor invasion and metastasis. Additionally, DHA has an
anticancer effect on lung, colorectal, pancreatic and breast
cancers (12). A previous study has
demonstrated that DHA inhibits the migration and invasion of HCC
cells and mediated skeleton remodeling by reducing the production
of ATP synthase (ATP1A1 and ATP5H) through the calcium/calmodulin
dependent protein kinase kinase 2 (CaMKK2)/solute carrier family 8
member B1 (NCLX) signaling pathway (13). However, it is not clear whether DHA
inhibits energy metabolism and transmission of HCC cells by
regulating ATP synthase activity through other calcium ion signals.
The present study aimed to elucidate the role of CANX in the
progression of HCC cells and the mechanism of DHA inhibition of
energy metabolism and transfer of HCC cells by regulating
mitochondrial function through CANX.
Materials and methods
Cell culture
Human HCC HuH-7 and Li-7 cell lines were purchased
from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd. (https://www.zqxzbio.com). After cell resuscitation,
the cells were inoculated in RPMI-1640 medium (cat. no.
C11875500BT; Thermo Fisher Scientific, Inc.) containing 10% FBS
(cat. no. G24-70500; Genial Biologicals Inc) and 1% double antibody
(cat. no. P1400; Beijing Solarbio Science & Technology Co.,
Ltd.), and cultured in a cell incubator at 37°C in 95% air
and 5% CO2. Culture medium was replaced every 2–3
days.
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA) database is an online analytical tool (http://gepia.cancer-pku.cn/index.html).
Based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue
Expression (GTEx) data, the expression difference of CANX liver
cancer samples (n=369) and normal samples (n=160) was further
validated through the ‘expression on box plots module’ of GEPIA.
P<0.05 was considered to indicate a statistically significant
difference.
cBioPortal (https://www.cbioportal.org/) is an online analytical
tool and platform designed to analyze cancer genomics data from
TCGA database, offering a user-friendly interface to explore
complex molecular data. In the present study, CANX mutation and
survival of patients analyses for CANX across liver cancer were
conducted using cBioPortal.
Small interfering (si)RNA transfection
and overexpression
Reagents were purchased from Sangon Biotech Co.,
Ltd. (https://www.sangon.com). The following
sequences were constructed: CANX siRNA1, sense:
5′-ACUGGUGCUUGGAACUGCUAUUGUU-3′ and antisense: 5′-
AACAAUAGCAGUUCCAAGCACCAGU-3′; CANX siRNA2, sense:
5′-CGAUGAUGAAAUUGCCAAAUATT-3′ and antisense:
5′-UAUUUGGCAAUUUCAUCAUCGTT-3′; and negative control (NC), sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense:
5′-ACGUGACACGUUCGGAATT-3′. A scrambled/non-targeting sequence acted
as the negative control. Full-length CANX (Sangon Biotech Co.,
Ltd.) was cloned into the pcDNA3.1(+) ZB02427 vector (cat. no.
240224HY4752-2; Sangon Biotech Co., Ltd.). The empty vector was the
control. HuH-7 and Li-7 cells in logarithmic growth phase were
seeded in 6-well plates with 1×105 cells per well. The
cells were divided into control, siRNA-NC, siRNA1 and siRNA2
groups, and control, pcDNA3.1 and pcDNA3.1-NC groups. When cell
fusion reached 70–80%, 100 pmol siRNA and 2.5 µg CANX
overexpression plasmids were transfected at 37°C for 24h according
to the Lipo8000™ Transfection Reagent (cat. no. C0533;
Beyotime Institute of Biotechnology) instructions. The transfection
effects of CANX siRNA and CANX overexpression plasmids were
detected by western blot analysis, as described below. Subsequent
experiments were performed 24 h after transfection.
Wound-healing assay
Using a marker pen, two horizontal lines spaced 1 cm
apart were drawn on the back of the 6-well plate. A total of
~5×105 Huh-7 and Li-7 cells were added into the hole.
Following a 24-h incubation at 37°C, a 200 µl pipette tip was used
to scratch the cells and cross the line vertically, and the cells
were washed with PBS three times. Serum-free medium was replenished
for an additional 24-h culture at 37°C. The cells were observed and
imaged using a light microscope (magnification, ×40; Olympus
Corporation). The migratory rate of the cells was calculated from
the images using Image-Pro Plus software (version 6.0; Media
Cybernetics, Inc.).
Transwell cell invasion assay
At 4°C, Matrigel (Corning, Inc.) was diluted with
PBS buffer, and 100 µl was evenly precoated at 37°C on the
polycarbonate membrane surface of the upper chamber for 2 h. HuH-7
and Li-7 cells (2×105) were seeded in the upper chambers
with serum-free medium. A total of 500 µl medium containing 10% FBS
with or without DHA was added in the lower chambers. The cells were
cultured in the incubator for 48 h at 37°C. Upon completion of
incubation, the cells were fixed with 4% paraformaldehyde for 30
min, stained with 0.1% crystal violet for 10 min at room
temperature and washed with PBS 3 times. A total of five fields of
vision were selected and counted under a light microscope
(magnification, 200×; Olympus Corporation).
JC-1 mitochondrial membrane potential
analysis
A total of 6×105 HuH-7 and Li-7 cells
were seeded in a 6-well plate. A total of 0.5 ml JC-1 staining
working fluid (Mitochondrial Membrane Potential Assay Kit with
JC-1; cat. no. M8650; Beijing Solarbio Science & Technology
Co., Ltd.) was added, reverse mixed and incubated at 37°C for 20
min. After incubation, the supernatant was discarded and the cells
were washed with the JC-1 Buffer (1X). Finally, the cells were
observed and imaged using a fluorescence microscope (Olympus
Corporation). The CCCP (10 mM) provided in the kit was added to the
cell culture medium at a ratio of 1:1,000 and diluted to 10 µM, and
was incubated with cells for 20 min. CCCP was the positive control
group. Image-Pro Plus software (version 6.0; Media Cybernetics,
Inc.) was used to analyze the data.
Mitochondrial fluorescence probe
HuH-7 and Li-7 cells (1×105) were
incubated with Mito-Tracker Red CMXRos working solution (cat. no.
C1049B; Beyotime Institute of Biotechnology) at 37°C for 30 min.
Subsequently, the Mito-Tracker Red CMXRos working solution was
removed, and fresh cell culture solution pre-warmed at 37°C was
added. Finally, the cells were observed and imaged using a
fluorescence microscope (Olympus Corporation). The average
fluorescence density was calculated using Image-Pro Plus software
(version 6.0, Media Cybernetics, Inc.).
Western blotting
HuH-7 and Li-7 cells were harvested and lysed with
RIPA lysis buffer (Nanjing KeyGen Biotech Co., Ltd.) on ice.
Protein concentration was determined using the BCA Protein Assay
Kit (cat. no. PT0001; Leagene; Beijing Regen Biotechnology Co.,
Ltd.). A total of 40 µg total protein was electrophoresed using
10–12% SDS-PAGE. The separated proteins were transferred onto a
PVDF membrane and the membranes were blocked with 5% skim milk
powder for 1 h at room temperature. Subsequently, primary
antibodies were added and incubated at 4°C overnight. The primary
antibodies used were CANX (1:5,000; cat. no. 66903-1-Ig;
Proteintech Group, Inc.), ATP6V0B (1:1,000; cat. no. 27671-1-AP;
Proteintech Group, Inc.) and ATP6V1F (1:1,000; ab190789; Abcam),
β-actin (1:2,000; cat. no. 20536-1-AP; Proteintech Group, Inc.).
The film was then washed three times with TBST (0.05% Tween 20),
and then incubated with goat anti-rabbit (1:5,000; cat. no.
ZB-2301; OriGene Technologies, Inc.) or anti-mouse IgG (1:5,000;
cat. no. ZB-2305; OriGene Technologies, Inc.) labeled with
horseradish peroxidase at room temperature for 1h. The film was
washed three times with TBST again, and ECL Hypersensitive
Luminescent Solution (cat. no. P1050; Applygen Technologies, Inc.)
was added. The film was then transferred to the Amersham Imager 600
Automatic Chemilescence Gel Imaging Analyzer (GE Healthcare) for
automatic exposure analysis. The gray values of proteins were
analyzed by ImageJ software (version 1.8.0; National Institutes of
Health).
ATP production
ATP content was detected using an ATP fluorescence
probe (pCMV-AT1.03; cat. no. D2604; Beyotime Institute of
Biotechnology). According to the user manual, the treated HuH-7 and
Li-7 cells (3×105) were seeded in 12-well plates,
incubated with 1 µg ATP fluorescence probe and Lipo8000™
(cat.no. C0533; Beyotime Institute of Biotechnology) for 24 h at
37°C, and then observed and imaged using a fluorescence microscope
(Olympus Corporation). Images were analyzed using Image-Pro Plus
software (version 6.0; Media Cybernetics, Inc.).
NAD+/NADH. The HuH-7 and
Li-7 cells (1×105) were inoculated on 6-well plates.
After the transfection and drug treatment in each group, the
culture medium was discarded and washed with PBS. After washing, 1
ml of NAD+/NADH extraction solution (cat. no. S0175;
Beyotime Institute of Biotechnology) was added to each well, and
the supernatant was obtained after centrifugation. A 100 µl sample
to be tested was transferred to the centrifuge tube and heated at
60°C for 30 min to decompose NAD+. Subsequently, the 20
µl supernatant was added into a 96-well plate. The samples were
incubated with 90 µl alcohol dehydrogenase working solution at 37°C
for 10 min for measuring the NADH content. They were then incubated
with 10 µl color developing solution at 37°C for 10 min. After
mixing, the absorbance at 450 nm was measured and the of
NAD+/NADH content was calculated according to the
standard curve.
Cell Counting Kit-8 (CCK-8) assay
HuH-7 and Li-7 cells (2×103) were seed in
96-well plates and then 10 µl CCK-8 reagent (cat. no. CK04; Dojindo
Laboratories, Inc.) was added to each well. After a 2-h incubation,
the absorbance at 450 nm was measured using a microplate
reader.
TUNEL assay
According to the manufacturer's instructions (cat.
no. C1090; Beyotime Institute of Biotechnology), cells were washed
with PBS then fixed with 4% paraformaldehyde for 30 min at room
temperature, following by PBS washing. Subsequently, cells were
incubated with 0.3% Triton X-100 for 5 min at room temperature.
Subsequently, 0.3% H2O2 in PBS was added and
incubated for 20 min at room temperature. After incubation, 50 µl
TUNEL working solution was added and incubated at 37°C for 1 h.
Finally, DAPI was used to stain the nucleus at room temperature for
3 min and seal the film, and then observed and imaged using a
fluorescence microscope (Olympus Corporation). 3 fields of view are
randomly selected. The cell death rate was calculated as
(TUNEL+ cells/total cells) ×100.
Reactive oxygen species (ROS)
detection
A Reactive Oxygen Species Detection Kit (cat. no.
S0033; Beyotime Institute of Biotechnology) was used to evaluate
intracellular ROS levels using a DCFH-DA probe. DCFH-DA was diluted
with serum-free culture medium at a ratio of 1:1,000 to attain a
final concentration of 10 µmol/l. Each group of HuH-7 and Li-7
cells (2.5×105) were incubated with the DCFH-DA working
solution at 37°C for 20 min. Rosup served as a positive control.
Following incubation, fluorescence intensity was visualized using a
fluorescence microscope (Olympus Corporation), imaged and analyzed
using Image-Pro Plus software (version 6.0; Media Cybernetics,
Inc.).
Statistical analysis
One-way analysis of variance and the Bonferroni post
hoc test were applied for three groups or multigroup analysis and
statistical analyses were performed using SPSS (version 26.0; IBM
Corp.); visualization was carried out using GraphPad Prism (version
5; Dotmatics). P<0.05 was considered to indicate a statistically
significant difference.
Results
CANX is highly expressed in liver
cancer and is associated with survival of patients with liver
cancer
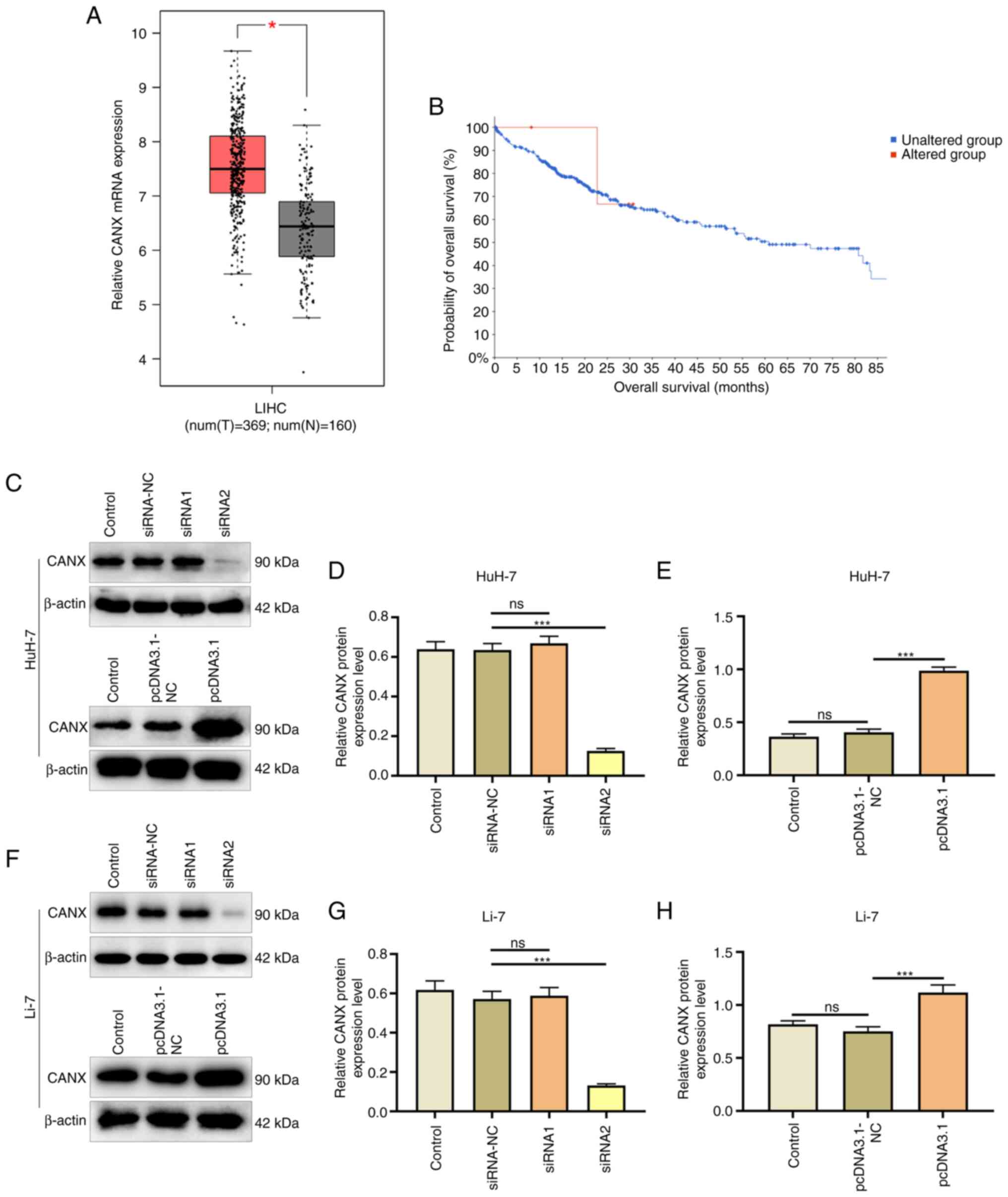
The expression of CANX in liver cancer and normal
tissue adjacent samples after TCGA and GTEx project data
integration was analyzed using the GEPIA database. The results
revealed that CANX had a significantly higher expression in the
liver cancer samples (n=369) compared with that in the normal
adjacent tissue samples (n=160; Fig.
1A). In addition, cBioPortal was used to analyze the effect of
CANX on patient survival, and the results demonstrated that CANX
gene expression was associated with survival and CANX mutations
were negatively correlated with survival (Fig. 1B). Furthermore, the effects of the
CANX overexpression plasmid and siRNA were assessed (Fig. 1C-H). Western blotting revealed that
Compared with the pcDNA3.1-NC group, the overexpression plasmid
(pcDNA3.1 group) significantly increased CANX expression.
Furthermore, the interference effect of CANX siRNA2 was significant
compared with siRNA-NC, and therefore it was chosen for use in
subsequent experiments.
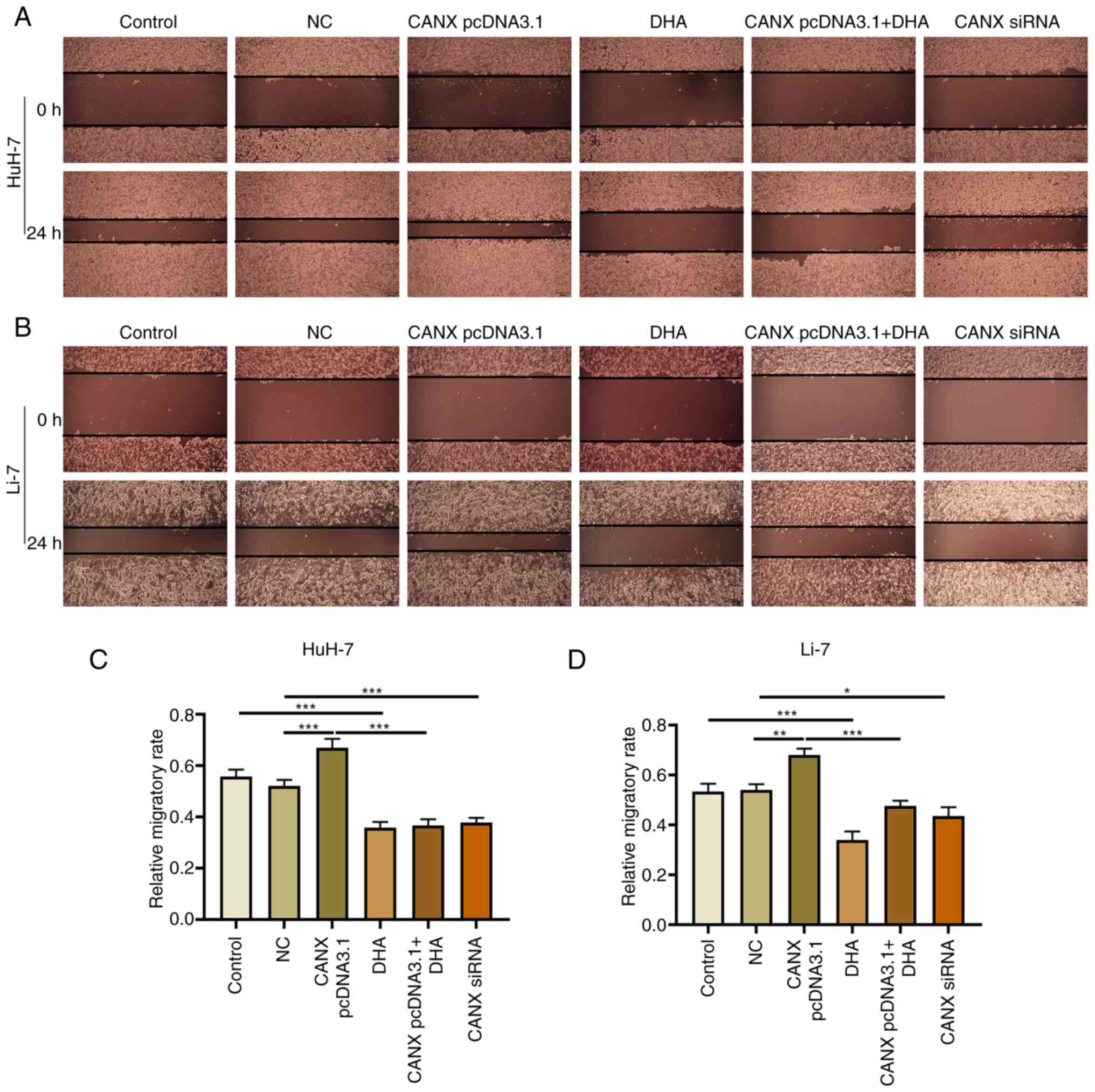
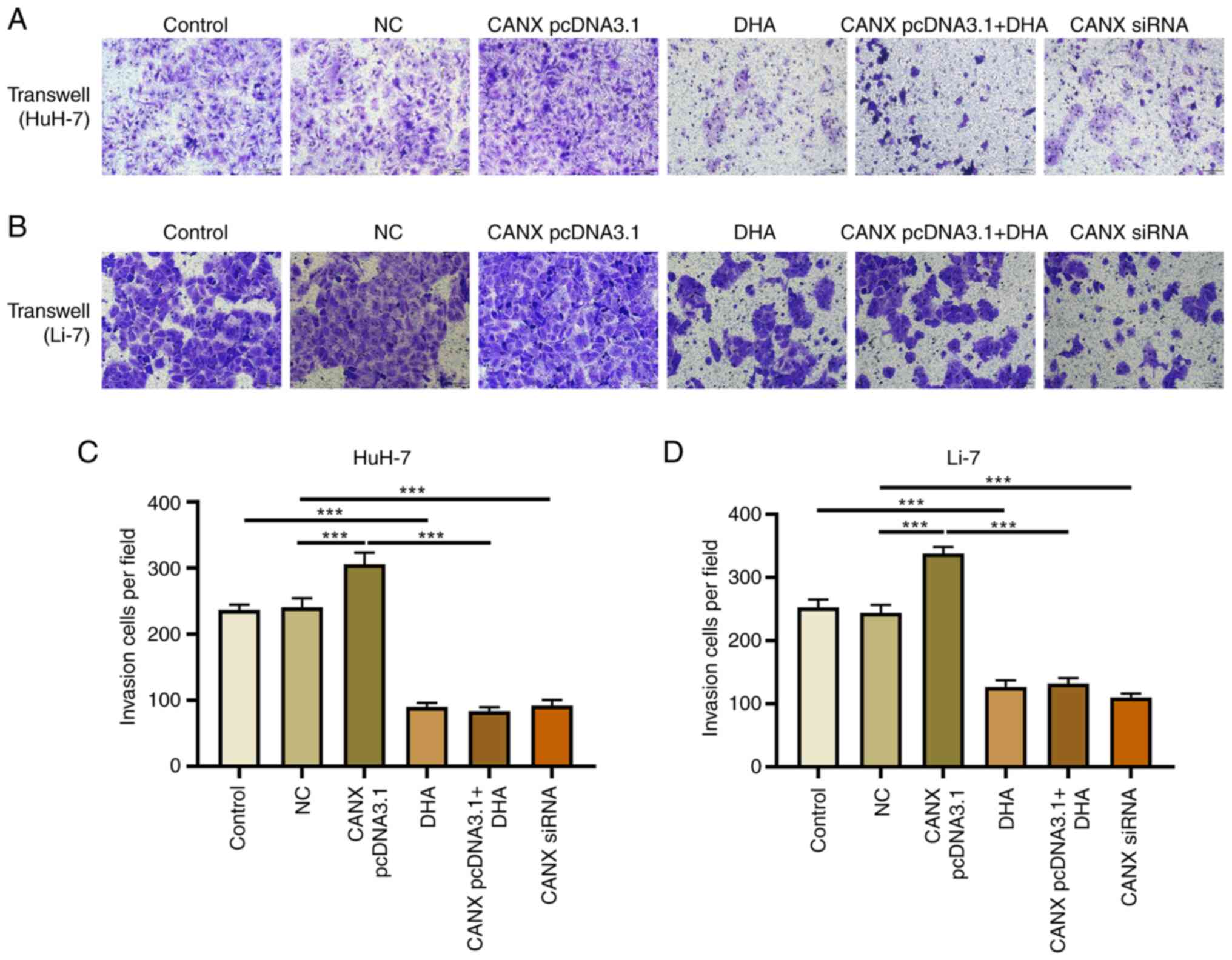
Effects of CANX and DHA on invasion
and migration of liver cancer cells
Cell scratch and Transwell experiments demonstrated
that overexpression of CANX significantly increased the migration
and invasion of liver cancer cells compared with the NC group.
Moreover, the results revealed that CANX siRNA significantly
inhibited the migration and invasion of liver cancer cells compared
with the NC group and DHA significantly inhibited the migration and
invasion of liver cancer cells compared with the control group
(Figs. 2 and 3). Notably, the combination of DHA and
CANX overexpression inhibited cell invasion and migration (Figs. 2 and 3). The results indicate that CANX can
mediate the metastasis of liver cancer cells, and this regulatory
effect can be inhibited by DHA.
Effects of CANX and DHA on
proliferation and apoptosis of liver cancer cells
The results of the cell proliferation experiment
demonstrated that overexpression of CANX significantly increased
the proliferation rate of cells compared with the NC group, whilst
knockdown of CANX (compared with the NC group) and DHA (compared
with the control group) administration exerted a significant
inhibitory effect (Fig. 4A and B).
Furthermore, the combination of DHA and CANX overexpression
significantly inhibited the proliferation of liver cancer cells
compared with the CANX pcDNA3.1 group (Fig. 4A and B). However, apoptosis
experiments revealed that there was no significant change in the
apoptosis rate of overexpressed CANX cells compared with the NC
group, whilst knockdown of CANX expression (compared with the NC
group) and DHA (compared with the control group) significantly
increased the apoptosis rate. Moreover, the combination of DHA and
CANX overexpression significantly increased the apoptosis rate
compared with the CANX pcDNA3.1 group (Fig. 4C-F). The aforementioned results
indicate that CANX can mediate apoptosis and proliferation;
however, DHA can inhibit the overexpression of CANX-mediated
regulation in addition to inhibiting the proliferation and
promoting apoptosis of HCC cells alone.
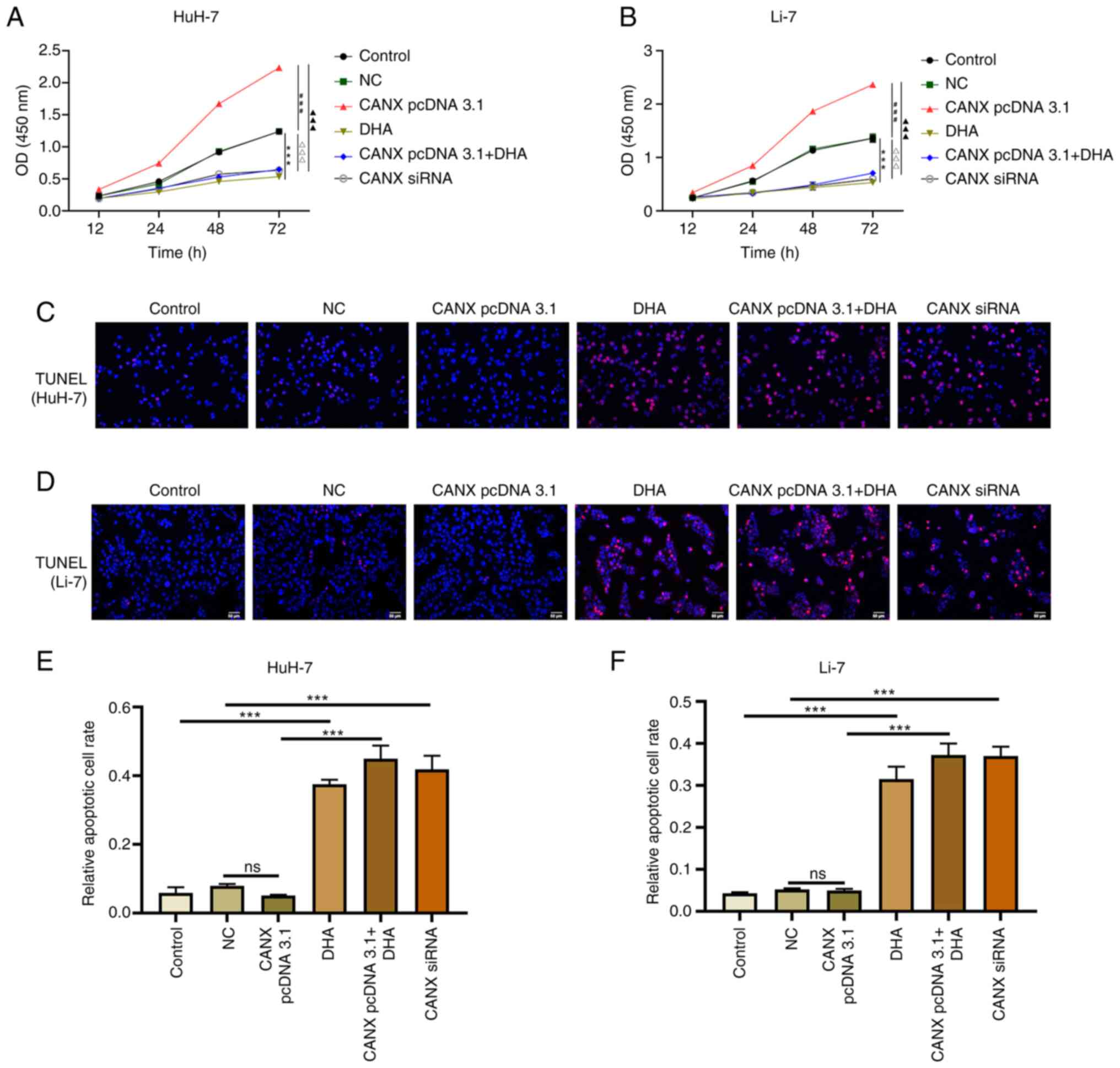 | Figure 4.Effects of CANX and DHA on the
proliferation and apoptosis of liver cancer cells. Cell Counting
Kit-8 assays detected the cell proliferation of (A) HuH-7 and (B)
Li-7 cells with CANX knockout, DHA or CANX overexpression.
***P<0.001, control vs. DHA; ###P<0.001, NC vs.
CANX pcDNA3.1; ∆∆∆P<0.001, NC vs. CANX siRNA;
▲▲▲P<0.001, CANX pcDNA3.1cCANX pcDNA 3.1+DHA. TUNEL
assays assessed the rate of apoptosis of (C) Huh-7 and (D) Li-7
cells after CANX knockout, DHA or CANX overexpression. The
apoptotic cell rate of (E) HuH-7 and (F) Li-7 cells (scale bar, 50
µm). ***P<0.001. CANX, calnexin; DHA, dihydroartemisinin; NC,
negative control; siRNA, small interfering RNA; ns, not
significant. |
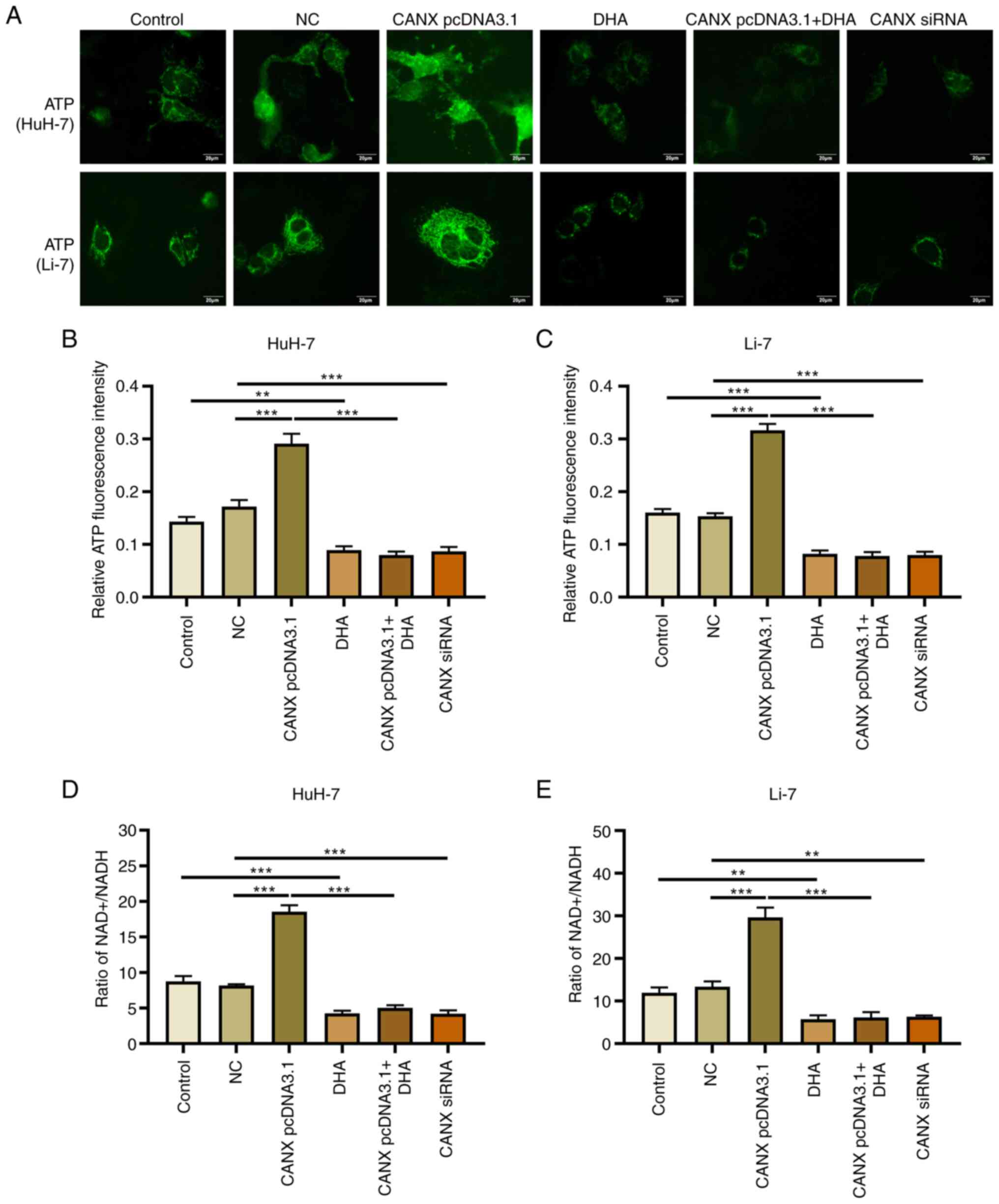
Effects of CANX and DHA on
mitochondrial ATP production and NAD+/NADH ratio
Mitochondrial ATP fluorescence probe results
revealed that overexpression of CANX significantly upregulated ATP
fluorescence intensity, whilst knockdown of CANX significantly
decreased it, compared with the NC group (Fig. 5A-C). The change in
NAD+/NADH ratio was also associated with energy
metabolism: Overexpression of CANX significantly increased the
NAD+/NADH ratio, whilst knockdown of CANX significantly
reduced the NAD+/NADH ratio compared with the NC group
(Fig. 5D and E). Furthermore,
treatment with DHA alone (compared with the control group) or in
combination with CANX overexpression (compared with the CANX
pcDNA3.1 group) significantly reduced ATP production and the
NAD+/NADH ratio in HCC cells (Fig. 5D and E). The results indicate that
CANX can mediate mitochondrial ATP production and the
NAD+/NADH ratio, thereby it participates in the energy
metabolism of HCC cells. However, the results also indicates that
DHA can inhibit CANX-mediated energy production.
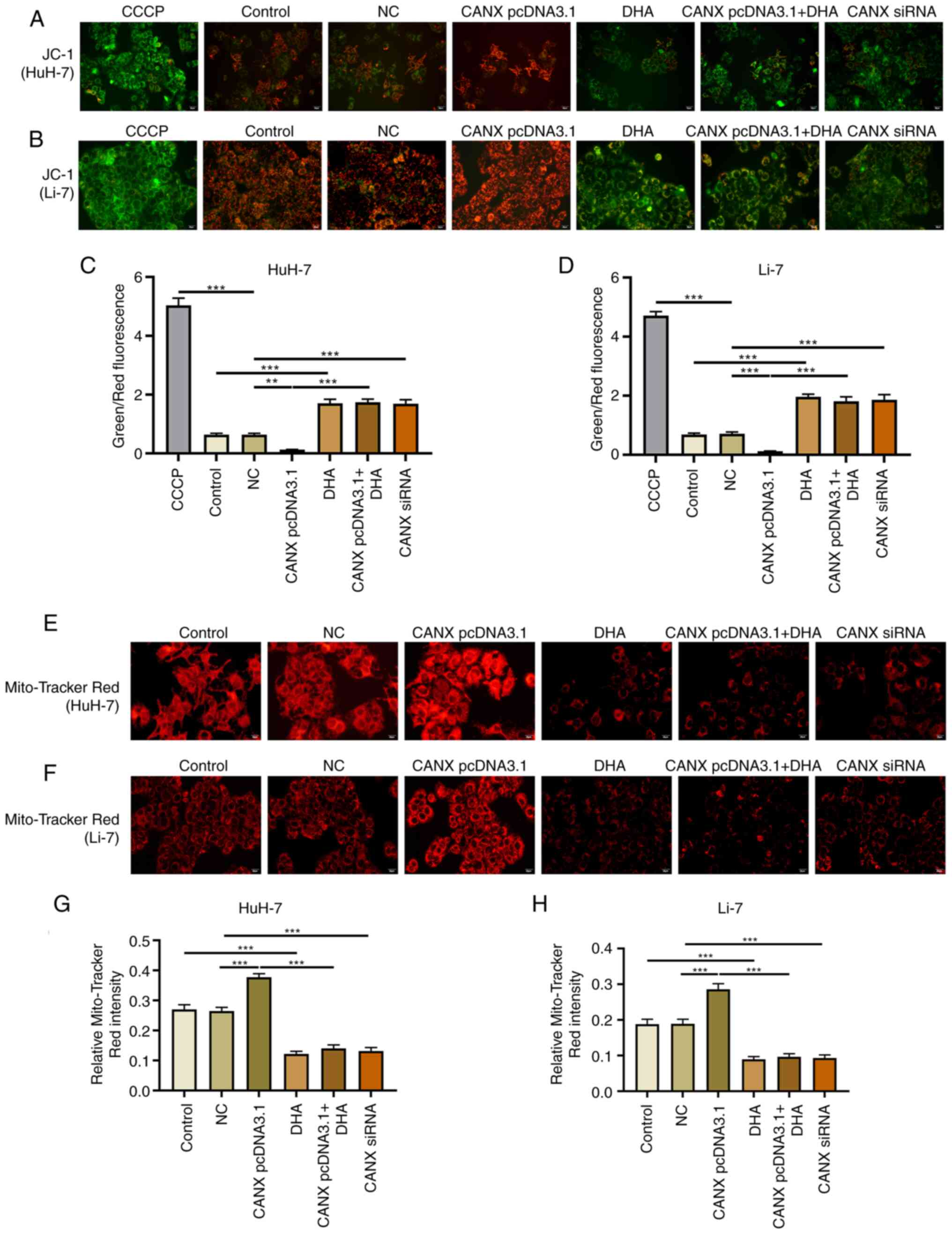
Effects of CANX and DHA on
mitochondrial membrane potential
Mitochondria play a central role in energy
metabolism (14). The mitochondrial
membrane potential and Mito-Tracker Red CMXRos assays revealed that
overexpression of CANX significantly upregulated the mitochondrial
membrane potential level compared with the NC group, and
aggregation or small flaky red fluorescence signals was observed.
Furthermore, knockdown of CANX and DHA administration significantly
reduced the fluorescence intensity of the mitochondrial membrane
potential, showing a sparse scattered pattern (Fig. 6). Notably, DHA inhibited the role of
CANX overexpression in mediating mitochondrial membrane
potential.
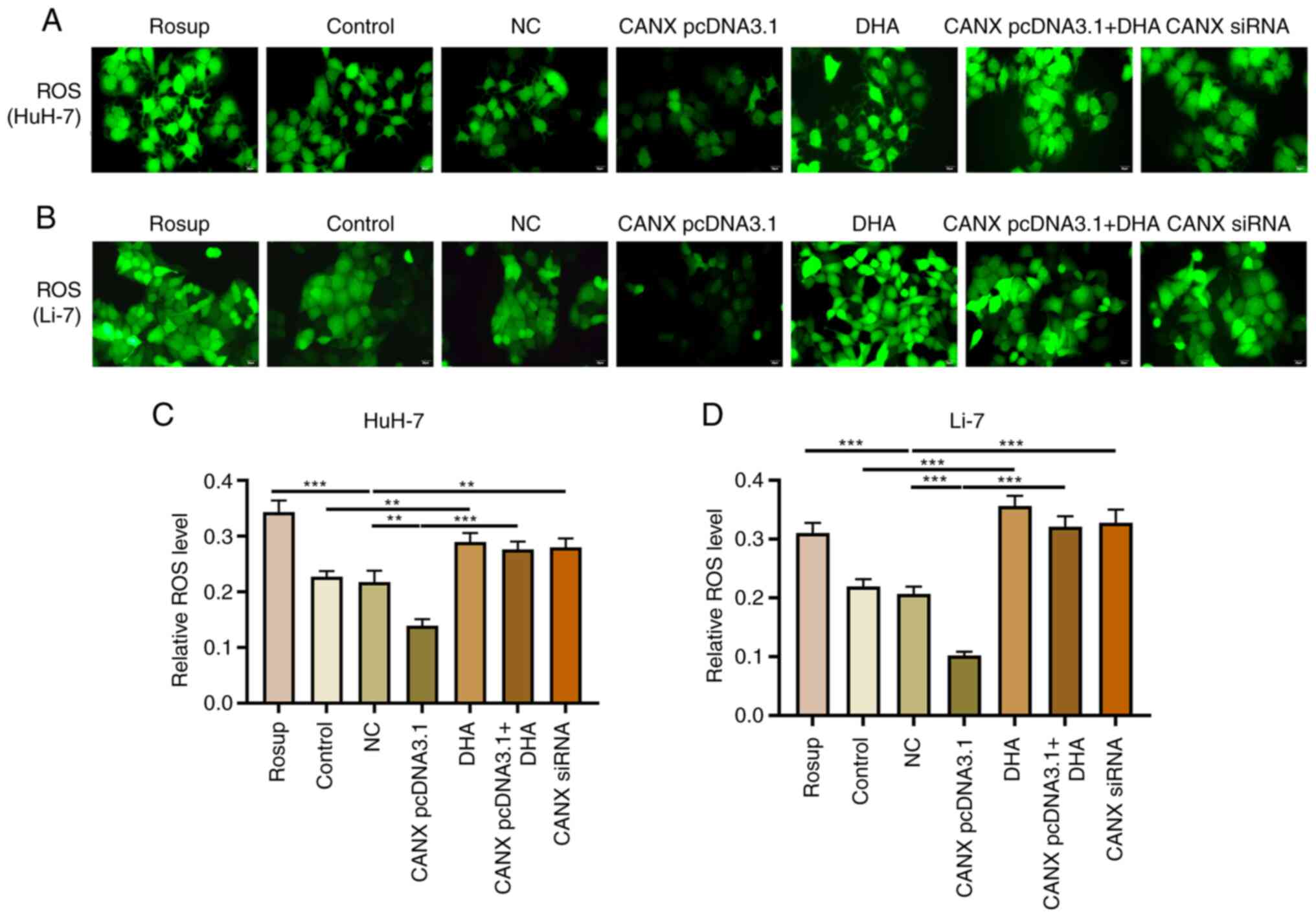
Effects of CANX on ROS production in
liver cancer cells
ROS serve a pivotal role in mitochondrial
dysfunction and decreased energy metabolism (15,16);
however, changes in ROS levels can have a direct impact on the
progression of liver cancer cells (17). Overexpression of CANX significantly
inhibited the production of ROS, compared with the NC group, with
decreased green fluorescence intensity observed. Moreover,
knockdown of CANX (compared with the NC group) and DHA (compared
with the control group) administration significantly promoted ROS
production, with enhanced green fluorescence intensity observed
(Fig. 7). The results indicate that
a combination of DHA and CANX overexpression can significantly
promote ROS production in HCC cells.
Effects of CANX and DHA on ATP6V0B and
ATP6V1F expression
ATP6V1 and V0 domain proteins participate in
acid-base balance and membrane transport, and serve an
indispensable role in the energy metabolism mechanism (18). The results of the present study
demonstrated that compared with the NC group, overexpression of
CANX significantly promoted the expression of ATP6V0B and ATP6V1F,
whereas the expression of ATP6V0B and ATP6V1F was significantly
downregulated by CANX siRNA compared with the NC group. Moreover,
compared with the control group, DHA administration significantly
decreased the expression of CANX. In addition, compared with the
CANX pcDNA3.1 group, the expression of ATP6V0B and ATP6V1F reduced
when cells were treated with a combination of DHA and CANX
overexpression (Fig. 8). These
findings indicate that CANX is involved in the regulation of the
molecular mechanism of energy metabolism in HCC cells, highlighting
that CANX may be a viable target of DHA.
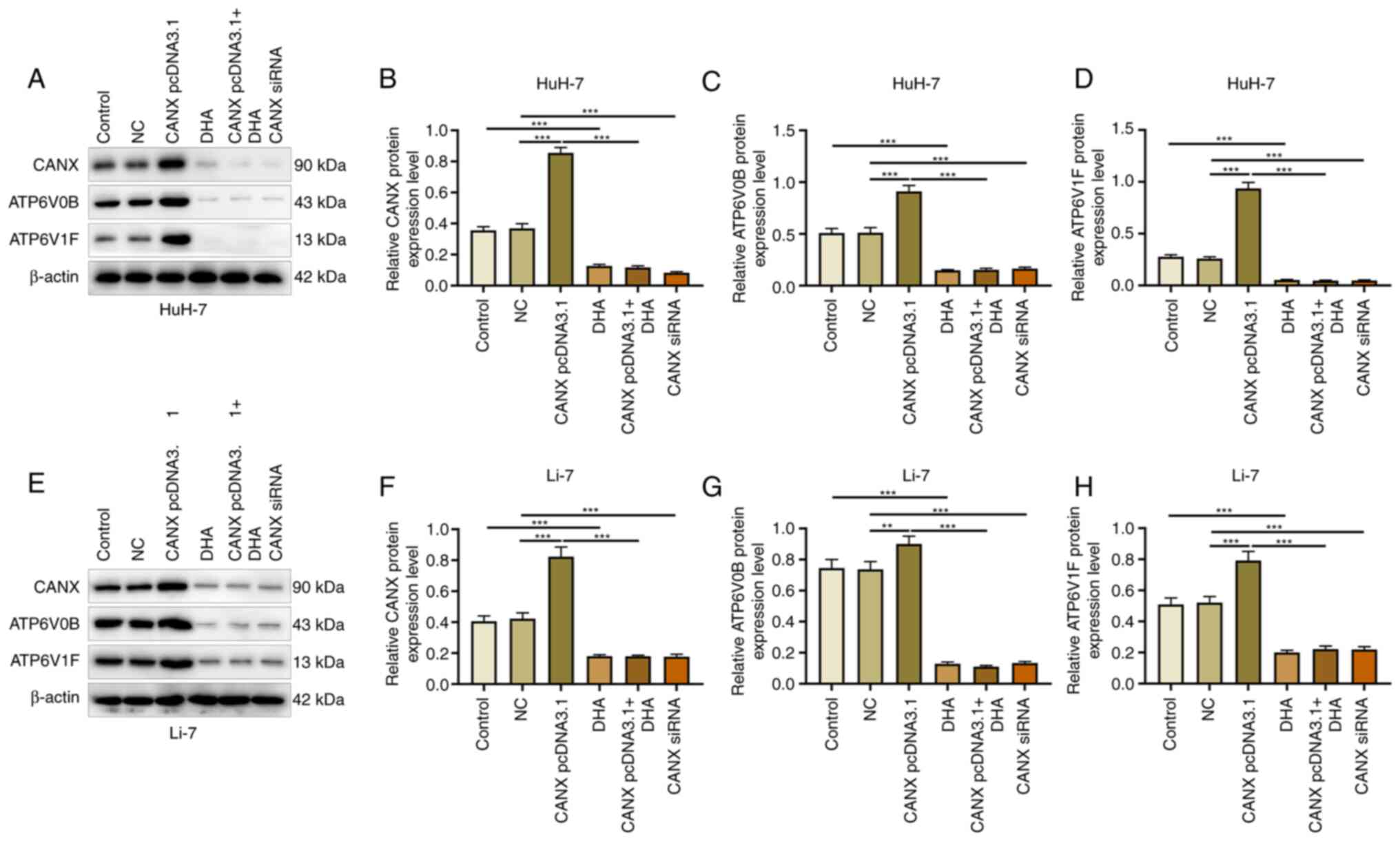 | Figure 8.Effects of CANX and DHA on ATP6V0B
and ATP6V1F expression. (A) Expression of (B) CANX, (C) ATP6V0B and
(D) ATP6V1F in HuH-7 cells, detected by western blotting after CANX
knockout, DHA or CANX overexpression. (E) Expression of (F) CANX,
(G) ATP6V0B and (H) ATP6V1F in Li-7 cells, detected by western
blotting after CANX knockout, DHA or CANX overexpression.
**P<0.01; ***P<0.001. CANX, calnexin; DHA,
dihydroartemisinin; NC, negative control; siRNA, small interfering
RNA. |
Discussion
Energy metabolism and mitochondrial function are
linked to the balance maintained by the calcium homeostatic control
system (19). A pivotal approach in
curtailing tumor cell energy metabolism involves targeting key
genes in calcium homeostasis regulation. CANX, a function-specific
ER chaperone protein, is implicated in the mechanism of cancer and
is pivotal in the storage and release of calcium ions in the ER
(7,20). It was previously discovered that DHA
prevented hepatoma cells from migrating and invading by decreasing
the synthesis of ATP synthase (ATP1A1 and ATP5H) via the
CaMKK2/NCLX signaling pathway (13). Furthermore, the present study
demonstrated that CANX serves a crucial role in liver cancer cell
proliferation, apoptosis, migration and invasion, energy
metabolism, mitochondrial function and ROS production. Knockdown of
CANX significantly inhibited liver cancer cell progression and more
significantly, DHA downregulated the expression of ATP6V0B and
ATP6V1F, and regulated the energy metabolism and metastasis of
liver cancer cells through CANX.
CANX is instrumental in binding, buffering and
storing Ca2+, thereby contributing to intracellular
calcium storage. It performs as a molecular chaperone for proteins,
regulates calcium homeostasis, enhances cell adhesion and preserves
RNA stability and gene expression with precision (21). CANX serves a crucial role in quality
control throughout the synthesis, maturation and transport of
secreted and membrane proteins. Dysregulation in calcium ion
homeostasis can trigger uncontrolled cell proliferation, ultimately
leading to the formation of tumors (22). Studies have reported that CANX is
extensively expressed in lung cancer tissues, and administration of
anti-CANX antibodies has proven efficacious in eliciting
substantial cytotoxic effects and suppressing tumor growth in
subcutaneous xenograft experiments (2,23).
This has validated CANX as a compelling therapeutic target. The
anti-calnexin antibody can detect calnexin protein via exosomes,
which can be applied to liquid biopsies (23). Moreover, prostate cancer cell
survival has been reported to be directly impacted by CANX
overexpression or knockdown in prostate cancer studies (24,25).
Through the CANX/calreticulin cycle, the mutant p53 ectonucleoside
triphosphate diphosphohydrolase 5 axis stimulates the production of
integrin-α5 and integrin-β1, which in turn promotes the growth,
metastasis and proliferation of tumor cells (26). Furthermore, in tumor cells, knocking
down CD317 lowered Ca2+ levels in the ER by targeting
calnexin and mediating tumor cell death (27). Furthermore, the results of the GEPIA
database retrieval in the present study indicated that CANX is
overexpressed in liver cancer tissue. The cell model of CANX
knockdown and overexpression validates the pivotal role of CANX in
regulating the proliferation and apoptosis of liver cancer cells,
which is consistent with prior research (28,29).
Moreover, the present study revealed that CANX knockdown effected
apoptosis. The findings further imply that, in addition to its
mediating function, CANX may serve as an independent regulatory
factor to influence the progression of cancer cells. Significantly,
DHA inhibits cell proliferation and apoptosis, and this effect
persists when DHA was applied to the CANX overexpression group.
This is in line with earlier findings (13) that DHA may impair the regulation of
calcium homeostasis, thereby having an anticancer effect.
CANX serves a crucial role in the interaction
between mitochondria and the smooth ER, and has a key role in the
mechanism of ATP production. Over the past decade, CANX and
BiP/78-kDa glucose-regulated protein, also referred to as BiP, have
been identified as multifunctional ER chaperone proteins. These
proteins can regulate ER and mitochondrial calcium ion homeostasis,
which in turn affects mitochondrial function, and controls the rate
of mitochondrial oxidative phosphorylation to regulate ATP input.
This indicates the mechanism by which ER chaperones regulate ATP
production and affect cellular energy metabolism (30,31).
From the experimental results of the present study, CANX is
involved in ATP production in liver cancer cells, regulating cell
proliferation and apoptosis. This also indicates its critical role
in liver cancer cell metastasis and suggests that CANX may serve a
‘bridging role’ in the pathway connecting the ER and the
mitochondria. However, further investigation using cryo-electron
microscopy is necessary. Previous research has also demonstrated
that DHA may influence the energy metabolism of liver cancer cells
by lowering ATP synthesis and influencing the formation of ROS via
CaMKK2/NCLX signaling (13). We
hypothesize that CANX represents a significant pathway through
which DHA reduces energy metabolism in liver cancer cells.
Abnormal mitochondrial function is a crucial factor
in mitochondria-mediated cell death, characterized by a decline in
ATP production and heightened ROS production (32,33).
Previous evidence has reported an interaction between acyl-CoA
synthetase long chain family member 4 and CANX, which collectively
modulates mitochondrial function and ATP production in cancer cells
(34). Additionally, the present
study demonstrated that CANX has a direct influence on
mitochondrial function, which may contribute to the reduction of
ATP production and promotion of ROS overproduction. Findings from a
previous study demonstrated that DHA can induce ROS production
(13), and in the present study, it
was observed that CANX regulates ROS production in HCC cells.
Notably, DHA can inhibit ROS production induced by the
overexpression of CANX.
ATP6V1 domain and V0 domain proteins serve crucial
roles in cancer cell progression mechanisms. ATP6V1F encodes a
constituent of V-ATPase and is essential for maintaining cancer
cell survival (35). ATP6V1
facilitates hydrogen ion transport, and its dysregulated expression
has been implicated in the clinicopathological features of several
malignancies, with notable associations observed in liver cancer
studies (10,36). ATP6V1F accelerates the invasive and
migratory capabilities of liver cancer cells whilst preventing cell
death, thereby fostering the progression and aggressiveness of
liver cancer (10). ATP6V1F is a
critical gene in cell proliferation, and miR-194 operates as a
tumor suppressor in digestive system cancers through the targeting
of ATP6V1F (37). The present study
demonstrated that CANX regulates the expression of ATP6V0B and
ATP6V1F, and it was hypothesized that CANX might affect calcium
homeostasis of hepatocellular carcinoma cells through ATP6. This is
consistent with the findings of Singh et al (38). The intricate regulation of
mitochondrial function relies on a network of ion signals that
safeguard the integrity of energy metabolism processes (39). Notably, NCLX and leucine zipper and
EF hand-containing transmembrane 1 are key players in this
regulatory machinery (40). In
light of the findings of earlier research (13), we hypothesize that NCLX is the
likely node where DHA controls calcium homeostasis and exerts its
anticancer effects. However, there could be alternative pathways
for ion exchange. In addition, DHA suppressed the expression of
ATP6V0B and ATP6V1F in the present study, suggesting that it may
prevent the synthesis of additional ATP synthases. The present
study demonstrated the regulatory role of CANX in liver cancer cell
progression, especially its effects on mitochondrial function and
energy metabolism. In addition, the potential anticancer effects of
DHA by inhibiting CANX expression was also assessed. Based on the
results of various functional experiments, the effects of the CANX
overexpression group and the DHA group are significantly opposite,
while the DHA group is similar to the CANX overexpression combined
with the DHA group. It is hypothesized that DHA may directly or
indirectly inhibit the function of CANX. Although overexpression of
CANX can produce a pro-oncogenic effect, DHA can significantly
inhibit this effect brought about by CANX. These results suggest
that DHA can significantly block or inhibit the action of CANX
pcDNA3.1, indicating that CANX is an effective target for DHA
(41,42).
Nevertheless, the present study has certain
limitations, for example it was not elucidated how CANX influences
calcium homeostasis through downstream genes, and the lack of
validation in animal models limited the comprehensive evaluation of
the therapeutic effects of DHA. Future studies should establish
mouse xenograft models to evaluate the antitumor activity of DHA
and enhance the understanding of its mechanisms.
In summary, CANX may be a crucial factor in the
progression, diagnosis, treatment and prognostic evaluation of
liver cancer. Its impact on the survival and metastasis of liver
cancer cells is mediated through mitochondrial function and energy
metabolism and the findings of the present study emphasize the
anticancer role of DHA by inhibiting CANX and downregulating
ATP6V0B and ATP6V1F expression.
Acknowledgements
Not applicable.
Funding
The present work was supported by the High-level Talents Project
of Hainan Natural Science Foundation of 2022 (grant no. 822RC830)
and the Hainan Health Industry Scientific Research Project of 2021
(grant no. 21A200072).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JC, QY, XL, WL and LG contributed to the study
conception and design. Material preparation, data collection and
analysis were performed by JC and QY. The first draft of the
manuscript was written by JC and XL. Data analysis, writing and
reviewing were performed by WL and LG. JC and LG confirm the
authenticity of all the raw data. All authors commented on previous
versions of the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu LC, Hsu CH, Hsu C and Cheng AL: Tumor
heterogeneity in hepatocellular carcinoma: Facing the challenges.
Liver Cancer. 5:128–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kobayashi M, Nagashio R, Jiang SX, Saito
K, Tsuchiya B, Ryuge S, Katono K, Nakashima H, Fukuda E, Goshima N,
et al: Calnexin is a novel sero-diagnostic marker for lung cancer.
Lung Cancer. 90:342–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ros M, Nguyen AT, Chia J, Le Tran S, Le
Guezennec X, Mcdowall R, Vakhrushev S, Clausen H, Humphries MJ,
Saltel F and Bard FA: ER-resident oxidoreductases are glycosylated
and trafficked to the cell surface to promote matrix degradation by
tumour cells. Nat Cell Biol. 22:1371–1381. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garbincius JF and Elrod JW: Mitochondrial
calcium exchange in physiology and disease. Physiol Rev.
102:893–992. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang LT, Lin MH, Liu KY, Chiou SS, Wang
SN, Chai CY, Tseng LW, Chiou HC, Wang HC, Yokoyama KK, et al:
WLS/wntless is essential in controlling dendritic cell homeostasis
via a WNT signaling-independent mechanism. Autophagy. 17:4202–4217.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng J, Yang T, Gao S, Cheng M, Shao Y,
Xi Y, Guo L, Zhang D, Gao W, Zhang G, et al: miR-148a-3p silences
the CANX/MHC-I pathway and impairs CD8(+) T cell-mediated immune
attack in colorectal cancer. FASEB J. 35:e217762021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lam STT and Lim CJ: Cancer biology of the
endoplasmic reticulum lectin chaperones calreticulin, calnexin and
PDIA3/ERp57. Prog Mol Subcell Biol. 59:181–196. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marshansky V, Rubinstein JL and Grüber G:
Eukaryotic V-ATPase: Novel structural findings and functional
insights. Biochim Biophys Acta. 1837:857–879. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Li H, Yang C, Liu L, Deng S and Li
M: Comprehensive analysis of ATP6V1s family members in renal clear
cell carcinoma with prognostic values. Front Oncol. 10:5679702020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu X, Li D, Zhu H, Yu T, Xiong X and Xu X:
ATP6V1F is a novel prognostic biomarker and potential immunotherapy
target for hepatocellular carcinoma. BMC Med Genomics. 16:1882023.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16:162016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen S, Du M, Liu Q, Gao P, Wang J, Liu S
and Gu L: Development of GLUT1-targeting alkyl glucoside-modified
dihydroartemisinin liposomes for cancer therapy. Nanoscale.
12:21901–21912. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang J, Xin C and Wang Y and Wang Y:
Dihydroartemisinin inhibits liver cancer cell migration and
invasion by reducing ATP synthase production through CaMKK2/NCLX.
Oncol Lett. 26:5402023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo Y, Ma J and Lu W: The significance of
mitochondrial dysfunction in cancer. Int J Mol Sci. 21:55982020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peoples JN, Saraf A, Ghazal N, Pham TT and
Kwong JQ: Mitochondrial dysfunction and oxidative stress in heart
disease. Exp Mol Med. 51:1–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang SW, Lee S and Lee EK: ROS and energy
metabolism in cancer cells: Alliance for fast growth. Arch Pharm
Res. 38:338–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Im H, Baek HJ, Yang E, Kim K, Oh SK, Lee
JS, Kim H and Lee JM: ROS inhibits RORα degradation by decreasing
its arginine methylation in liver cancer. Cancer Sci. 114:187–200.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brown D, Paunescu TG, Breton S and
Marshansky V: Regulation of the V-ATPase in kidney epithelial
cells: Dual role in acid-base homeostasis and vesicle trafficking.
J Exp Biol. 212:1762–1772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Molinari M, Eriksson KK, Calanca V, Galli
C, Cresswell P, Michalak M and Helenius A: Contrasting functions of
calreticulin and calnexin in glycoprotein folding and ER quality
control. Mol Cell. 13:125–135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kozlov G and Gehring K: Calnexin cycle -
structural features of the ER chaperone system. FEBS J.
287:4322–4340. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lüningschrör P, Andreska T, Veh A, Wolf D,
Giridhar NJ, Moradi M, Denzel A and Sendtner M: Calnexin controls
TrkB cell surface transport and ER-phagy in mouse cerebral cortex
development. Dev Cell. 58:1733–1747.e6. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yi YC, Liang R, Chen XY, Fan HN, Chen M,
Zhang J and Zhu JS: Dihydroartemisinin suppresses the tumorigenesis
and cycle progression of colorectal cancer by targeting
CDK1/CCNB1/PLK1 signaling. Front Oncol. 11:7688792021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lim S, Ha Y, Lee B, Shin J and Rhim T:
Calnexin as a dual-role biomarker: Antibody-based diagnosis and
therapeutic targeting in lung cancer. BMB Rep. 57:155–160. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo L, Li P, Xie Q, Wu Y, Qin F, Liao D,
Zeng K and Wang K: n6-methyladenosine-modified circular RNA family
with sequence similarity 126, member A affects cholesterol
synthesis and malignant progression of prostate cancer cells by
targeting microRNA-505-3p to mediate calnexin. J Cancer.
15:966–980. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ruiz C, Alborelli I, Manzo M, Calgua B,
Keller EB, Vuaroqueaux V, Quagliata L, Rentsch CA, Spagnoli GC,
Diener PA, et al: Critical evaluation of transcripts and long
noncoding RNA expression levels in prostate cancer following
radical prostatectomy. Pathobiology. 90:400–408. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pavlakis E, Neumann M, Merle N, Wieboldt
R, Wanzel M, Ponath V, Pogge Von Strandmann E, Elmshäuser S and
Stiewe T: Mutant p53-ENTPD5 control of the calnexin/calreticulin
cycle: A druggable target for inhibiting integrin-α5-driven
metastasis. J Exp Clin Cancer Res. 42:2032023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng J, Zhang G, Deng T, Liu Z, Zhang M,
Zhang P, Adeshakin FO, Niu X, Yan D, Wan X and Yu G: CD317
maintains proteostasis and cell survival in response to proteasome
inhibitors by targeting calnexin for RACK1-mediated autophagic
degradation. Cell Death Dis. 14:3332023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ryan D, Carberry S, Murphy ÁC, Lindner AU,
Fay J, Hector S, Mccawley N, Bacon O, Concannon CG, Kay EW, et al:
Calnexin, an ER stress-induced protein, is a prognostic marker and
potential therapeutic target in colorectal cancer. J Transl Med.
14:1962016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Okayama A, Miyagi Y, Oshita F, Nishi M,
Nakamura Y, Nagashima Y, Akimoto K, Ryo A and Hirano H: Proteomic
analysis of proteins related to prognosis of lung adenocarcinoma. J
Proteome Res. 13:4686–4694. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stahon KE, Bastian C, Griffith S, Kidd GJ,
Brunet S and Baltan S: Age-related changes in axonal and
mitochondrial ultrastructure and function in white matter. J
Neurosci. 36:9990–10001. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gutiérrez T and Simmen T: Endoplasmic
reticulum chaperones tweak the mitochondrial calcium rheostat to
control metabolism and cell death. Cell Calcium. 70:64–75. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du C, Guo X, Qiu X, Jiang W, Wang X, An H,
Wang J, Luo Y, Du Q, Wang R, et al: Self-reinforced bimetallic
mito-jammer for Ca(2+) overload-mediated cascade mitochondrial
damage for cancer cuproptosis sensitization. Adv Sci (Weinh).
11:e23060312024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim S, Ramalho TR and Haynes CM:
Regulation of proteostasis and innate immunity via
mitochondria-nuclear communication. J Cell Biol.
223:e2023100052024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Radif Y, Ndiaye H, Kalantzi V, Jacobs R,
Hall A, Minogue S and Waugh MG: The endogenous subcellular
localisations of the long chain fatty acid-activating enzymes ACSL3
and ACSL4 in sarcoma and breast cancer cells. Mol Cell Biochem.
448:275–286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haugen ØP, Khuu C, Weidemann HM, Utheim TP
and Bergersen LH: Transcriptomic and functional studies reveal
miR-431-5p as a tumour suppressor in pancreatic ductal
adenocarcinoma cells. Gene. 822:1463462022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen F, Kang R, Liu J and Tang D: The
V-ATPases in cancer and cell death. Cancer Gene Ther. 29:1529–1541.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang P, Xia L, Guo Q, Huang C, Wang Z,
Huang Y, Qin S, Leng W and Li D: Genome-wide association studies
identify miRNA-194 as a prognostic biomarker for gastrointestinal
cancer by targeting ATP6V1F, PPP1R14B, BTF3L4 and SLC7A5. Front
Oncol. 12:10255942022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Singh J, Meena A and Luqman S: New
frontiers in the design and discovery of therapeutics that target
calcium ion signaling: A novel approach in the fight against
cancer. Expert Opin Drug Discov. 18:1379–1392. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kannurpatti SS: Mitochondrial calcium
homeostasis: Implications for neurovascular and neurometabolic
coupling. J Cereb Blood Flow Metab. 37:381–395. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Marchi U, Santo-Domingo J, Castelbou C,
Sekler I, Wiederkehr A and Demaurex N: NCLX protein, but not LETM1,
mediates mitochondrial Ca2+ extrusion, thereby limiting
Ca2+-induced NAD(P)H production and modulating matrix redox state.
J Biol Chem. 289:20377–20385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma Y, Zhang P, Zhang Q, Wang X, Miao Q,
Lyu X, Cui B and Ma H: Dihydroartemisinin suppresses proliferation,
migration, the Wnt/β-catenin pathway and EMT via TNKS in gastric
cancer. Oncol Lett. 22:6882021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang X, Guo P, Tian J, Li J, Yan N, Zhao X
and Ma Y: LncRNA GAS5 participates in childhood pneumonia by
inhibiting cell apoptosis and promoting SHIP-1 expression via
downregulating miR-155. BMC Pulm Med. 21:3622021. View Article : Google Scholar : PubMed/NCBI
|