Introduction
Endometrial carcinoma (EC) is one of the most common
gynecological malignancies in developed countries (1). Notably, the incidence of EC has been
increasing in a number of countries, including the United States,
as well as in Europe and East Asia, which may be due to a greater
exposure to environmental risk factors, such as obesity, increasing
age (≥55 years) and a shift in female reproductive patterns
(2–4). Moreover, the incidence of EC is
estimated to increase by 55% from 2010 to 2030 (5).
Current clinical data have indicated that the
prognosis of EC is closely related to the time of diagnosis, with
an earlier diagnosis associated with a better prognosis
[International Federation of Gynecology and Obstetrics (FIGO) stage
I–II]; for example, the 5-year survival rate has been reported to
decrease from 85% for stage I disease to 25% for stage IV disease
(6–8). Currently, EC is diagnosed by a
combination of transvaginal ultrasound (TVUS) and endometrial
biopsy; however, there is marked heterogeneity in the accuracy of
TVUS for detecting malignancies, as its sensitivity ranges from 0.5
to 0.8 for gynecologic oncological diseases (9). On the other hand, endometrial biopsy
is invasive and uncomfortable for the patient, and pathological
assessment sometimes cannot be carried out due to the failure of
the sampling owing to the pain of the sampling process or problems
of cervical stenosis (10).
Furthermore, the role of test results in guiding personalized
treatment plans requires more research support. There are still
unanswered questions regarding EC, including a number in the
domains of treatment toxicity, diagnostic procedures and adjuvant
therapy (11,12). Therefore, reliable detection of EC
is necessary to ensure adequate treatment and reduce EC-associated
mortality.
With recent advancements in technology, researchers
have focused on developing robust and sensitive detection methods,
such as circulating tumor cell (CTC) detection, as well as methods
involving genomics, epigenomics and transcriptomics (13,14).
CTCs disseminate into the bloodstream from either the primary tumor
site or metastatic sites (15).
Consequently, the number of CTCs is higher in patients with various
types of cancer, such as lung cancer and breast cancer, than in
healthy volunteers (16,17). In a study of the detection of EC
CTCs, aminopeptidase N (CD13) was identified as an alternative
prognostic marker for both cervical and endometrial cancer, as its
expression was detected in patients with EC before surgery and
after recurrence (18).
Long noncoding RNAs (lncRNAs) have an important role
in the epigenetic regulatory network, and can regulate gene
expression and post-transcriptional processes by influencing the
structures of protomers, chromatin and transcription factors
(19). A number of studies have
reported that some lncRNAs affect various hallmarks of human
cancer, such as replicative immortality, antagonism of cell death
and evasion of immunosurveillance (20,21);
therefore, lncRNAs, such as RP4-616B8.5, RP11-389G6.3,
carboxy-terminal domain (CTD)-2377D24.6, AC138904.1 and AC099329.2,
are used as biomarkers in numerous cancer diagnoses (22,23).
Ding et al (24) reported
that the combination of lncRNAs RP4-616B8.5, RP11-389G6.3 and
CTD-2377D24.6 had good performance (P<0.0001) in EC diagnosis.
Xin et al (25) reported
that low RP11-395G23.3 expression was significantly associated with
advanced histological grade and lymphovascular space invasion in
patients with EC, and that RP11-395G23.3 may be a target for the
diagnosis and treatment of EC.
Cytosine methylation of DNA within CpG dinucleotides
is the most well-researched epigenetic alteration in humans
(26). Hypermethylation of the CpG
islands of gene promoters can silence genes, and this is the basis
of the clinical use of a number of biomarkers (27,28).
DNA methylation is a highly stable molecular feature that can be
detected in tumor tissues and cells (29,30).
During malignant transformation, EC cells acquire two main types of
aberrant DNA methylation patterns: Local DNA hypermethylation and
global DNA hypomethylation (31).
Qi et al (32) reported that
hypermethylated cysteine dioxygenase type 1 (CDO1) and CUGBP
Elav-like family member 4 (CELF4) could serve as triage
strategy biomarkers in the non-invasive examination of endometrial
malignant lesions, and the sensitivity and specificity of
CDO1/CELF4 dual-gene methylation assay for
endometrial atypical hyperplasia and endometrial cancer reached
84.9 and 86.6%, respectively. CDO1 and zinc finger protein
454 hypermethylation has also been verified in histological samples
from patients with EC and atypical hyperplasia (AH) compared with
those from patients with benign and normal endometria (P<0.001)
(33).
To increase the efficacy of EC screening, a
combination of major biomarkers, namely, CTCs, lncRNAs
(RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6) and DNA methylation
(CDO1 and CELF4), was evaluated in the present study
to construct a better diagnostic model for EC.
Materials and methods
Specimens
A total of 85 patients, including 71 with EC, and 14
without EC (NO-EC) but with uterine fibroids or polyps, were
enrolled from The First Affiliated Hospital of Soochow University
(Suzhou, China) between March 2023 and March 2024. All enrolled
patients were female; aged 32–84 years (mean age, 57.75 years); and
had provided written informed consent before participation in the
present study, with permission given for sample collection and
analysis. The present study was approved by the Ethics Committee of
The First Affiliated Hospital of Soochow University (approval no.
2021.351). The diagnostic criteria for EC were based on the 2014
World Health Organization Classification of Tumours of the Female
Reproductive Organs. The samples were collected before any
anticancer drug treatment. The specific clinical information of the
subjects is shown in Table I and
the groups are shown in Fig. 1.
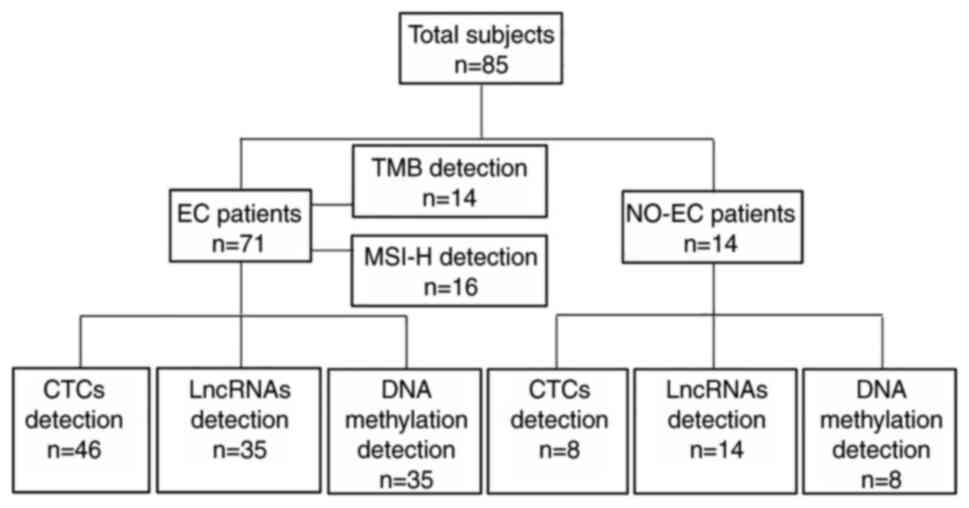 | Figure 1.Patient groups. A total of 85
patients, including 71 patients with EC and 14 NO-EC patients with
uterine fibroids or polyps, were included. A total of 46 patients
with EC and 8 NO-EC patients underwent CTC detection. A total of 35
patients with EC and 14 NO-EC patients underwent RP4-616B8.5,
RP11-389G6.3 and carboxy-terminal domain-2377D24.6 lncRNA
detection. A total of 35 patients with EC and 8 NO-EC patients
underwent cysteine dioxygenase type 1 and CUGBP Elav-like family
member 4 DNA methylation analysis. Of the patients with EC, 14
underwent TMB analysis and 16 underwent MSI-H analysis. CTC,
circulating tumor cell; EC, endometrial carcinoma; lncRNA, long
noncoding RNA; MSI-H, microsatellite instability-high; NO-EC,
without EC; TMB, tumor mutational burden. |
 | Table I.Patient information. |
Table I.
Patient information.
|
| Patients with
ECa (n=71) | NO-ECa patients (n=14) | EC vs. NO-EC |
|---|
|
|
|
|
|
|---|
| Characteristic | Endometrioid
adenocarcinoma (n=55) | Serous
adenocarcinoma (n=9) | Other EC (n=7) | Uterine fibroids
(n=10) | Polyps (n=4) | P-value |
|---|
| Mean ± SD age,
years | 57.80±10.20 | 59.11±8.824 | 57.8±9.822 | 53.70±8.001 | 64.00±10.00 | 0.6448 |
| Hypertension |
|
|
|
|
|
|
|
Yes | 33/55 (60.00%) | 2/9 (22.22%) | 3/7 (42.86%) | 2/10 (20.00%) | 1/4 (25.00%) | 0.0281 |
| No | 22/55 (40.00%) | 7/9 (77.78%) | 4/7 (57.14%) | 8/10 (80.00%) | 3/4 (75.00%) |
|
| Diabetes |
|
|
|
|
|
|
|
Yes | 13/55 (23.64%) | 2/9 (22.22%) | 1/7 (14.29%) | 0/10 (0.00%) | 0/4 (0.00%) | 0.0494 |
| No | 42/55 (76.36%) | 7/9 (77.78%) | 6/7 (85.71%) | 10/10
(100.00%) | 4/4 (100.00%) |
|
| Fatty liver |
|
|
|
|
|
|
|
Yes | 19/55 (34.55%) | 3/9 (33.33%) | 0/7 (0.00%) | 1/10 (10.00%) | 2/4 (50.00%) | 0.4791 |
| No | 36/55 (65.45%) | 6/9 (66.67%) | 7/7 (100.00%) | 9/10 (90.00%) | 2/4 (50.00%) |
|
| LDLa |
|
|
|
|
|
|
| High
(>3.4 mmol/l) | 19/55 (34.55%) | 5/9 (55.56%) | 5/7 (71.43%) | 1/10 (10.00%) | 2/4 (50.00%) | 0.1746 |
| Normal
(≤3.4 mmol/l) | 36/55 (65.45%) | 4/9 (44.44%) | 2/7 (28.57%) | 9/10 (90.00%) | 2/4 (50.00%) |
|
| HDLa |
|
|
|
|
|
|
| Low
(<1.0 mmol/l) | 16/55 (29.09%) | 1/9 (11.11%) | 2/7 (28.57%) | 1/10 (10.00%) | 0/4 (0.00%) | 0.1165 |
| Normal
(≥1.0 mmol/l) | 39/55 (70.91%) | 8/9 (88.89%) | 5/7 (71.43%) | 9/10 (90.00%) | 4/4 (100.00%) |
|
| TAGa |
|
|
|
|
|
|
| High
(>1.7 mmol/l) | 20/55 (36.36%) | 5/9 (55.56%) | 5/7 (71.43%) | 3/10 (30.00%) | 1/4 (25.00%) | 0.8599 |
| Normal
(≤1.7 mmol/l) | 35/55 (63.64%) | 4/9 (44.44%) | 2/7 (28.57%) | 7/10 (70.00%) | 3/4 (75.00%) |
|
| Cholesterol |
|
|
|
|
|
|
| High
(>5.2 mmol/l) | 19/55 (34.55%) | 5/9 (55.56%) | 3/7 (42.86%) | 2/10 (20.00%) | 1/4 (25.00%) | 0.2399 |
| Normal
(≤5.2 mmol/l) | 36/55 (65.45%) | 4/9 (44.44%) | 4/7 (57.14%) | 8/10 (80.00%) | 3/4 (75.00%) |
|
| HE4a |
|
|
|
|
|
|
| High
(>70 pmol/l, before menopause; >140 pmol/l,
post-menopause) | 18/55 (32.73%) | 1/9 (11.11%) | 2/7 (28.57%) | 0/10 (00.00%) | 0/4 (0.00%) | 0.0188 |
| Normal
(≤70 pmol/l, before menopause; ≤140 pmol/l, post-menopause) | 37/55 (67.27%) | 8/9 (88.89%) | 5/7 (71.43%) | 10/10
(100.00%) | 4/4 (100.00%) |
|
| Glucose |
|
|
|
|
|
|
| Normal
(3.9-6.1 mmol/l) | 16/55 (29.09%) | 1/9 (11.11%) | 1/7 (14.29%) | 1/10 (10.00%) | 0/4 (0.00%) | 0.1165 |
| High
(>6.1 mmol/l) | 39/55 (70.91%) | 8/9 (88.89%) | 6/7 (85.71%) | 9/10 (90.00%) | 4/4 (100.00%) |
|
| Number of
pregnancies |
|
|
|
|
|
|
| 0 | 3/55 (5.45%) | 0/9 (0.00%) | 0/7 (0.00%) | 0/10 (0.00%) | 0/4 (0.00%) | 0.4257 |
| 1 | 12/55 (21.82%) | 2/9 (22.22%) | 0/7 (0.00%) | 3/10 (30.00%) | 1/4 (25.00%) |
|
| 2 | 14/55 (25.45%) | 3/9 (33.33%) | 4/7 (57.14%) | 4/10 (40.00%) | 2/4 (50.00%) |
|
| 3 | 12/55 (21.82%) | 3/9 (33.33%) | 1/7 (14.29%) | 1/10 (10.00%) | 0/4 (0.00%) |
|
| ≥4 | 14/55 (25.45%) | 1/9 (11.11%) | 2/7 (28.57%) | 2/10 (20.00%) | 1/4 (25.00%) |
|
| Stage |
|
|
|
|
|
|
| Stage
I | 48/55 (87.27%) | 5/9 (55.56%) | 3/7 (42.86%) |
|
|
|
| Stage
II | 4/55 (7.27%) | 1/9 (11.11%) | 3/7 (42.86%) |
|
|
|
| Stage
III | 3/55 (5.45%) | 1/9 (11.11%) | 1/7 (14.29%) |
|
|
|
| Stage
IV | 0/55 (0.00%) | 2/9 (22.22%) | 0/7 (0.00%) |
|
|
|
| Muscular layer
infiltration depth |
|
|
|
|
|
|
|
<1/2 | 44/55 (80.00%) | 6/9 (66.67%) | 4/7 (57.14%) |
|
|
|
|
≥1/2 | 11/55 (20.00%) | 3/9 (33.33%) | 3/7 (42.86%) |
|
|
|
| Tumor size, cm |
|
|
|
|
|
|
|
<2 | 16/55 (29.09%) | 3/9 (33.33%) | 2/7 (28.57%) |
|
|
|
| ≥2 | 39/55 (70.91%) | 6/9 (66.67%) | 5/7 (71.43%) |
|
|
|
| HPVa |
|
|
|
|
|
|
|
Positive | 6/31 (19.35%) | 4/8 (50.00%) | 2/6 (33.33%) |
| 0/3 (0.00%) | 0.3695 |
|
Negative | 25/31 (80.65%) | 4/8 (50.00%) | 4/6 (66.67%) |
| 3/3 (100.00%) |
|
| CEAa |
|
|
|
|
|
|
| High
(>5 ng/ml, no smoking; >10 ng/ml, smoking) | 1/39 (2.57%) | 0/7 (0.00%) | 0/3 (0.00%) | 0/10 (0.00%) | 1/3 (33.33%) | 0.3131 |
| Normal
(0–5 ng/ml, no smoking; 0–10 ng/ml, smoking) | 38/39 (97.43%) | 7/7 (100.00%) | 3/3 (100.00%) | 10/10
(100.00%) | 2/3 (66.67%) |
|
| CA19-9a |
|
|
|
|
|
|
| High
(>37 U/ml) | 9/51 (17.65%) | 1/8 (12.50%) | 0/6 (0.00%) | 1/10 (10.00%) | 0/3 (0.00%) | 0.4734 |
| Normal
(0–37 U/ml) | 42/51 (82.35%) | 7/8 (87.50%) | 6/6 (100.00%) | 9/10 (90.00%) | 3/3 (100.00%) |
|
| CA125a |
|
|
|
|
|
|
| High
(>35 U/ml) | 9/53 (16.98%) | 2/9 (22.22%) | 0/6 (0.00%) | 2/10 (20.00%) | 0/2 (0.00%) | 0.9667 |
| Normal
(0–35 U/ml) | 44/53 (83.02%) | 7/9 (77.78%) | 6/6 (100.00%) | 8/10 (80.00%) | 2/2 (100.00%) |
|
| ERa |
|
|
|
|
|
|
|
Positive | 44/50 (88.00%) | 7/7 (100.00%) | 4/4 (100.00%) |
|
|
|
|
Negative | 6/50 (12.00%) | 0/7 (0.00%) | 0/4 (0.00%) |
|
|
|
| PRa |
|
|
|
|
|
|
|
Positive | 42/54 (77.78%) | 6/6 (100.00%) | 5/5 (100.00%) |
|
|
|
|
Negative | 12/54 (22.22%) | 0/6 (0.00%) | 0/5 (0.00%) |
|
|
|
| Ki67a |
|
|
|
|
|
|
|
Positive | 55/55
(100.00%) | 7/7 (100.00%) | 5/5 (100.00%) |
|
|
|
|
Negative | 0/55 (0.00%) | 0/7 (0.00%) | 0/5 (0.00%) |
|
|
|
| MSH2a |
|
|
|
|
|
|
|
Positive | 28/28
(100.00%) | 4/4 (100.00%) | 3/3 (100.00%) |
|
|
|
|
Negative | 0/28 (0.00%) | 0/4 (0.00%) | 0/3 (0.00%) |
|
|
|
| CTCa |
|
|
|
|
|
|
|
Positive | 31/36 (86.11%) | 4/8 (50.00%) | 2/2 (100.00%) | 0/8 (0.00%) |
| 0.0098 |
|
Negative | 5/36 (13.89%) | 4/8 (50.00%) | 0/2 (0.00%) | 8/8 (100.00%) |
|
|
| CDO1 DNA
methylationa |
|
|
|
|
|
|
|
Positive | 7/29 (24.14%) | 0/5 (0.00%) | 0/1 (0.00%) | 0/8 (0.00%) |
| 0.1748 |
|
Negative | 22/29 (75.86%) | 5/5 (100.00%) | 1/1 (100.00%) | 8/8 (100.00%) |
|
|
| CELF4 DNA
methylationa |
|
|
|
|
|
|
|
Positive | 2/29 (6.90%) | 0/5 (0.00%) | 0/1 (0.00%) | 0/8 (0.00%) |
| 0.5004 |
|
Negative | 27/29 (93.10%) | 5/5 (100.00%) | 1/1 (100.00%) | 8/8 (100.00%) |
|
|
CTC enrichment and detection
A total of 46 patients with EC and 8 NO-EC patients
underwent CTC detection. Peripheral blood (PB) samples (4
ml/patient) were collected before surgery or treatment, stored in
EDTA tubes (Becton, Dickinson and Company) and CTCs were detected
within 6 h using the CytoBot® 2000 system (Holosensor
Medical Technology Ltd.). Before CTC detection, PB mononuclear
cells (PBMCs) were isolated from the PB. Briefly, 4 ml density
gradient separation solution (Shenzhen DAKEWE Bio-engineering Co.,
Ltd.) and a diluted blood sample (4 ml PB with an equal volume of
phosphate buffer, pH 7.0; Biological Industries) were added
sequentially to a sterile 15-ml centrifuge tube and centrifuged at
700 × g for 20 min at room temperature. The PBMCs were then
carefully pipetted into a new 15-ml centrifuge tube, washed twice
with 5–10 ml PBS (pH 7.2) and centrifuged at 500 × g for 5 min at
25°C.
CTCs were detected using the CytoBot 2000 system, a
novel CTC platform based on advanced technology, including
microfluidics and immunoenrichment. Briefly, CTC capture chips were
manufactured using a metal mesh with pores measuring 15 µm in
diameter, and gold-covered polymers and the purified anti-human
CD326 (Ep-CAM) capture antibody (cat. no. 324202; BioLegend, Inc.)
were seeded onto the surface to form a capture chip with unique
functionality. In the present study, the PBMCs were resuspended in
PBS (pH 7.2) to a volume of 300 µl and were loaded onto the capture
chip. CTCs were captured and stained by the CytoBot®
2000 system using the preset procedures and pre-prepared reagents
from the CTCs detection kit (Holosensor, Inc.).
The immunofluorescence staining was carried out
using the CytoBot 2000 system and the CTCs detection kit
(Holosensor, Inc.), and the indicators used were Alexa
Fluor® 488 anti-pancytokeratin (CK), PE anti-human CD45
antibodies and DAPI. The cell types were determined under a
fluorescence microscope [RX50M; Sunny Optical Technology (Group)
Company Limited]. The evaluation criteria of CTCs was
CK+CD45−DAPI+, and the threshold
for CTC positivity was a
CK+CD45−DAPI+ CTC number of
≥2.
Tissue sample collection, RNA
isolation and reverse transcription-quantitative PCR (RT-qPCR)
analysis
In total, 35 patients with EC and 14 NO-EC patients
underwent RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6 lncRNA
detection. Tumor tissues, paracancerous tissues (at a 1-cm distance
from tumor tissues), uterine fibroid and polyp tissues were
obtained during surgery before treatment. Total RNA was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and RNA was subsequently reverse transcribed into
cDNA with a PrimeScript RT reagent kit (Takara Biotechnology, Ltd.)
according to the manufacturer's protocol. The expression levels of
RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6 lncRNAs were measured
by qPCR using the Hiff qPCR SYBR Green Master Mix (Shanghai Yeasen
Biotechnology Co., Ltd.) and the QuantStudio 6 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The qPCR conditions
were as follows: Pre-denaturation at 95°C for 10 min; followed by
40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for
10 sec and extension at 70°C for 30 sec; and a hold at 95°C for 15
sec, 60°C for 1 min. The positive standard was a Cq value of ≤40.
The expression levels were normalized to the levels of GAPDH
mRNA and were calculated using the ΔCq method (34). The primers used for analysis are
listed in Table II.
 | Table II.Primer sequences used for reverse
transcription-quantitative PCR. |
Table II.
Primer sequences used for reverse
transcription-quantitative PCR.
| Primer | Sequence,
5′-3′ |
|---|
|
CTD-2377D24.6-F |
TTCCGGTGTCCAGATGTTCA |
|
CTD-2377D24.6-R |
AAGGTGAGTTGGGGAGGATG |
| RP4-616B8.5-F |
ATGAGTGTGGCAGCCTATGT |
| RP4-616B8.5-R |
AACTCCTGACCTCGTGATCC |
| RP11-389G6.3-F |
GGCCTTGAGAGATAGAGGGG |
| RP11-389G6.3-R |
ATACGTCCTTCCCATCCTGC |
| GAPDH-F |
GCACAGTCAAGGCTGAGAATG |
| GAPDH-R |
ATGGTGGTGAAGACGCCAGTA |
CDO1 and CELF4 DNA methylation
analysis
In total, 35 patients with EC and 8 NO-EC patients
were included in this analysis. For clinical testing, cervical
epithelial cells and endocervix cells were collected using a
cervical brush or cervical epidermal cell sampler. In this
experiment, the cervical epidermal cell samples were scraped from
subjects with endometrial cell collectors (SAP-I) and were placed
in sample preservation solution (cat. no. AM7020; Thermo Fisher
Scientific, Inc.). Genomic DNA was extracted using the TIANamp
Genomic DNA Kit (cat. no. DP304; Tiangen Biotech Co., Ltd.) and 20
µl DNA eluent was obtained.
A custom-developed bisulfite conversion kit
(methylation detection sample pretreatment kit; Holosensor Medical
Technology Ltd.) was used to convert the extracted DNA into
bisulfite and obtain the transformed bis-DNA. Finally, CDO1
and CELF4 amplification was performed on an ABI 7500 device
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction
mixture consisted of the PCR solution and primer probes, and the
transformed bis-DNA samples were added to the mixture. The reaction
conditions were as follows: Pre-denaturation at 96°C for 5 min,
followed by 45 cycles of denaturation at 94°C for 15 sec and
annealing at 60°C for 35 sec, and a hold at 25°C for 10 min. The
positive standard was a Cq value of ≤38. The primers used for the
analysis are listed in Table
III.
 | Table III.Primer sequences used for DNA
methylation detection. |
Table III.
Primer sequences used for DNA
methylation detection.
| Primer | Sequence,
5′-3′ |
|---|
| CDO1 F |
ATCAACGTTTATATTTTTAAGTTATCG |
| CDO1 R |
GACTTAGACCCTCTACTAATCCG |
| CDO1 FP |
FAM-CATTCTATTTCGGGCGCGGAGATGCGG-BHQ1 |
| CELF4 F |
ATCTCCATGTATATAAAGATGGITACG |
| CELF4 R |
GATATAAGAACTATAACTTAATCCG |
| CELF4
FP |
ROX-ATACCTATAACGGGTTCGGTAGTAGTT-BHQ2 |
Statistical analysis
Statistical analyses, including receiver operating
characteristic (ROC) curve analysis, paired Student's t-test and
unpaired Student's t-test, were performed using GraphPad Prism
10.1.2 software (Dotmatics). P<0.05 was considered to indicate a
statistically significant difference.
Results
Diagnostic value of CTC detection
CTC detection, a classic screening method for
tumors, has been applied effectively in numerous types of cancer
(35–37). In the present study, CTC detection
was used to evaluate patients with EC and NO-EC patients. The
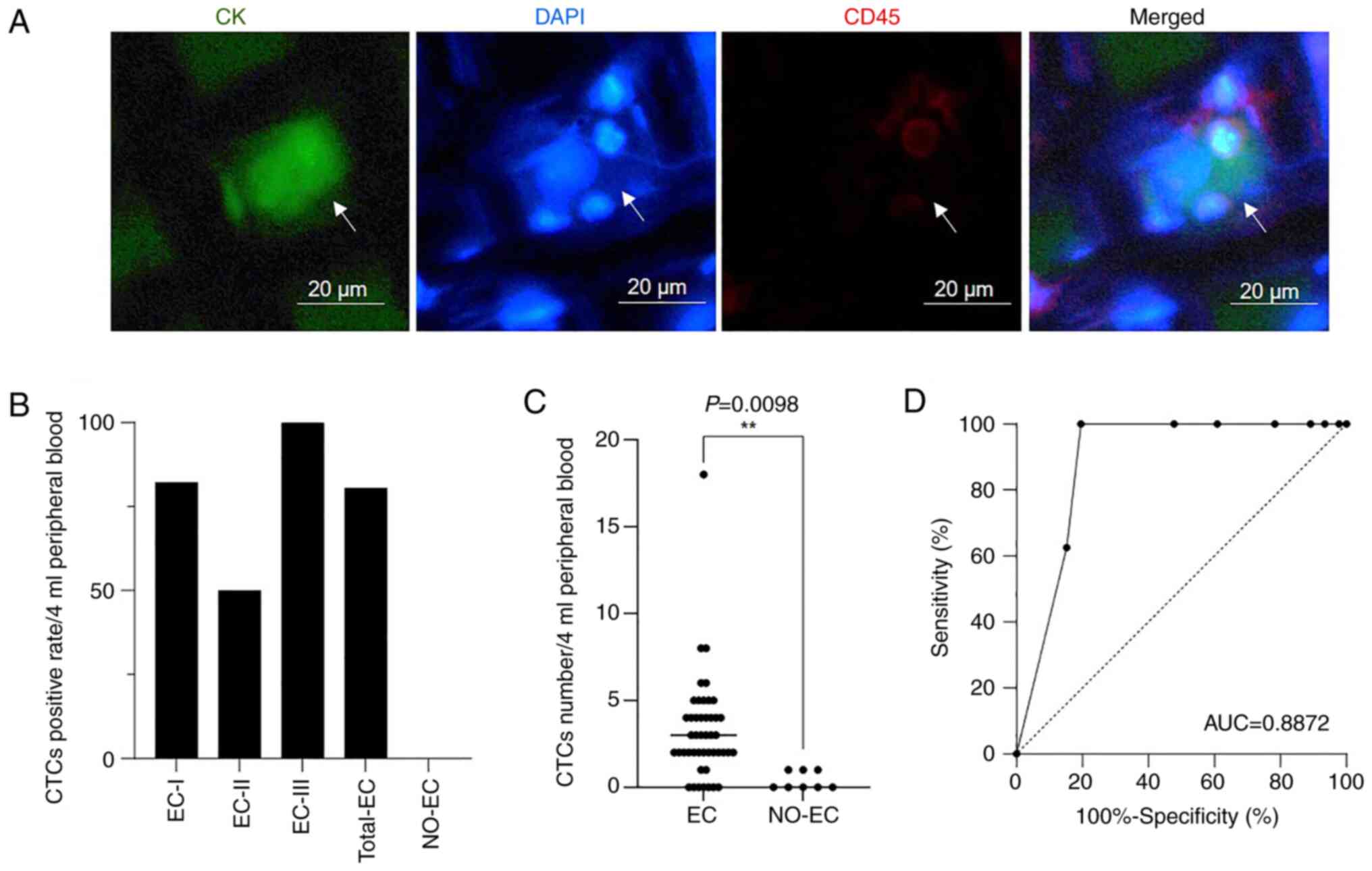
results of CTC enrichment and detection are shown in Fig. 2 and Table I. The classic staining
characteristics of the CTCs were
CK+CD45−DAPI+ (Fig. 2A).
A total of 54 subjects, including 46 patients with
EC and 8 NO-EC patients, underwent CTC detection. The total CTC
positivity rates for all patients with EC, those with stage I EC,
those with stage II EC and those with stage III EC were 80.43%
(37/46), 82.05% (32/39), 50% (2/4) and 100% (3/3), respectively
(Fig. 2B). In the present study, no
patients with stage IV EC underwent CTC detection. Among the 8
NO-EC patients, the CTC positivity rate was 0% (0/8), and the
threshold for CTC-positive patients was a
CK+CD45−DAPI+ CTC number of ≥2.
The number of CTCs was significantly increased in patients with EC
compared with in NO-EC patients (Fig.
2C). In addition, CTCs performed well in distinguishing between
the EC and NO-EC groups, with an area under the curve (AUC) value
of 0.8872 (Fig. 2D). These findings
indicated that CTCs had a good effect on EC diagnosis.
Diagnostic value of lncRNA detection
in EC
Ding et al (24) measured the lncRNAs RP4-616B8.5,
RP11-389G6.3 and CTD-2377D24.6 in clinical samples, and reported
that they had good diagnostic performance regarding histological
subtype (P=0.0001), advanced clinical stage (P=0.011) and clinical
grade (P<0.0001) in patients with EC. The present study
evaluated the lncRNAs RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6
in patients with EC and NO-EC patients by RT-qPCR analysis;
however, the results obtained were different from the results of
the previous study (24). The
expression levels of the RP4-616B8.5, RP11-389G6.3 and
CTD-2377D24.6 lncRNAs were not significantly different between
tumor (n=35) and paracancerous (n=35) tissues according to the
results of RT-qPCR (P=0.2730, 0.0517 and 0.5180, respectively;
Fig. 3A, E and I) and ROC curve
analyses (AUC=0.5380, 0.5747 and 0.5192, respectively; Fig. 3C, G and K). However, the performance
of RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6 in distinguishing
the EC group (n=35) from the NO-EC group (n=14) was good (Fig. 3B, F and J), with AUC values of
0.8184, 0.8347 and 0.8265, respectively (Fig. 3D, H and L).
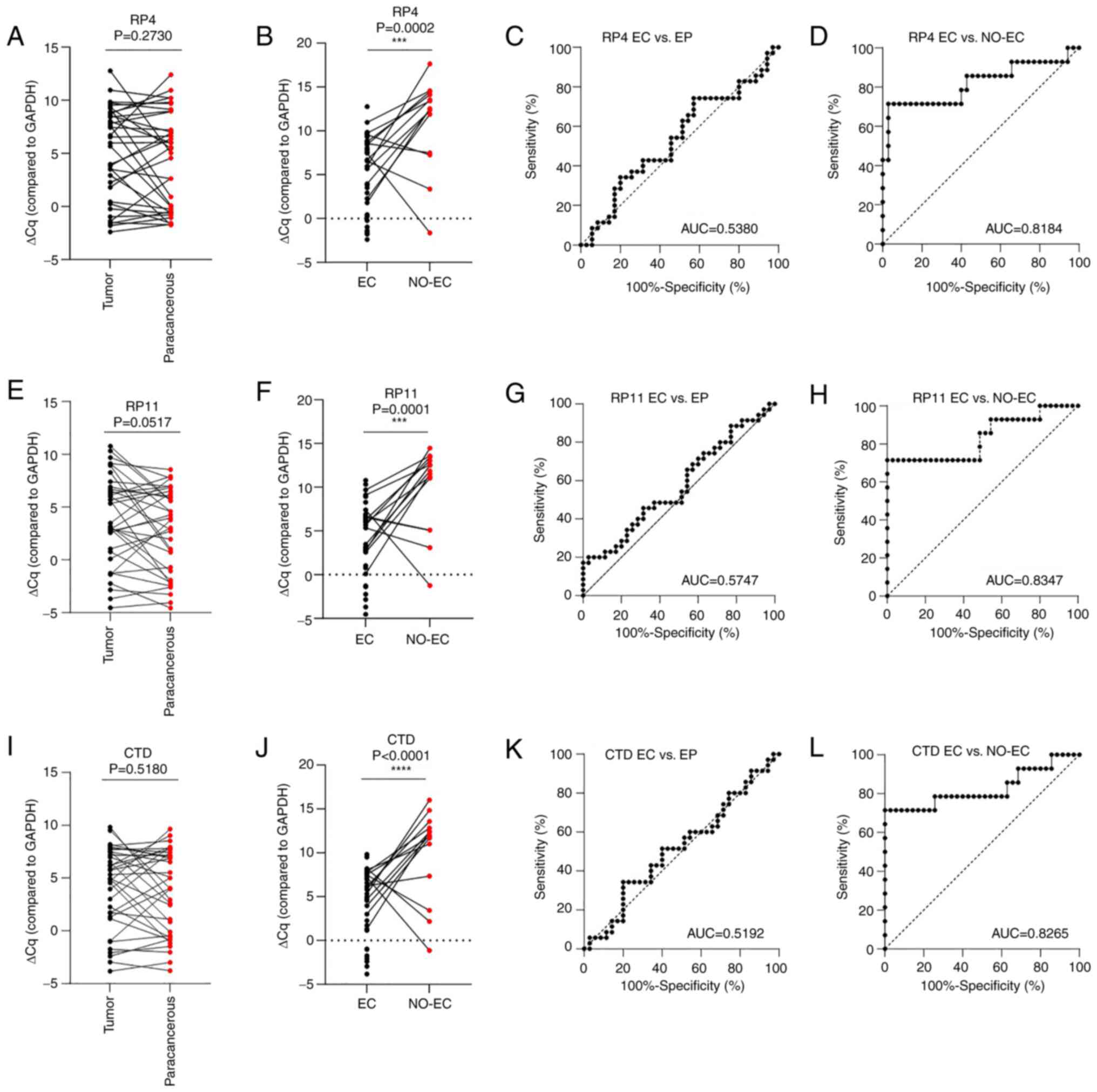 | Figure 3.Diagnostic performance of the
RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6 lncRNAs in EC. ΔCq of
RP4-616B8.5 in (A) tumor (n=35) and paracancerous (n=35) tissues,
and (B) EC (n=35) and NO-EC (n=14) tissues, as determined by
RT-qPCR; GAPDH was used as the reference gene.
***P<0.001. ROC analysis of RP4-616B8.5 lncRNA expression
between (C) tumor (n=35) and paracancerous (n=35) tissues, and (D)
EC (n=35) and NO-EC (n=14) tissues. ΔCq of RP11-389G6.3 lncRNA in
(E) tumor (n=35) and paracancerous (n=35) tissues, and (F) EC
(n=35) and NO-EC (n=14) tissues, as determined by RT-qPCR;
GAPDH was used as the reference gene. ***P<0.001. ROC
curve analysis of RP11-389G6.3 lncRNA expression between (G) tumor
(n=35) and paracancerous (n=35) tissues, and (H) EC (n=35) and
NO-EC (n=14) tissues. ΔCq of CTD-2377D24.6 lncRNA in (I) tumor
(n=35) and paracancerous (n=35) tissues, and (J) EC (n=35) and
NO-EC (n=14) tissues, as determined by RT-qPCR; GAPDH was
used as the reference gene. ****P<0.0001. ROC curve analysis of
CTD-2377D24.6 lncRNA expression between (K) tumor (n=35) and
paracancerous (n=35) tissues, and (L) EC (n=35) and NO-EC (n=14)
tissues. AUC, area under the curve; CTD, carboxy-terminal domain;
EC, endometrial carcinoma; EP, paracancerous tissue; lncRNA, long
noncoding RNA; NO-EC, without EC; ROC, receiver operating
characteristic. |
Diagnostic value of CDO1 and CELF4 DNA
methylation
DNA methylation detection has been widely used in
cancer screening studies (38–40).
Huang et al (41) reported
that a panel comprising any two of the three hypermethylated genes,
BHLHE22, CDO1 and CELF4, reached a sensitivity of
91.8% and specificity of 95.5%. In view of the good performance
that has previously been reported, the present study performed a
CDO1 and CELF4 DNA methylation analysis. CDO1
and CELF4 were detected in 35 EC samples and 8 NO-EC samples
(Fig. 1; Table I). The positive rates of CDO1
and CELF4 methylation were 20% (7/35) and 5.71% (2/35) in
EC, respectively, and these values were lower than those reported
in other studies (32,33,40–42).
CDO1 and CELF4 DNA methylation did not significantly
differ between the EC (n=35) and NO-EC (n=8) groups
(P=0.1748 and 0.5004, respectively; Table I). In addition, the AUC values were
only 0.6000 and 0.5286 for CDO1 and CELF4
methylation, respectively (Fig. 4A and
B).
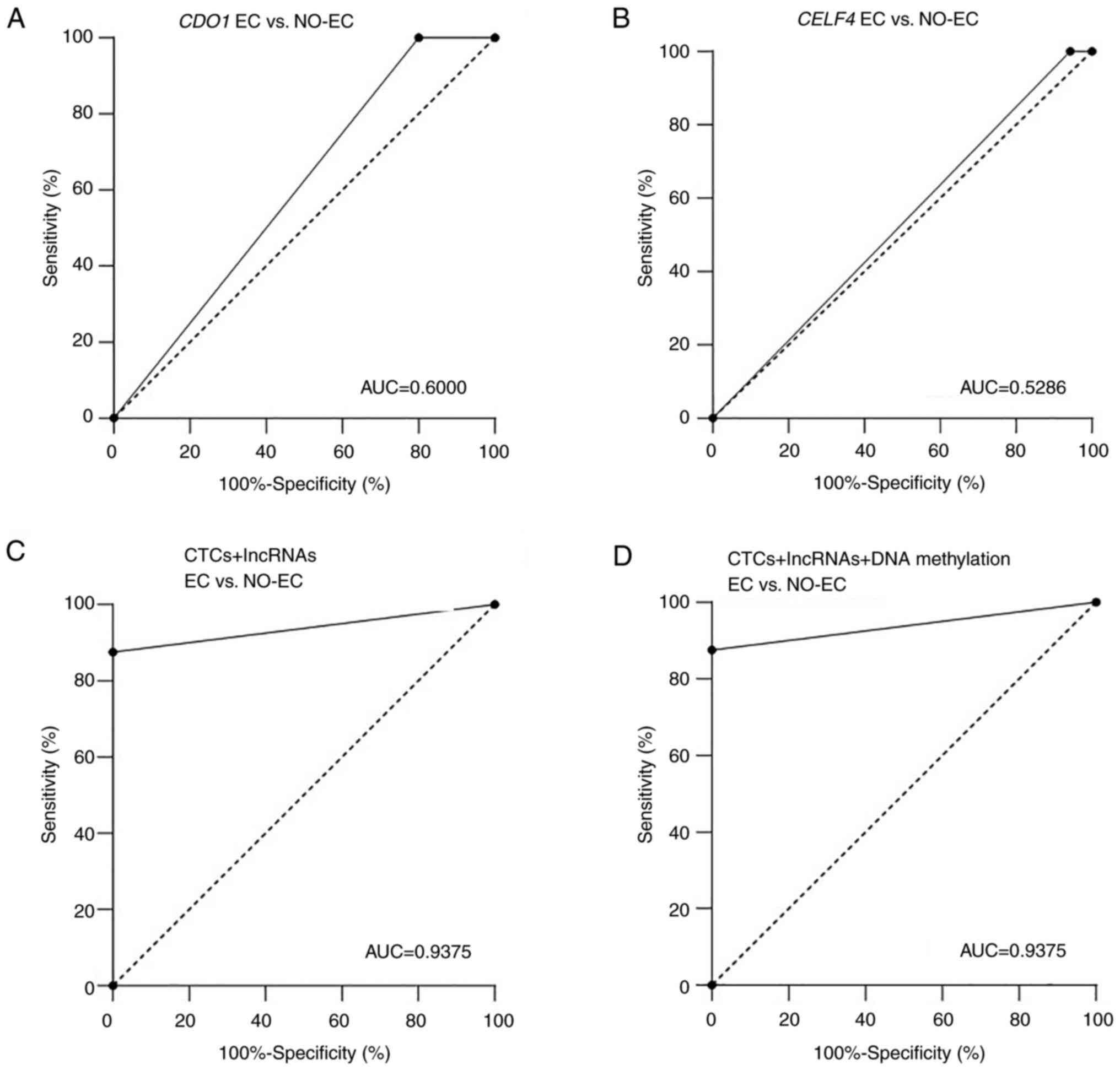 | Figure 4.Diagnostic performance of lncRNAs in
EC. ROC curve analysis of (A) CDO1 and (B) CELF4 DNA
methylation in EC (n=35) and NO-EC (n=8) tissue samples. (C) ROC
curve analysis of CTCs + lncRNAs (RP4, RP11 and CTD) between EC
(n=19) and NO-EC (n=8) samples. (D) ROC curve analysis of CTCs +
DNA methylation (CDO1 and CELF4) + lncRNAs (RP4, RP11
and CTD) between EC (n=13) and NO-EC (n=8) samples. AUC, area under
the curve; CDO1, cysteine dioxygenase type 1; CELF4,
CUGBP Elav-like family member 4; CTD, carboxy-terminal
domain; EC, endometrial carcinoma; lncRNA, long noncoding RNA;
NO-EC, without EC; ROC, receiver operating characteristic. |
To better understand the diagnostic performance of
these biomarkers, CTCs and lncRNAs (RP4-616B8.5, RP11-389G6.3 and
CTD-2377D24.6) were combined, and the AUC value reached 0.9375
(Fig. 4C), thus indicating that
CTCs and these lncRNAs had good performance in distinguishing the
EC and NO-EC groups. When all three groups of tumor markers were
combined, the AUC value was also 0.9375 (Fig. 4D). These results revealed that the
diagnostic performance was not markedly improved after adding
methylated genes.
Associations of microsatellite
instability-high (MSI-H) status or tumor mutational burden (TMB)
with tumor invasiveness and tumor volume
MSI-H and TMB are predictive biomarkers for immune
checkpoint inhibitors (43). MSI is
an indicator of DNA instability and represents a novel cascade in
the carcinogenesis of EC in which MSI mutates hMSH6 (C8), increases
gene instability, and leads to the accumulation of mutations in
other cancer-related genes (44).
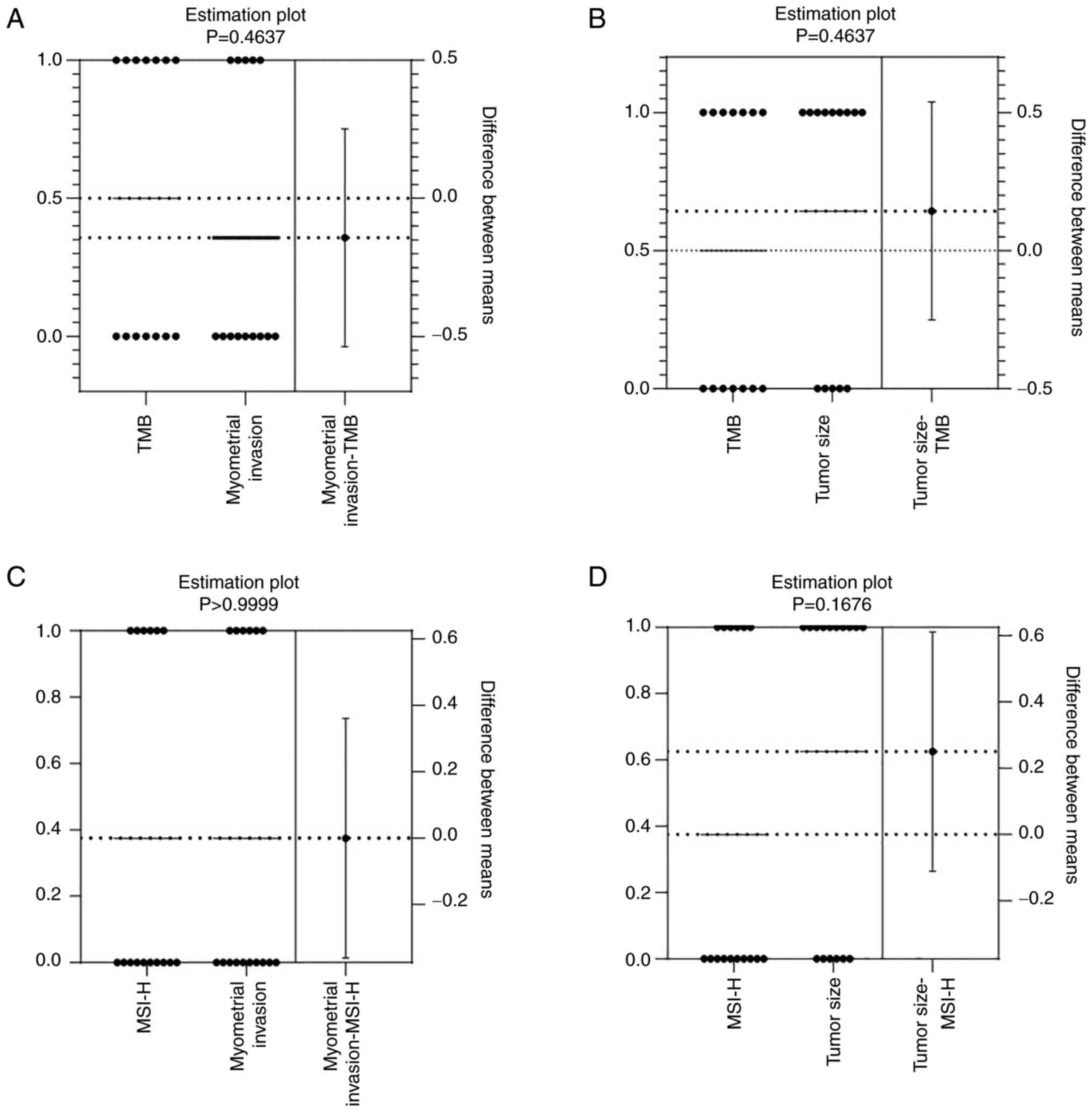
The present study investigated the effects of MSI-H status and TMB
on EC invasion and tumor volume. TMB was detected in 14 patients
with EC, and its association with the depth of muscle infiltration
(<1/2 or ≥1/2) or tumor volume (<2 cm or ≥2 cm) was
evaluated. The results revealed that high TMB was not significantly
correlated with muscle infiltration (P=0.4637; Fig. 5A) or tumor volume (P=0.4637;
Fig. 5B). Moreover, MSI-H status
was detected in 16 patients with EC, and muscle infiltration
(P>0.9999; Fig. 5C) and tumor
volume (P=0.1676; Fig. 5D) were not
significantly correlated with MSI-H status. These findings
indicated that MSI-H status and TMB were not significantly
associated with tumor invasion and tumor volume in EC.
Discussion
In the bloodstream of patients with solid tumors,
the ratio of CTCs to white blood cells has been reported to be
1:106−1:107, thus these cells are considered
quite rare. Even so, the prognostic role of CTCs has been clearly
demonstrated in numerous types of cancer (45). Magbanua et al (46) reported that the CTC trajectory
pattern over the course of treatment was a good predictor of
progression-free survival (PFS) and overall survival (OS). CTC
counts of ≥2 and 5 per 7.5 ml have also been shown to be associated
with reduced PFS and OS in patients with non-small cell lung cancer
(47). Chen et al (48) reported that the CTC test accurately
identified patients who were at a high risk for prostate cancer,
allowing for the early intervention and effective treatment of
patients. There are a number of types of sorting methods for CTCs,
including the dielectrophoretic DLD method (49), the DEPArray™ system (50), emerging microfluidic technologies
(51), dielectrophoretic enrichment
(52) and the negative-selection
enrichment method (48,53,54).
In the present study, CTCs were detected using the
CytoBot® 2000 system, which works based on microscale
meshes with a nanofunctionalized coating that enables the efficient
capture of CTCs (55). The results
revealed that the positive rate for CTCs in patients with EC was
80.43%. The AUC value between EC and NO-EC groups was 0.8872,
indicating that CTC detection had a good screening performance on
EC.
lncRNAs are a class of RNA transcripts that are
>200 nucleotides long (56). It
has been reported that some lncRNAs have specific effects on tumor
screening. For example, risk scores have been obtained for lncRNAs
RP4-792G4.2 and RP11-325122.2 in glioblastoma, and their scores can
be used for risk assessment (57).
Additionally, RP11-54H7.4 is a possible prognostic target for
tongue squamous cell carcinoma (58). Furthermore, the performance of the
three-lncRNA signature comprising RP4-616B8.5, RP11-389G6.3 and
CTD-2377D24.6 has been reported to be higher in EC than in
paracancerous tissue (24). On the
basis of existing studies, the present study explored whether
combining more indicators could improve the performance of a
diagnostic model for EC.
In the present study, the levels RP4-616B8.5,
RP11-389G6.3 and CTD-2377D24.6 were measured in tumor tissues
(n=35) and matched paracancerous tissues (n=35). However, these
three indicators did not significantly distinguish tumor tissue
from paracancerous tissue (P=0.2730, 0.0517 and 0.5180). The
present study further evaluated whether these three indicators were
effective in distinguishing between the EC (n=35) and NO-EC (n=14)
groups. Notably, the performance of these indicators in
differentiating the EC group (n=35) from the NO-EC group (n=14) was
good (P=0.0002, 0.0001 and P<0.0001, respectively). Therefore,
the lncRNAs RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6 may be
suitable for distinguishing between the EC and NO-EC groups.
Qi et al (32) reported that the
CDO1/CELF4 dual-gene methylation assay had high
sensitivity and specificity for AH and EC. Similarly, Krasnyi et
al (42) reported that
CDO1 and CDH13 gene methylation could predict early
EC treatment outcomes. In the present study, methylated CDO1
and CELF4 were used to distinguish the EC group from the
NO-EC group; however, the AUC values were only 0.6000 and 0.5286,
respectively. Notably, when methylated CDO1 and CELF4
were added into the CTCs and lncRNAs panel, these two indicators
could not improve the screening performance. The reason for this
result may be only two methylated genes (CDO1 and
CELF4) were assessed. In addition, due to limited cell
samples, the present study could not simultaneously conduct a
number of molecular biology experiments. In the future, more gene
indicators could be added and next generation sequencing may be
used to improve EC screening performance.
MSI-H status and TMB are cancer-related conditions
(43,44). The present study investigated
whether these two indicators were related to the depth of muscle
infiltration or EC tumor volume; however, the results revealed no
significant correlation.
In conclusion, in the differentiation between EC and
NO-EC groups, the performance of the combined model comprising CTCs
and three lncRNAs (RP4-616B8.5, RP11-389G6.3 and CTD-2377D24.6) was
promising.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Suzhou Science and
Technology Plan Project (grant no. SKY2021035).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HD, YC and JY contributed to the study design, data
analysis and writing of the manuscript. BL, JW and XZ contributed
to the study design and writing of the manuscript. SX and HC
contributed to CTC detection and statistics. JY and XZ provided the
CTC detection instruments and chips. JM and LF contributed to
experimental system verification and DNA methylation detection. JZ
contributed to patient clinical information arrangement and data
analysis. FS and HZ contributed to sample collection, lncRNA qPCR
detection and statistics. HD provided funding. HD and JY confirm
the authenticity of all the raw data. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Soochow University
(approval no. 2021.351). The enrolled patients provided written
informed consent before participation in this study, with
permission for sample collection and analysis.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li W, Xu Y, Zeng X, Tan J, Wang Y, Wu H,
Li M and Yi C: Etiological relationship between lipid metabolism
and endometrial carcinoma. Lipids Health Dis. 22:1162023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kokts-Porietis RL, Elmrayed S, Brenner DR
and Friedenreich CM: Obesity and mortality among endometrial cancer
survivors: A systematic review and meta-analysis. Obes Rev.
22:e133372021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang X, Glubb DM and O'Mara TA: Dietary
factors and endometrial cancer risk: A mendelian randomization
study. Nutrients. 15:6032023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Makker V, MacKay H, Ray-Coquard I, Levine
DA, Westin SN, Aoki D and Oaknin A: Endometrial cancer. Nat Rev Dis
Primers. 7:882021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sheikh MA, Althouse AD, Freese KE, Soisson
S, Edwards RP, Welburn S, Sukumvanich P, Comerci J, Kelley J,
LaPorte RE and Linkov F: USA endometrial cancer projections to
2030: Should we be concerned? Future Oncol. 10:2561–2568. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Van Wijk F, Huikeshoven F, Abdulkadir L,
Ewing P and Burger C: Stage III and IV endometrial cancer: A
20-year review of patients. Int J Gynecol Cancer. 16:1648–1655.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vermij L, Jobsen JJ, León-Castillo A,
Brinkhuis M, Roothaan S, Powell ME, de Boer SM, Khaw P, Mileshkin
LR, Fyles A, et al: Prognostic refinement of NSMP high-risk
endometrial cancers using oestrogen receptor immunohistochemistry.
Br J Cancer. 128:1360–1368. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Crosbie EJ, Kitson SJ, McAlpine JN,
Mukhopadhyay A, Powell ME and Singh N: Endometrial cancer. Lancet.
399:1412–1428. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian Y and Luo H: Diagnostic accuracy of
transvaginal ultrasound examination for local staging of cervical
cancer: A systematic review and meta-analysis. Med Ultrason.
24:348–355. 2022.PubMed/NCBI
|
|
10
|
Elmstrøm-Christensen LB and Lauszus FF:
Diagnostic delay of gynaecological cancer in women with
postmenopausal bleeding. Dan Med J. 69:A092107442022.PubMed/NCBI
|
|
11
|
Oaknin A, Bosse TJ, Creutzberg CL,
Giornelli G, Harter P, Joly F, Lorusso D, Marth C, Makker V, Mirza
MR, et al: Endometrial cancer: ESMO clinical practice guideline for
diagnosis, treatment and follow-up. Ann Oncol. 33:860–877. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tronconi F, Nero C, Giudice E, Salutari V,
Musacchio L, Ricci C, Carbone MV, Ghizzoni V, Perri MT, Camarda F,
et al: Advanced and recurrent endometrial cancer: State of the art
and future perspectives. Crit Rev Oncol Hematol. 180:1038512022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vermij L, Smit V, Nout R and Bosse T:
Incorporation of molecular characteristics into endometrial cancer
management. Histopathology. 76:52–63. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kasius JC, Pijnenborg JMA, Lindemann K,
Forsse D, van Zwol J, Kristensen GB, Krakstad C, Werner HMJ and
Amant F: Risk stratification of endometrial cancer patients: FIGO
stage, biomarkers and molecular classification. Cancers (Basel).
13:58482021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kiss I, Kolostova K, Pawlak I and Bobek V:
Circulating tumor cells in gynaecological malignancies. J BUON.
25:40–50. 2020.PubMed/NCBI
|
|
16
|
Hu X, Zang X and Lv Y: Detection of
circulating tumor cells: Advances and critical concerns. Oncol
Lett. 21:4222021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Castro-Giner F and Aceto N: Tracking
cancer progression: From circulating tumor cells to metastasis.
Genome Med. 12:312020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Law KS, Huang CE and Chen SW: Detection of
circulating tumor cell-related markers in gynecologic cancer using
microfluidic devices: A pilot study. Int J Mol Sci. 24:23002023.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin W, Zhou Q, Wang CQ, Zhu L, Bi C, Zhang
S, Wang X and Jin H: LncRNAs regulate metabolism in cancer. Int J
Biol Sci. 16:1194–1206. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Park EG, Pyo SJ, Cui Y, Yoon SH and Nam
JW: Tumor immune microenvironment lncRNAs. Brief Bioinform.
23:bbab5042022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xing C, Sun SG, Yue ZQ and Bai F: Role of
lncRNA LUCAT1 in cancer. Biomed Pharmacother. 134:1111582021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang G, Sun J and Zhang X: A novel
cuproptosis-related LncRNA signature to predict prognosis in
hepatocellular carcinoma. Sci Rep. 12:113252022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding H, Jiang F, Deng L, Wang J, Wang P,
Ji M, Li J, Shi W, Pei Y, Li J, et al: Prediction of clinical
outcome in endometrial carcinoma based on a 3-lncRNA signature.
Front Cell Dev Biol. 9:8144562021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xin W, Gao X, Zhao S, Zhao P, Yu H, Wu Q
and Hua K: LncRNA RP11-395G23.3 suppresses the endometrial cancer
progression via regulating microRNA-205-5p/PTEN axis. Am J Transl
Res. 12:4422–4433. 2020.PubMed/NCBI
|
|
26
|
Esteller M: Aberrant DNA methylation as a
cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 45:629–656.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Q, Xiong F, Wu G, Liu W, Chen J, Wang
B and Chen Y: Gene body methylation in cancer: Molecular mechanisms
and clinical applications. Clin Epigenetics. 14:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Papanicolau-Sengos A and Aldape K: DNA
methylation profiling: An emerging paradigm for cancer diagnosis.
Annu Rev Pathol. 17:295–321. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jamshidi A, Liu MC, Klein EA, Venn O,
Hubbell E, Beausang JF, Gross S, Melton C, Fields AP, Liu Q, et al:
Evaluation of cell-free DNA approaches for multi-cancer early
detection. Cancer Cell. 40:1537–1549.e12. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Caplakova V, Babusikova E, Blahovcova E,
Balharek T, Zelieskova M and Hatok J: DNA methylation machinery in
the endometrium and endometrial cancer. Anticancer Res.
36:4407–4420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi B, Sun Y, Lv Y, Hu P, Ma Y, Gao W, Li
S, Zhang X, Jin X, Liou Y, et al: Hypermethylated CDO1 and CELF4 in
cytological specimens as triage strategy biomarkers in endometrial
malignant lesions. Front Oncol. 13:12893662023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Dong L, Xu J, Guo L, Wang Y, Wan
K, Jing W, Zhao L, Feng X, Zhang K, et al: Hypermethylated CDO1 and
ZNF454 in cytological specimens as screening biomarkers for
endometrial cancer. Front Oncol. 12:7146632022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lawrence R, Watters M, Davies CR, Pantel K
and Lu YJ: Circulating tumour cells for early detection of
clinically relevant cancer. Nat Rev Clin Oncol. 20:487–500. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao H, Wen L, Li Z and Xia C: Analysis of
diagnostic value of CTC and CTDNA in early lung cancer. Cell Mol
Biol (Noisy-le-grand). 69:57–62. 2023. View Article : Google Scholar
|
|
37
|
Francini S, Duraes M, Rathat G, Macioce V,
Mollevi C, Pages L, Ferrer C, Cayrefourcq L and Alix-Panabières C:
Circulating tumor cell detection by liquid biopsy during
early-stage endometrial cancer surgery: A pilot study.
Biomolecules. 13:4282023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Fan Z, Meng Y, Liu S and Zhan H:
Blood-based DNA methylation signatures in cancer: A systematic
review. Biochim Biophys Acta Mol Basis Dis. 1869:1665832023.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Harada H, Hosoda K, Moriya H, Mieno H, Ema
A, Ushiku H, Washio M, Nishizawa N, Ishii S, Yokota K, et al:
Cancer-specific promoter DNA methylation of cysteine dioxygenase
type 1 (CDO1) gene as an important prognostic biomarker of gastric
cancer. PLoS One. 14:e02148722019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kong LH, Xiao XP, Wan R, Chao XP, Chen XJ,
Wang J, Wu HW and Li L: The role of DNA methylation in the
screening of endometrial cancer in postmenopausal women. Zhonghua
Yi Xue Za Zhi. 103:907–912. 2023.(In Chinese). PubMed/NCBI
|
|
41
|
Huang RL, Su PH, Liao YP, Wu TI, Hsu YT,
Lin WY, Wang HC, Weng YC, Ou YC, Huang TH and Lai HC: Integrated
epigenomics analysis reveals a DNA methylation panel for
endometrial cancer detection using cervical scrapings. Clin Cancer
Res. 23:263–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Krasnyi AM, Gadzhieva LT, Kokoeva DN,
Kosenko MG, Yarotskaya EL, Pavlovich SV, Ashrafyan LA and Sukhikh
GT: Analysis of CDO1, PITX2, and CDH13 gene methylation in early
endometrial cancer for prediction of medical treatment outcomes.
Int J Mol Sci. 25:48922024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Salem ME, Bodor JN, Puccini A, Xiu J,
Goldberg RM, Grothey A, Korn WM, Shields AF, Worrilow WM, Kim ES,
et al: Relationship between MLH1, PMS2, MSH2 and MSH6 gene-specific
alterations and tumor mutational burden in 1057 microsatellite
instability-high solid tumors. Int J Cancer. 147:2948–2956. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kawaguchi M, Banno K, Yanokura M,
Kobayashi Y, Kishimi A, Ogawa S, Kisu I, Nomura H, Hirasawa A,
Susumu N and Aoki D: Analysis of candidate target genes for
mononucleotide repeat mutation in microsatellite instability-high
(MSI-H) endometrial cancer. Int J Oncol. 35:977–982.
2009.PubMed/NCBI
|
|
45
|
Vasseur A, Kiavue N, Bidard FC, Pierga JY
and Cabel L: Clinical utility of circulating tumor cells: An
update. Mol Oncol. 15:1647–1666. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Magbanua MJM, Hendrix LH, Hyslop T, Barry
WT, Winer EP, Hudis C, Toppmeyer D, Carey LA, Partridge AH, Pierga
JY, et al: Serial analysis of circulating tumor cells in metastatic
breast cancer receiving first-line chemotherapy. J Natl Cancer
Inst. 113:443–452. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Krebs MG, Sloane R, Priest L, Lancashire
L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G,
et al: Evaluation and prognostic significance of circulating tumor
cells in patients with non-small-cell lung cancer. J Clin Oncol.
29:1556–1563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen J, Xie T, Yang J, Lin X, Huang L, Su
S and Deng J: Feasibility study of expressing epcam +/vimentin +
CTC in prostate cancer diagnosis. J Cancer Res Clin Oncol.
149:8699–8709. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rahmati M and Chen X: Separation of
circulating tumor cells from blood using dielectrophoretic DLD
manipulation. Biomed Microdevices. 23:492021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Di Trapani M, Manaresi N and Medoro G:
DEPArray™ system: An automatic image-based sorter for
isolation of pure circulating tumor cells. Cytometry A.
93:1260–1266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wei X, Chen K, Guo S, Liu W and Zhao XZ:
Emerging microfluidic technologies for the detection of circulating
tumor cells and fetal nucleated red blood cells. ACS Appl Bio
Mater. 4:1140–1155. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
S Iliescu F, Sim WJ, Heidari H, P Poenar
D, Miao J, Taylor HK and Iliescu C: Highlighting the uniqueness in
dielectrophoretic enrichment of circulating tumor cells.
Electrophoresis. 40:1457–1477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Burr R, Edd JF, Chirn B, Mishra A, Haber
DA, Toner M and Maheswaran S: Negative-selection enrichment of
circulating tumor cells from peripheral blood using the
microfluidic CTC-iChip. Methods Mol Biol. 2471:309–321. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Andree KC, van Dalum G and Terstappen LW:
Challenges in circulating tumor cell detection by the CellSearch
system. Mol Oncol. 10:395–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang J, Dallmann R, Lu R, Yan J and
Charmet J: Flow rate-independent multiscale liquid biopsy for
precision oncology. ACS Sens. 8:1200–1210. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Paul Y, Thomas S, Patil V, Kumar N, Mondal
B, Hegde AS, Arivazhagan A, Santosh V, Mahalingam K and
Somasundaram K: Genetic landscape of long noncoding RNA (lncRNAs)
in glioblastoma: Identification of complex lncRNA regulatory
networks and clinically relevant lncRNAs in glioblastoma.
Oncotarget. 9:29548–29564. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang M, Chen Z, Zhang S, Wu L, Jie Y,
Liao Y, Huang Y, Chen J and Shi B: Analysis of differentially
expressed long non-coding RNAs and the associated TF-mRNA network
in tongue squamous cell carcinoma. Front Oncol. 10:14212020.
View Article : Google Scholar : PubMed/NCBI
|