Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common of all tumors worldwide and has the third highest mortality
rate of all tumors. The curative effect of surgery, chemotherapy
and radiotherapy was not satisfactory (1) and Few patients with advanced stages
survive more than five years (2).
HCC is the most common form of liver cancer,
accounting for approximately 75–85% of cases worldwide. The
etiology of HCC is multifactorial, with chronic hepatitis B virus
(HBV) infection identified as a major risk factor, especially in
endemic areas. Non-alcoholic fatty liver disease (NAFLD) has a
global prevalence of 25% and is the leading cause of cirrhosis and
hepatocellular carcinoma (3).
Metabolic disorders, insulin resistance, and lipid deposition also
contribute to the progression from NAsh to HCC cirrhosis (3,4).
Alcohol can enhance the risk of liver cancer by inducing direct
hepatocyte toxicity, resulting in cellular damage and apoptosis
(5), promoting oxidative stress and
inflammatory response within the liver (6), interfering with carbon monoxide
metabolism, thereby causing DNA damage and dysregulation of repair
mechanisms (7) and other ways.
Early diagnosis of HCC remains a major challenge, mainly due to the
asymptomatic nature of the disease in its initial stages and the
lack of specific biomarkers (8,9).
NFκB is inactive when it binds to inhibitor of NFκB
at rest and is found in the cytoplasm. Activated NFκB protein
enters the nucleus and participates in inflammatory and immune
responses (10) and NFκB inhibitors
are involved in the complex regulation of NFκB (11). However, little is currently known
about NFκB inhibitors in HCC and their carcinogenic mechanism
(12).
Deficiency of nuclear factor of κ light polypeptide
gene enhancer in B-cells inhibitor (NFKBI)E leads to
hyperproliferation of a specific subset of B1 B-cells and results
in increased activation of NF-κB in these cells upon stimulation by
Toll-like receptors. Furthermore, the deficiency of NFKBIE
synergizes with mutant MYD88 signaling, thereby enhancing B-cell
proliferation in vitro (13). NFKBIE mutations can facilitate the
progression of chronic lymphocytic leukemia through various
mechanisms, including bidirectional communication with the
microenvironment and reduced responsiveness to BTK inhibitor
therapy (14). However, the role of
NFKBIE in HCC and its mechanism have not been reported.
Based on information from multiple open databases,
the present study evaluated the NFκB inhibitor family at multiple
levels, including gene expression, prognosis, clinical features,
and immune infiltration, to elucidate a new prognostic marker and
treatment strategy for HCC.
Materials and methods
The cancer genome atlas (TCGA)
database
All RNA expression patterns of NFκB inhibitors in
HCC and pan-cancer were taken from The University of Alabama at
Birmingham Cancer Data Analysis Portal (UALCAN) and the Tumour
Immune Estimation Resource (TIMER; http://cistrome.shinyapps.io/timer/), whose data were
obtained from the TCGA database. TIMER (15,16)
predicts tumour-infiltrating immune cells using gene expression
patterns. In the present study, it was used it to assess NFKBIE and
immunological infiltration. UALCAN (https://ualcan.path.uab.edu/index.html) (17) is a resource that is available for
free that can be used to evaluate TCGA gene expression data. In the
present study, UALCAN was used to compare the NFκB inhibitor family
and its relationship with clinical and pathological characteristics
and its prognostic value.
Kyoto encyclopedia of genes and
genomes (KEGG)
KEGG (18) is the
main public database on pathways. Pathway analysis of differential
protein coding genes was performed using the KEGG database, and the
significance of differential gene enrichment in each pathway entry
was calculated using the hypergeometric distribution test. The fold
change (FC) was set to 1.5, whilst adjusted P<0.05 was
statistically significant.
Survival analysis
Kaplan-Meier plots/Liver cancer mRNA-seq/Auto select
best cutoff (http://kmplot.com/analysis) were used to estimate HCC
survival rates at 7/5/2024 (19).
Metabolic gene rapid visualizer
(MERAV)
Data from >4,000 microarrays are combined in
MERAV (http://merav.wi.mit.edu/), enabling
reliable cross-sectional comparisons of several tissues and cell
lines (20).
The human protein atlas (HPA)
NFKBIE protein expression information was obtained
from the HPA (https://www.proteinatlas.org/search/NFKBIE). The HPA
is a database based on proteomics data that contains protein
expression in both normal and malignant tissues (21).
LinkedOmics
LinkedOmics (https://linkedomics.org/) (22) is a web-based analytical tool for
comparing multiomics cancer datasets. This database was used for
KEGG analysis and analysis of the TOP50 genes positively and
negatively associated with NFKBIE.
STRING
STRING (https://string-db.org/. version 12.0) database is a
protein interaction network database based on public database and
literature information. It gathers several public databases,
including UniProt, KEGG, NCBI, and Gene Ontology, to integrate
these data and generate a comprehensive protein interaction network
database (23–25).
The genomics of drug sensitivity in
cancer
The Genomics of Drug Sensitivity in Cancer (GDSC,
version 8.5, www.cancerRxgene.org) database currently encompasses
data on drug sensitivity from nearly 75,000 experiments, detailing
the responses of approximately 700 cancer cell lines to 138
anti-cancer drugs. Graphical representations of the data are
provided throughout, along with links to related resources, and all
datasets are fully downloadable. GDSC offers a unique resource,
comprising an extensive collection of drug sensitivity and genomic
datasets, to facilitate the discovery of new therapeutic biomarkers
for cancer treatment (26).
The cancer therapeutics response
portal (CTRP)
CTRP (https://portals.broadinstitute.org/ctrp. version 2.1)
integrates the intricate relationship between cancer cell line
genetics, lineage, and other cellular characteristics with small
molecule sensitivity to expedite the discovery of personalized
cancer treatments. By seamlessly integrating comprehensive drug
sensitivity data with detailed genomic information, CTRP serves as
a robust resource for gaining profound insights into the complex
interplay between cancer genetics and drug responses (27–29).
Hepatocellular carcinoma database
(HCCDB)
The gene expression data of ~4,000 clinical samples
are contained in the HCCDB (version, http://lifeome.net/database/hccdb/about.html), which
is a database that provides accurate and reproducible differential
expression profiles for HCC (30).
The present study used this database to look at NFKBIE gene
networks co-expressed with HCC as well as with normal tissues.
Gene set cancer analysis
(GSCALite)
GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/.) is web
server for dynamic analysis and visualization of cancer gene sets
and drug sensitivity correlations that will be of wide practical
value to cancer researchers. The present study used this database
to analyze the signaling pathways of NFKBIE effects and studies
related to drug resistance (31).
Tissue microarray and
immunohistochemistry (IHC)
An HCC tissue microarray (cat. no. HLivH160CS02)
consisting of 80 matched tumours and normal samples was purchased
from Shanghai Outdo Biotech Co., Ltd. Following the dewaxing of
paraffin-embedded sections, antigens were repaired (20X Tris-EDTA
antigen repair solution; Wuhan Servicebio Technology Co., Ltd.;
cat. no. G1203), and endogenous peroxidase activity was quenched
(3% hydrogen peroxide solution, incubated at room temperature for
25 min away from light). Subsequent steps included 3% BSA (Wuhan
Servicebio Technology Co., Ltd.; cat. no. GC305010) at room
temperature for 30 min blocking to prevent non-specific binding,
incubation with the primary overnight at 4°C and (NFKBIE polyclonal
antibodies 1:200; cat no. 11273-1-AP; Proteintech Group, Inc.)
secondary antibodies at room temperature (Wuhan Servicebio
Technology Co., Ltd.; cat. no. GB23303; 1:500) and development of
the signal using DAB chromogen (Wuhan Servicebio Technology Co.,
Ltd.; cat. no. G1212). Finally, hematoxylin stained nucleus was
used for comparison. The prepared slides were then digitized using
a Pannoramic MIDI scanner (3DHISTECH), ensuring high-resolution
imaging of the histological details.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was isolated from cell samples with a
total of 3×105 cells using the RNeasy Mini Kit (Qiagen
GmbH) according to the manufacturer's instructions. RNA purity and
quantity were assessed using a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc.). Complementary DNA (cDNA) was synthesized
from the isolated RNA using the PrimeScript RT Reagent Kit (Takara
Biotechnology Co., Ltd.), followed by cDNA amplification with the
SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.). The
amplification reactions were conducted on an ABI StepOne Plus
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). NFKBIE-Forward primer sequence: TCTGGCATTGAGTCTCTGCG;
NFKBIE-Reversed primer sequence: AGGAGCCATAGGTGGAATCAG.
GAPDH-Forward primer sequence: GAGAAGTATGACAACAGCCTCAA.
GAPDH-Reversed primer sequence: GCCATCACGCCACAGTTT. The
2−ΔΔCq method was used for quantitative analysis
(32). The PCR cycling conditions
were set as follows: initial denaturation at 94–95°C for 1–3 min;
25–35 cycles of denaturation at 94°C to 95°C for 15–30 sec,
annealing at 4–5°C below the primer's melting temperature
(~50–65°C) for 20–40 sec and extension at 72°C for ~1 min per 1 kb
of target DNA. This was followed by a final extension at 72°C for
5–10 min, and the samples were then held at 4°C. All protocols for
RNA extraction, cDNA synthesis, and qPCR were conducted strictly
following the manufacturers' guidelines. These experiments were
replicated three times to ensure reliability and reproducibility of
the results.
Cell culture and transfection
HEP-3B and PLC/PRF/5 cells were purchased from the
American Type Culture Collection. Huh-7 cells were purchased from
JCRB Cell Bank. HCCLM3 and MHCC97H cells were purchased from the
Shanghai Institute of Biochemistry and Cell Biology (Chinese
Academy of Sciences; Shanghai, China). All cells were cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) or RPMI 1640 (Gibco;
Thermo Fisher Scientific, Inc.) with 10% (Bioind) and 1% ampicillin
(Biosharp Life Sciences) at 37°C in a 5% CO2
incubator.
Short-interfering (si)RNA technology was used to
knockdown NFKBIE. The sequences of the siRNAs [General Biol (Anhui)
Co., Ltd.] used were as follows: NFKBIE scrambled siControl:
5′-UUCUCCGAACGUGUCACGUTT-3′ and 3′-ACGUGACACGUUCGGAGAATT-5′; NFKBIE
siRNA-1: 5′-UCAAGGAACCACAGGAGAATT-3′ and
3′-UUCUCCUGUGGUUCCUUGATT-5′; NFKBIE siRNA-2:
5′-GGACCGGCAUGGUGACACATT-3′ and 3′-UGUGUCACCAUGCCGGUCCTT-5′; and
NFKBIE siRNA-3: 5′-GGAAACUGCUGCUGUGUACTT-3′ and
3′-GUACACAGCAGCAGUUUCCTT-5′. Briefly, HCCLM3 and Huh-7 were first
digested with Trypsin-EDTA Solution (Biosharp Life Sciences; cat.
no. BL512A), then ~2.5×105 cells were counted in each
6-well plate and, after the cells were attached to the wall,
pre-configured NFKBIE-siControl (100 nM), NFKBIE-si1 (100 nM), and
NFKBIE-si2 (100 nM), were added into the corresponding 6-well
plates for 8 h at 37°C. The co-transfection reagent used was
Lipofectamine® 2000 Transfection Reagent (Thermo Fisher
Scientific, Inc.). The culture medium was changed 8 h after
transfection, and the NFKBIE protein level was measured by western
blotting after 48 h. HCCLM3 and Huh-7 cells were transfected with
the method and the NFKBIE siRNA knockdown effect was verified by
western blotting, and then RNA-sequencing (RNA-seq) was performed
by Shanghai OE Biotech Co., Ltd. (Shanghai, China). Wound healing
and migration experiments were performed on HCCLM3 and Huh-7 cells
after NFKBIE knockdown.
Western blotting
The same number of cells (1×106) were
treated with cell lysate (Biosharp Life Sciences), the sample
buffer (Bio-Rad 2X Laemmli Sample Buffer; Bio-Rad Laboratories,
Inc.; cat no. 1610737) was added and the proteins were denatured
and stored at −80°C. The 5×105 cells yielded 100 ul
protein solution, 10 µl per well. The proteins were separated on
10% SDS-PAGE gel and transferred onto a PVDF membrane
(MilliporeSigma; 0.45 µm). The membrane was blocked in skimmed milk
at room temperature for 30–60 min and incubated with the primary
antibody followed by the second antibody (OriGene Technologies,
Inc.; cat. no. ZF-0516; 1:5,000) for 40–60 min at room temperature.
ECL developer (Beyotime Institute of Biotechnology; cat. no.
P0018FS) was used. In this study, protein concentration was
determined using the BL367A assay kit provided by Biosharp Life
Sciences. Initially, BCA Reagent A and BCA Reagent B were mixed in
specified ratios to prepare the working solution. Protein samples
and a series of BSA standards with known concentrations were then
prepared and added to a 96-well plate. The BCA working solution was
added to each well, and the plate was incubated at room temperature
for 30 min to 2 h. Following incubation, absorbance at 562 nm was
measured using a spectrophotometer. Protein concentrations were
calculated by comparing these measurements against a standard
curve. ImageJ software (National Institutes of Health, version
1.48) was used to calculate grayscale values. NFKBIE polyclonal
antibodies (1:1,000; cat no. 11273-1-AP; Proteintech Group, Inc.)
and GAPDH monoclonal antibodies (1:10,000; cat no. 60004-1-Ig;
Proteintech Group, Inc.) were used as primary antibodies.
Wound healing assays
A total of 3×105 HCCLM3/Huh-7 cells were
initially cultured in Dulbecco's Modified Eagle Medium (DMEM)
supplemented with 10% fetal bovine serum (FBS) and incubated at
37°C in a 5% CO2 atmosphere until reaching 90%
confluence. A sterile 200 µl pipette tip was employed to create a
consistent scratch across the cell monolayer at near full
confluence, focusing on maintaining uniform scratch width. After
scratching, cells were washed with phosphate-buffered saline (PBS)
to remove dislodged cells and debris, followed by the replacement
of the culture medium with DMEM containing 1% FBS, to minimize
proliferation and concentrate on migration. Photographic
documentation was conducted immediately post-scratch (0 h) and at
specified intervals (48 h), utilizing an inverted microscope with
consistent magnification and focus. The extent of wound closure was
assessed by measuring scratch width changes using ImageJ software
(National Institutes of Health, version 1.48). The assay was
replicated a minimum of three times to ensure data reliability.
Statistical differences between groups were analysed using
Student's t-test or one-way ANOVA with Tukey's multiple comparisons
test, to determine the impact of the experimental conditions on the
cells' migratory capabilities.
Migration experiment
The complete medium was prepared using DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Bioind)
and 1% penicillin-streptomycin (Biosharp). For both the
experimental and control groups, cells were digested with trypsin
and counted to achieve a concentration of 2×104 cells
per 200 µl. These cells were then seeded into culture chambers
(Falcon) with serum-free medium inside the chambers and complete
medium outside. The cells were incubated at 4°C in an atmosphere of
5% CO2 for 48 h. After incubation, cells were fixed with
4% paraformaldehyde (Biosharp) and stained with crystal violet.
Subsequently, the upper layer of cells in the chamber was gently
wiped away, and the cells were observed and photographed under an
inverted microscope. This experiment was repeated at least three
times. Cell counting was performed using Image J, and data analysis
was conducted using GraphPad Prism 8.0 software. Statistical
significance between the experimental and control groups was
assessed using t-tests or analysis of variance (ANOVA).
RNA-seq and differentially expressed
genes (DEGs) analysis
The libraries were sequenced (TruSeq Stranded mRNA
LTSample Prep Kit, Illumina, RS-122-2101; Agencourt AMPure XP,
BECKMAN COULTER, A63881; Qubit RNA Assay Kit, Life Technologies,
Q32852; Qubit dsDNA Assay Kit, Life Technologies, Q328520;
Bioanalyzer 2100 RNA-6000 Nano Kit, Agilent, 5067-1511; Bioanalyzer
2100 DNA-1000 Kit Agilent, 5067-1504; SuperScript II Reverse
Transcriptase, Invitrogen, 18064014;) on an Illumina HiSeq X Ten
platform (cat. no. DOE20221816; Illumina, Inc.) and 150 bp
paired-end reads were generated. The Agilent 2100 bioanalyzer was
used to measure the length and quality of the library. The base
error rate in Illumina sequencing was determined by the Phred
score, which was calculated by the model to predict the probability
of error in base discrimination. Raw data (raw reads) in fastq
format were firstly processed using Trimmomatic (version 0.36)
(33) and the low-quality reads
were removed to obtain the clean reads. Subsequently, ~14,000 clean
reads for each sample were retained for analysis. The clean reads
were mapped to the human genome (GRCh38) using HISAT2 (version
2.2.1.0) (34). The fragments per
kilobase of transcript per million mapped reads (35) of each gene was calculated using
Cufflinks (version 2.2.1) (36),
and the read counts of each gene were obtained by HTSeq count
(version 0.9.1) (37). The
concentration of RNA samples can be quickly measured using NanoDrop
spectrophotometers and the purity of RNA can be assessed by
absorption ratios (A260/A280 and A260/A230). An A260/A280 ratio
close to 2.0 usually indicates less protein contamination, while an
A260/A230 ratio greater than 2.0 indicates less pollution from
organic solvents or other impurities.
Differential expression analysis was performed using
the DESeq (version 1.18.0) R package (38). P<0.05 and FC >1.5 or <0.5
were set as the threshold for significantly differential
expression. Hierarchical cluster analysis of DEGs was performed to
demonstrate the expression pattern of genes in different groups and
samples. Gene Ontology (http://geneontology.org) (39) enrichment and KEGG (18) pathway enrichment analysis of DEGs
were performed respectively using R (version 4.3.3, Comprehensive R
Archive Network) based on the hypergeometric distribution.
Statistical analysis
Kaplan-Meier plots were used to establish survival
curves. Correlations between gene expression levels were assessed
using Spearman's correlation coefficient. An unpaired Student's t
test was used to evaluate the statistical significance of
differences between two independent groups. Ordinary one-way ANOVA
with Tukey's multiple comparisons test was used to assess the
statistical significance between three independent groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Transcriptional levels of the NFκB
inhibitor family in HCC and across cancers
A total of five NFκB inhibitors were identified and
their transcript levels were compared at the pan-cancer level:
NFKBIB, NFKBID and NFKBIE were significantly overexpressed in HCC
tissue compared with normal tissue, and NFKBIA and NFKBIZ
expression was not significantly different between HCC and normal
tissues (Figs. 1 and S1).
Correlation between NFκB inhibitor
expression and prognosis
The predictive ability of NFκB inhibitors for HCC
were analysed using Kaplan-Meier plots. According to the results,
there was no significant association between NFKBIA, NFKBIB, NFKBID
and NFKBIZ expression and prognosis (Fig. 2A). However, a low expression of
NFKBIE was significantly associated with a better prognosis for
patients with HCC, regardless of overall survival, disease specific
survival and relapse-free survival, compared with a high expression
(Fig. 2B). Significant correlations
were demonstrated between the RNA expression level of NFKBIE and
several clinical characteristics, including the stage of cancer and
tumour protein P53 (TP53) mutation status, cancer stages, tumour
grade, nodal metastasis status, patient's weight, and patient's age
(Fig. 2C). Data from MERAV and the
HPA database also demonstrated that HCC had significantly higher
transcriptional and protein levels of NFKBIE than normal tissue
samples (Fig. 2D and E).
 | Figure 2.Low mRNA expression of NFKBIE in HCC
is associated with a favourable prognosis compared with high
expression. (A) Prognostic value of NFKBIA, NFKBIB, NFKBID and
NFKBIZ in patients with HCC at the mRNA level. (B) Survival curve
of NFKBIE in HCC (overall survival, disease specific survival,
relapse-free survival, and progression-free survival). (C) NFKBIE
mRNA expression is positively correlated with tumour grade, TP53
mutation status, tumour stage, lymph node metastatic status,
patient weight, and age. (D) NFKBIE is overexpressed in HCC,
according to data from the Metabolic Gene Rapid Visualizer. (E)
NFKBIE expression in HCC and normal liver tissue was analysed using
data from the Human Protein Atlas database. NFKBIE expression was
higher in HCC than in normal liver tissue. *P<0.05; **P<0.01;
***P<0.001. NFKBI, nuclear factor of κ light polypeptide gene
enhancer in B-cells inhibitor; HCC, hepatocellular carcinoma; TP53,
tumour protein P53; LIHC, liver HCC; TCGA, The Cancer Genome Atlas;
HR, hazard ratio. |
KEGG analyses and co-expressed
genes
The 50 genes with the strongest associations were
mainly enriched in the ‘Ribosome’, ‘NFκB signalling pathway’ and
‘Primary immunodeficiency’ (Fig.
3A). Heatmaps were created to demonstrate all 50 genes that
exhibited the strongest associations with NFKBIE using the
LinkedOmics database (Fig. 3B and
C). Furthermore, the HCCDB indicated notable differences in the
co-expression gene network between HCC and normal tissue (Fig. 3D and E).
Drug sensitivity, cancer pathway
activity and the protein-protein interaction network
V-rel avian reticuloendotheliosis viral oncogene
homolog A (RELA), v-rel avian reticuloendotheliosis viral oncogene
homolog (REL), conserved helix-loop-helix ubiquitous kinase (CHUK),
inhibitor of kappa light polypeptide gene enhancer in B-cells,
kinase gamma (IKBKG), inhibitor of kappa light polypeptide gene
enhancer in B-cells, kinase beta (IKBKB), and v-rel avian
reticuloendotheliosis viral oncogene homolog B (RELB) were mainly
enriched in ‘Shigellosis’, ‘Kaposi sarcoma-associated herpesvirus
infection’, ‘C-type lectin receptor signaling pathway’, ‘NFκB
signaling pathway’, and ‘antifolate resistance’ (Fig. 4A). These proteins also had the
strongest interactions with NFKBIE protein (Fig. 4B). In addition, data from Gene Set
Cancer Analysis (GSCALite) indicated that NFKBIE suppression
notably reduced the PI3K/AKT, TSC/mTOR, and other signaling
pathways (Fig. 4C). Moreover, a
drug sensitivity analysis using the Genomics of Drug Sensitivity in
Cancer and Cancer Therapeutics Response Portal databases revealed
that low expression of NFKBIE was correlated with resistance to
Z-LLNle-Cho (Fig. 4D) and
dabrafenib (Fig. 4E). These results
provide new therapeutic ideas for patients with HCC.
Correlation between NFKBIE and tumour
immune infiltration
In HCC, memory B cells had the greatest expression
of NFKBIE, whereas numerous T cells had the lowest expression of
NFKBIE (Fig. 5A) (14). Using the TIMER database, B cells and
macrophage cells with TP53 mutation had higher levels of immune
infiltration. (Fig. 5B). NFKBIE was
also significantly negatively associated with endothelial cell and
haematopoietic stem cell immune infiltration. Conversely, B cell,
M1 macrophage and myeloid dendritic cell immune infiltration were
found to be significantly positively correlated with NFKBIE
(Fig. 5C-G). Low expression of
NFKBIE combined with immune cell infiltration has a better
prognosis (Fig. S2).
Silencing NFKBIE regulates HCC through
‘antigen processing and presentation’. To assess the role
that NFKBIE serves in HCC, siRNA was used to knockdown NFKBIE
expression and then RNA-seq analysis was performed, and a volcanic
map of DEGs was shown (Fig. 6A).
Among the top 20 elevated genes, the ‘P53 signalling pathway’ was
significantly enriched (Fig. 6B).
It was demonstrated that knockdown of NFKBIE expression
significantly affected ‘antigen presentation’ (HSA04612) and
‘hepatocellular carcinoma’ (HSA05225) (Fig. 6C). Additionally, among the top 20
elevated genes, the ‘longevity-regulating pathway’ was
significantly enriched (Fig. 6D).
Similar results were demonstrated using data from the HPA database,
and NFKBIE was notably associated with ‘antigen presentation’ with
high confidence (Fig. 6E).
Silencing NFKBIE affects the
proliferation and migration of HCC cells
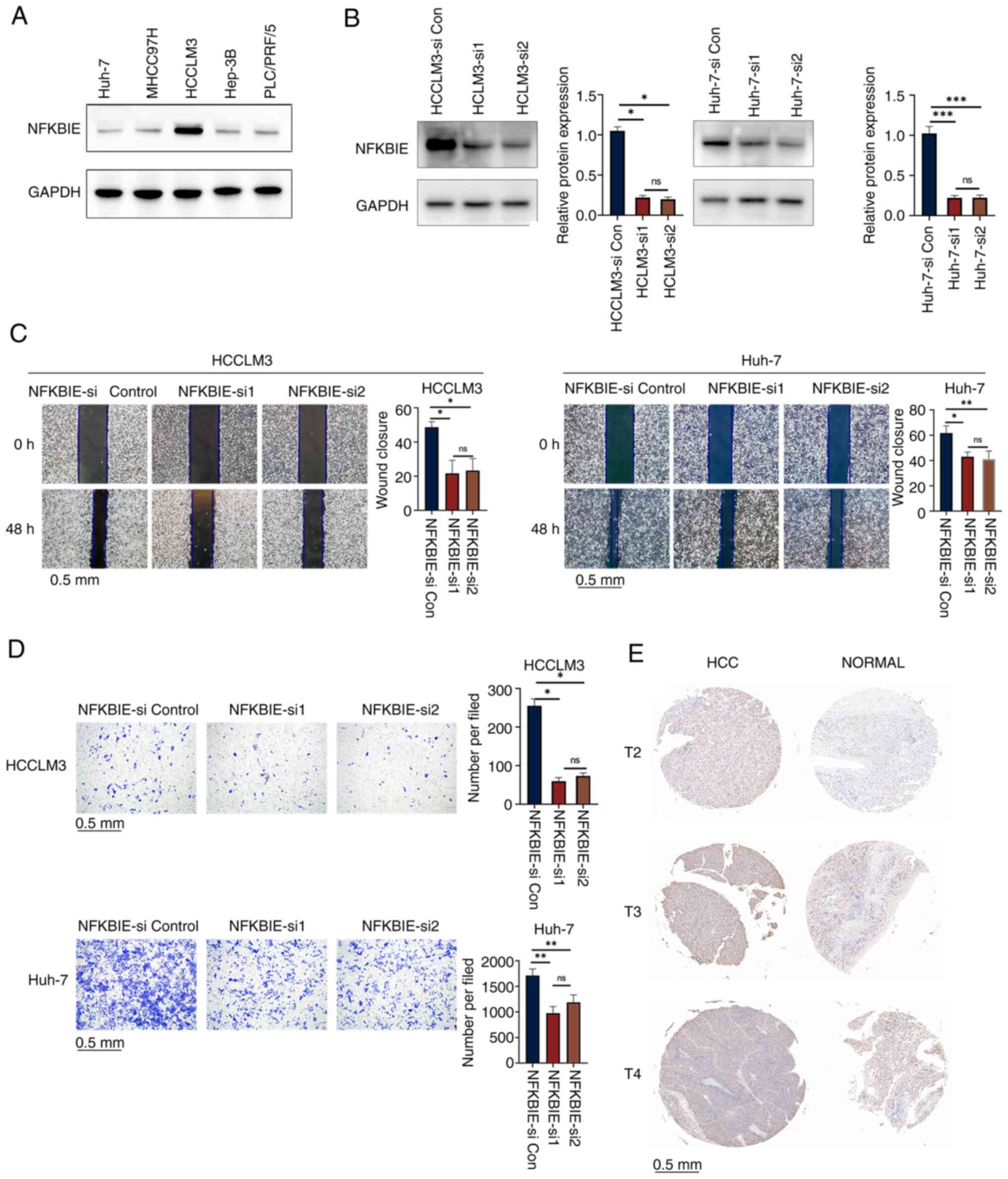
The expression of NFKBIE was notably the lowest in
Huh-7 cells and the highest in HCCLM3 cells (Fig. 7A). After siRNA-mediated reduction in
NFKBIE expression (Fig. 7B), the
proliferation and migration of HCCLM3 and Huh-7 cells were
significantly reduced, compared with the control (Fig. 7C and D). Furthermore, IHC of tissue
microarray demonstrated NFKBIE overexpression in different stages
of HCC (Figs. 7E and S3).
Discussion
Liver cancer is the fourth leading cause of
cancer-related deaths worldwide and a global health threat. The
main risk factors include infection, behavioural factors, metabolic
factors, and aflatoxin (40).
The activation of NFκB is necessary for the
formation and progression of HCC. In addition, the NFκB protein
complex is essential for both conventional and atypical
inflammatory activation and immunological development pathways
(10,41,42).
The NFκB inhibitor family regulates NFκB. Biologically, proteins
encoded by the NFκB inhibitor family bind to a part of NFκB,
blocking it from entering the nucleus and activating genes there
(43,44). However, the role of the NFκB
inhibitor family and associated carcinogenic processes in HCC are
poorly understood. In the present study, most malignant tissues
expressed more NFκB inhibitors than normal tissues and unpaired HCC
samples had higher NFKBIB, NFKBID and NFKBIE expression than normal
tissues (Fig. 1B-D).
The mechanism of NFKBIB, NFKBID and NFKBIE in HCC
has not been reported to the best of the authors' knowledge;
however, the RS3138053 NFKBIA polymorphism has been linked to an
increased risk of HCC (45). NFKBID
modulates the B-cell response, providing an antiparasitic antibody
response (46). Targeting the
NFKBID gene, lung tumour-derived exosome mir-3473b promotes the
NFκB pathway in local lung fibroblasts, therefore promoting the
pulmonary colonization of lung tumour cells (47). NFKBID encodes IκBNS, and its
deletion affects B-cell development and function (48,49).
Moreover, NFKBIE deficiency increases NFκB activation in cells and
increases in vitro B-cell proliferation (13). In the present study, it was
demonstrated that NFKBIB, NFKBID and NFKBIE were overexpressed in a
range of malignancies. Furthermore, using Kaplan-Meier plots, the
present study found that NFKBIA, NFKBID and NFKBIZ had no
significant effect on prognosis. However, patients with HCC with
low NFKBIE expression had a better prognosis; thus, NFKBIE was
selected for future research.
NFKBIE is an NFκB suppressor, and it has been linked
to several clinical characteristics of HCC. In the blood, deletion,
and mutation of NFKBIE has been associated with several leukaemias
(14,50). However, similar conditions have not
been observed in HCC. Through the LinkedOmics database, it was
demonstrated that NFKBIE was positively associated with the NFκB
signalling pathway and primary immunodeficiency. The STRING
database also verified the positive correlation between NFKBIE and
the NFκB pathway. The NFκB family is known to comprise P50, P52,
P65, C-REL, and RelB (51). From
the STRING database, it was demonstrated that RELA, RELB, REL,
NFKB1 and other genes interact with NFKBIE. RELA regulates its
downstream target genes by activating apoptosis, and RELA/NFκB may
be an important new therapeutic target for human liver cancer
(52). In prostate cancer, RELB is
essential for the differential radiosensitization of ascorbic acid
(53). Moreover, according to GSCA
database analysis, NFKBIE suppressed the PI3K/AKT, RAS/MAPK, RTK
and TSC/mTOR signalling pathways.
Inflammation is a local protective response of the
body to injury or infection, but chronic inflammation serves a
pathogenic role in cancer (54).
HCC is a useful tumour type to study the association between
tumours and inflammation as most HCC occurs in an inflamed liver
(55). According to the results of
the present study, the expression of NFKBIE was higher in B cells
and lower in T cells. TP53 mutant samples also had higher levels of
B-cell infiltration than wild-type TP53 samples. NFKBIE was
negatively correlated with endothelial cells and haematopoietic
stem cells, but positively correlated with B cells, M1 macrophages
and myeloid dendritic cells. Furthermore, it was demonstrated that
low levels of NFKBIE combined with High T cell CD4+
effector memory, macrophages and common lymphoid progenitors
indicated a good prognosis. After the knockdown of NFKBIE, ‘antigen
processing and presentation’ and the ‘hepatocellular carcinoma’
pathways were significantly affected, which is consistent with data
from the Human Protein Atlas. Moreover, the results also
demonstrated that the genes with increased expression were enriched
in the P53 pathway and the longevity-regulating pathway.
Additionally, the genes with reduced expression were enriched in
non-alcoholic liver disease, which eventually progresses to liver
cancer (56). These findings
suggest that NFKBIE may have potential significance in treating
this disease as the inhibition of NFKBIE significantly decreased
both the proliferation and migration of HCC cells. In conclusion,
NFKBIE is a promising immunotherapeutic target for HCC.
In the present study, the mechanism of NFKBIE was
not deeply studied. In the future, the specific mechanism of NFKBIE
promoting the proliferation and migration of liver cancer will be
further explored.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of City Cancer
Early Diagnosis and Treatment, which is supported by the Ministry
of Finance and National Health Commission of the People's Republic
of China, the Natural Science Foundation of Anhui Province (grant
no. 2108085QH339) and the Youth Research Fund of Anhui Cancer
Hospital (grant no. 2022YJQN019 and 2022YJQN020).
Availability of data and materials
The data generated in the present study may be found
in the NCBI repository at the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA893825.
Authors' contributions
LQ, YZ and HL designed the research. JT, JW, LZ, TD,
SS and CT helped with the analysis. ZY wrote the manuscript. ZY and
QL confirm the authenticity of all the raw data. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The experimental protocol of the present study was
established according to the ethical guidelines of the Helsinki
Declaration and was approved by the Human Ethics Committee of The
First Affiliated Hospital of University of Science and Technology
of China [approval no. 2020-SN(H)-023]. Written informed consent
was obtained from individual participants or the guardians of
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Chemoembolization for intermediate HCC: Is there proof of survival
benefit? J Hepatol. 56:984–986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louafi S, Boige V, Ducreux M, Bonyhay L,
Mansourbakht T, de Baere T, Asnacios A, Hannoun L, Poynard T and
Taïeb J: Gemcitabine plus oxaliplatin (GEMOX) in patients with
advanced hepatocellular carcinoma (HCC): Results of a phase II
study. Cancer. 109:1384–1390. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Powell EE, Wong VW and Rinella M:
Non-alcoholic fatty liver disease. Lancet. 397:2212–2224. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han SK, Baik SK and Kim MY: Non-alcoholic
fatty liver disease: Definition and subtypes. Clin Mol Hepatol. 29
(Suppl):S5–S16. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu SY, Tsai IT and Hsu YC:
Alcohol-related liver disease: Basic mechanisms and clinical
perspectives. Int J Mol Sci. 22:51702021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Allameh A, Niayesh-Mehr R, Aliarab A,
Sebastiani G and Pantopoulos K: Oxidative stress in liver
pathophysiology and disease. Antioxidants (Basel). 12:16532023.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seitz HK, Bataller R, Cortez-Pinto H, Gao
B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G and Tsukamoto
H: Alcoholic liver disease. Nat Rev Dis Primers. 4:162018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan EE, Hopkins RA, Lim CK, Jamuar SS, Ong
C, Thoon KC, Koh MJ, Shin EM, Lian DW, Weerasooriya M, et al:
Dominant-negative NFKBIA mutation promotes IL-1β production causing
hepatic disease with severe immunodeficiency. J Clin Invest.
130:5817–5832. 2020. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo
X, Zhang J, Ji L, Ren T, An J, et al: Increased mitochondrial
fission promotes autophagy and hepatocellular carcinoma cell
survival through the ROS-modulated coordinated regulation of the
NFKB and TP53 pathways. Autophagy. 12:999–1014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Della-Valle V, Roos-Weil D, Scourzic L,
Mouly E, Aid Z, Darwiche W, Lecluse Y, Damm F, Mémet S, Mercher T,
et al: Nfkbie-deficiency leads to increased susceptibility to
develop B-cell lymphoproliferative disorders in aged mice. Blood
Cancer J. 10:382020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bonato A, Chakraborty S, Bomben R,
Canarutto G, Felician G, Martines C, Zucchetto A, Pozzo F, Vujovikj
M, Polesel J, et al: NFKBIE mutations are selected by the tumor
microenvironment and contribute to immune escape in chronic
lymphocytic leukemia. Leukemia. Mar 15–2024.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li B, Severson E, Pignon JC, Zhao H, Li T,
Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al: Comprehensive
analyses of tumor immunity: Implications for cancer immunotherapy.
Genome Biol. 17:1742016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36((Database Issue)): D480–D484.
2008.PubMed/NCBI
|
|
19
|
Menyhárt O, Nagy A and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shaul YD, Yuan B, Thiru P, Nutter-Upham A,
McCallum S, Lanzkron C, Bell GW and Sabatini DM: MERAV: A tool for
comparing gene expression across human tissues and cell types.
Nucleic Acids Res. 44(D1): D560–D566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Asplund A, Edqvist PHD, Schwenk JM and
Pontén F: Antibodies for profiling the human proteome-the human
protein atlas as a resource for cancer research. Proteomics.
12:2067–2077. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46(D1): D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Kirsch R, Koutrouli M,
Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT,
Pyysalo S, et al: The STRING database in 2023: Protein-protein
association networks and functional enrichment analyses for any
sequenced genome of interest. Nucleic Acids Res. 51(D1): D638–D646.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49(D1): D605–D612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41((Database Issue)): D808–D815. 2013.PubMed/NCBI
|
|
26
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
27
|
Seashore-Ludlow B, Rees MG, Cheah JH,
Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK,
Gould J, et al: Harnessing connectivity in a large-scale
small-molecule sensitivity dataset. Cancer Discov. 5:1210–1223.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rees MG, Seashore-Ludlow B, Cheah JH,
Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL,
Bodycombe NE, et al: Correlating chemical sensitivity and basal
gene expression reveals mechanism of action. Nat Chem Biol.
12:109–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Basu A, Bodycombe NE, Cheah JH, Price EV,
Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al:
An interactive resource to identify cancer genetic and lineage
dependencies targeted by small molecules. Cell. 154:1151–1161.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lian Q, Wang S, Zhang G, Wang D, Luo G,
Tang J, Chen L and Gu J: HCCDB: A database of hepatocellular
carcinoma expression atlas. Genomics Proteomics Bioinformatics.
16:269–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q and
Guo AY: GSCALite: A web server for gene set cancer analysis.
Bioinformatics. 34:3771–3772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roberts A, Trapnell C, Donaghey J, Rinn JL
and Pachter L: Improving RNA-Seq expression estimates by correcting
for fragment bias. Genome Biol. 12:R222011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Anders S and Huber W: Differential
expression of RNA-Seq data at the gene level-the DESeq package.
European Molecular Biology Laboratory (EMBL); 2012
|
|
39
|
The Gene Ontology Consortium: The gene
ontology resource: 20 Years and still GOing strong. Nucleic Acids
Res. 47(D1): D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Global Burden of Disease Liver Cancer
Collaboration, . Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu
MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, et al: The
burden of primary liver cancer and underlying etiologies from 1990
to 2015 at the global, regional, and national level: Results from
the global burden of disease study 2015. JAMA Oncol. 3:1683–1691.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hunter JE, Leslie J and Perkins ND: c-Rel
and its many roles in cancer: An old story with new twists. Br J
Cancer. 114:1–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun SC: The noncanonical NF-κB pathway.
Immunol Rev. 246:125–140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Oeckinghaus A and Ghosh S: The NF-kappaB
family of transcription factors and its regulation. Cold Spring
Harb Perspect Biol. 1:a0000342009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hayden MS and Ghosh S: NF-κB, the first
quarter-century: Remarkable progress and outstanding questions.
Genes Dev. 26:203–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang M, Huang J, Tan X, Bai J, Wang H, Ge
Y, Xiong H, Shi J, Lu W, Lv Z and Liang C: Common polymorphisms in
the NFKBIA gene and cancer susceptibility: A meta-analysis. Med Sci
Monit. 21:3186–3196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Souza SP, Splitt SD, Sànchez-Arcila JC,
Alvarez JA, Wilson JN, Wizzard S, Luo Z, Baumgarth N and Jensen
KDC: Genetic mapping reveals Nfkbid as a central regulator of
humoral immunity to Toxoplasma gondii. PLoS Pathog.
17:e10100812021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du C, Duan X, Yao X, Wan J, Cheng Y, Wang
Y, Yan Y, Zhang L, Zhu L, Ni C, et al: Tumour-derived exosomal
miR-3473b promotes lung tumour cell intrapulmonary colonization by
activating the nuclear factor-κB of local fibroblasts. J Cell Mol
Med. 24:7802–7813. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Arnold CN, Pirie E, Dosenovic P, McInerney
GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB and Beutler
B: A forward genetic screen reveals roles for Nfkbid, Zeb1, and
Ruvbl2 in humoral immunity. Proc Natl Acad Sci USA.
109:12286–12293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Touma M, Keskin DB, Shiroki F, Saito I,
Koyasu S, Reinherz EL and Clayton LK: Impaired B cell development
and function in the absence of IkappaBNS. J Immunol. 187:3942–3952.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mansouri L, Noerenberg D, Young E, Mylonas
E, Abdulla M, Frick M, Asmar F, Ljungström V, Schneider M, Yoshida
K, et al: Frequent NFKBIE deletions are associated with poor
outcome in primary mediastinal B-cell lymphoma. Blood.
128:2666–2670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen T, Li J, Xu M, Zhao Q, Hou Y, Yao L,
Zhong Y, Chou PC, Zhang W, Zhou P and Jiang Y: PKCε phosphorylates
MIIP and promotes colorectal cancer metastasis through inhibition
of RelA deacetylation. Nat Commun. 8:9392017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chiao PJ, Na R, Niu J, Sclabas GM, Dong Q
and Curley SA: Role of Rel/NF-kappaB transcription factors in
apoptosis of human hepatocellular carcinoma cells. Cancer.
95:1696–1705. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wei X, Xu Y, Xu FF, Chaiswing L, Schnell
D, Noel T, Wang C, Chen J, St Clair DK and St Clair WH: RelB
expression determines the differential effects of ascorbic acid in
normal and cancer cells. Cancer Res. 77:1345–1356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Karin M, Lawrence T and Nizet V: Innate
immunity gone awry: Linking microbial infections to chronic
inflammation and cancer. Cell. 124:823–835. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Greten TF, Wang XW and Korangy F: Current
concepts of immune based treatments for patients with HCC: From
basic science to novel treatment approaches. Gut. 64:842–848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Buzzetti E, Pinzani M and Tsochatzis EA:
The multiple-hit pathogenesis of non-alcoholic fatty liver disease
(NAFLD). Metabolism. 65:1038–1048. 2016. View Article : Google Scholar : PubMed/NCBI
|