Introduction
Colorectal cancer (CRC) is a major threat to life,
and is a prevalent disease, with eating habits and lifestyle
patterns contributing to its high incidence rate, which stands at
9.8 cases per 100,000 individuals (1). The onset of CRC is a complex
biological process characterized by various genomic and epigenomic
alterations. The increasing occurrence and poor outcomes of CRC
have prompted extensive scientific research and ongoing trials to
uncover the underlying pathological processes of CRC progression,
halt these processes and prevent further progression (2–4).
Transforming growth factor-β (TGF-β) and E-cadherin
are biomarkers associated with epithelial-mesenchymal transition
(EMT), a process that plays a crucial role in driving cellular
events, resulting in the loss of cell-cell contact and increased
cell motility (5). The TGF-β
superfamily regulates multiple cellular processes, including
migration, apoptosis, proliferation and EMT (6). Paradoxically, TGF-β exhibits both
tumor-suppressive and tumor-promoting effects in cancer, depending
on the molecular and cellular pathways that it influences (7). Although TGF-β signaling pathways can
contribute to tumor progression, their role in carcinogenesis
remains unclear. Cadherins are a key group of adhesion proteins
that are crucial in facilitating cellular interactions by binding
to calcium ions (8). Abnormalities
in E-cadherin molecules have been shown to contribute to the
progression of neoplastic disease in the stomach, pancreas and
large intestine (9).
Sirtuin 1 (SIRT1) is one of the seven isoforms of
the SIRT family, which bind to various histone and non-histone
proteins. The functions of SIRT proteins differ according to their
substrates, with some acting as lysine deacetylases (SIRT1-3, 5, 6
and 7), ADP ribosyl transferases (SIRT4 and 6) and deacetylases
(SIRT5) (10). SIRTs are crucial in
the maintenance of normal cellular balance as they participate in
the regulation of metabolism, autophagy and preservation of genetic
stability (10). SIRTs are involved
in various age-related illnesses, including metabolic syndrome,
cardiovascular disease, neurodegeneration and cancer (11). It is imperative to note that SIRT1
is a multifaceted protein with a pivotal function in multiple
pathways (12). Nevertheless, its
involvement in cancer is yet to be decisively established.
The identification of reliable and non-invasive
biomarkers, such as long chain noncoding ribonucleic acids
(lncRNAs) and microRNAs (miRNAs/miRs) for CRC should facilitate the
early detection of this cancer, and thereby enable prompt
intervention to prevent its progression. miRNAs contribute to the
regulation of gene expression by binding to target mRNA (13). miRNAs regulate the transcripts of
intestinal barrier proteins, which contributes to gastrointestinal
pathologies, and these regulatory roles are associated with
inflammation and colon cancer (14). Regulation of the hypoxia response,
immune cell performance and mesenchymal differentiation have all
been shown to be associated with the expression of miR-130 in CRC
(15). However, knowledge of the
involvement of miR-130 in carcinogenesis is limited.
lncRNAs are noncoding RNA transcripts. They are a
key area of research, as they have been shown to be associated with
carcinogenesis and metastasis in various human cancers, including
breast (16), liver (17) and gastric (18) cancer. Numerous studies have linked
the prognosis of patients with cancer to the expression of specific
lncRNAs. One notable example is HOX transcript antisense intergenic
RNA (HOTAIR), which has been shown to be highly oncogenic in
various malignancies, including breast (16), colon (19) and gastric (20) cancer. HOTAIR is a 2,158-nucleotide
lncRNA located on chromosome 12q13.13 within the homeobox C gene
locus (19). To the best of our
knowledge, the difference in the serum levels of HOTAIR and
miR-130a according to the grade of CRC (low and high) and their
correlation with TGF-β-1, SIRT1 and cadherin levels are unclear.
Therefore, the present study was undertaken to evaluate the
correlations between the serum levels of TGF-β-1, SIRT1 and
E-cadherin and those of HOTAIR and miR-130a in individuals with CRC
in order to explore their associations and diagnostic potential for
CRC.
Materials and methods
Characteristics of participants
In the present retrospective cross-sectional study,
70 patients with pathologically diagnosed CRC and complete clinical
records during the period from October 2023 to May 2024 were
recruited from Fayoum University Hospital (Fayoum, Egypt). The
protocol was approved by the Medical Ethics and Human Clinical
Trial Committee of the Faculty of Medicine, Fayoum University
(approval no. R492; date of approval, September 17, 2023),
following the ethical principles of the Declaration of Helsinki.
Written informed consent was obtained from all subjects prior to
participation in the study; all subjects signed a consent form
after being briefed on the objectives of the study. The range age
of the patients was 37–61 years (mean age, 49.65±11.98 years) and
38.4% of participants were female, while 61.6% were male. The CRC
was present in a variety of locations, including the sigmoid,
ascending, transverse and rectosigmoid colon. All patients were
newly diagnosed with CRC by colonoscopy and confirmed by pathology.
A colonoscopy was recommended for individuals with a positive fecal
occult blood test, hemorrhoids, unexplained abdominal pain or
visible bleeding. After surgery, a definitive pathology diagnosis
and tumor grade were obtained. The CRC was precisely classified
using the World Health Organization classification system (21) into low-grade (well-differentiated to
moderately differentiated) and high-grade (poorly differentiated to
undifferentiated) categories. None of the patients had received
chemotherapy or radiotherapy before the collection of blood
samples. Patients who had a history of secondary or recurring
tumors were excluded from the study. A total of 30 healthy control
participants (mean age, 46.97±9.50 years; 40.4% female and 59.6%
male) who had negative colonoscopy results for malignancy or
inflammatory bowel disease and had no history of familial
adenomatous polyposis or hereditary non-polyposis CRC were also
recruited.
Data and sample collection
Relevant medical history data were collected from
all subjects, including age and body mass index. A 10-ml venous
blood sample was taken from each participant. After allowing the
blood sample to coagulate, serum was extracted by centrifugation at
1,000-2,000 × g for 10 min in a refrigerated centrifuge and frozen
at −70°C for subsequent biochemical and molecular analysis.
Enzyme-linked immunosorbent
assays
Serum E-cadherin (cat. no. DCADE0B), SIRT1 (cat. no.
201-12-2558) and TGF-β1 (cat. no. MBS2501101) were determined using
quantitative sandwich enzyme immunoassay kits from R&D Systems
Europe, Ltd., Shanghai Sun Red Biological Technology Co., Ltd. and
BioSource Europe SA, respectively. The assays were performed
according to the manufacturers' instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
The circulating RNA levels of miRNA-130a and HOTAIR
in the study participants were determined using RT-qPCR. Briefly,
RNA was isolated from the serum samples using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The isolated
RNA was then reverse transcribed using the miScript II RT kit
(Qiagen, Inc.), according to the manufacturer's instructions. The
miScript SYBR® Green PCR Kit (cat. no. 218073; Qiagen
GmbH) was used for qPCR, along with the target-specific miScript
primer assay for miRNA-130a (cat. no. MS00003444) compared with the
reference gene RUN U6B (cat. no. MS00033740). In addition, GAPDH
was used as the reference gene for HOTAIR. The primer sequences
were as follows: miRNA-130a forward, 5′-GTCAGTGCTAAAAGGGCAT-3′ and
reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; and U6 forward,
5′-GCTTCGGCAGCACTATAAT-3′ and reverse, 5′-CGCTTCACGAATTGCTGTCAT-3′;
HOTAIR forward, 5′-GGTAGAAAAAGCAACCACGAAGC-3′, and reverse,
5′-ACATAAACCTCTGTCTGTGAGTGCC-3′; GAPDH forward,
5′-GAAGGTCGGAGTCAACGGATT-3′, and reverse,
5′-CGCTCCTGGAAGATGGTGAT-3′. The Rotor-Gene Q System (Qiagen, Inc.)
was programmed as follows: Heating at 95°C for 10 min, followed by
45 cycles of denaturation at 95°C for 15 sec, and annealing and
extension at 60°C for 60 sec. The data were analyzed using the
2−ΔΔCq method (22).
Statistical analysis
Data are presented as the mean ± standard error or
standard deviation. Differences between two groups were analyzed
using unpaired Student's t-test for continuous data and Chi-square
test for categorical data. One-way ANOVA was used to examine the
differences among multiple groups. When the ANOVA indicated a
significant difference, Tukey's multiple range test was utilized to
conduct pairwise analysis of the groups. The normality assumptions
for each variable were verified using the Shapiro test. The
correlations between variables were evaluated using Pearson's
correlation analysis. The diagnostic value of miR-130a and HOTAIR
was assessed using receiver operating characteristic (ROC) curve
analysis. The analyses were performed using SPSS version 22
software (IBM Corp). P<0.05 was considered to indicate a
statistically significant result.
Results
Patient characteristics
The study included 70 patients with CRC, with an
average age of 49.65±11.98 years. Of these, 40 (57.14%) were
non-obese and the remaining 30 (42.86%) were obese. The most common
presenting symptoms were weight loss, which was exhibited by 40
patients (57.14%), and constipation, which affected 45 patients
(64.30%). Based on colonoscopic findings, the most common locations
of CRC were the sigmoid colon (n=20; 28.57%), rectum (n=15;
21.43%), rectosigmoid region (n=14; 20%) and transverse colon
(n=10; 14.29%). Less commonly, CRC was found in the cecum (n=4;
5.71%) and ascending colon (n=7; 10%) (Table I).
 | Table I.Demographic characteristics of the
study groups. |
Table I.
Demographic characteristics of the
study groups.
| Variables | Healthy
participants (n=30) | Patients with CRC
(n=70) | P-value |
|---|
| Mean age ± SD,
years | 46.97±9.65 | 49.65±11.98 | >0.05 |
| BMI, n (%) |
|
| 0.141 |
|
Non-obese | 22 (73.33) | 40 (57.14) |
|
|
Obese | 8 (26.67) | 30 (42.86) |
|
| Weight loss, n
(%) |
|
|
|
|
Yes |
| 40 (57.14) |
|
| No |
| 30 (42.86) |
|
| Constipation, n
(%) |
|
|
|
|
Yes |
| 45 (64.30) |
|
| No |
| 25 (35.70) |
|
| Location, n
(%) |
|
|
|
| Sigmoid
colon |
| 20 (28.57) |
|
|
Ascending colon |
| 7 (10) |
|
|
Transverse colon |
| 10 (14.29) |
|
|
Rectosigmoid region |
| 14 (20) |
|
|
Rectum |
| 15 (21.43) |
|
|
Cecum |
| 4 (5.71) |
|
Comparison of serum analyte
levels
Regarding the serum levels of TGF-β1 and SIRT1, all
patients with CRC had significantly higher levels than the healthy
individuals (P<0.05). Moreover, patients with high-grade CRC had
significantly higher levels of TGF-β1 than those with low-grade CRC
(P<0.05). Additionally, a notable difference in SIRT1 levels was
observed between the low-grade and high-grade CRC groups (Table II; Fig.
1A and B).
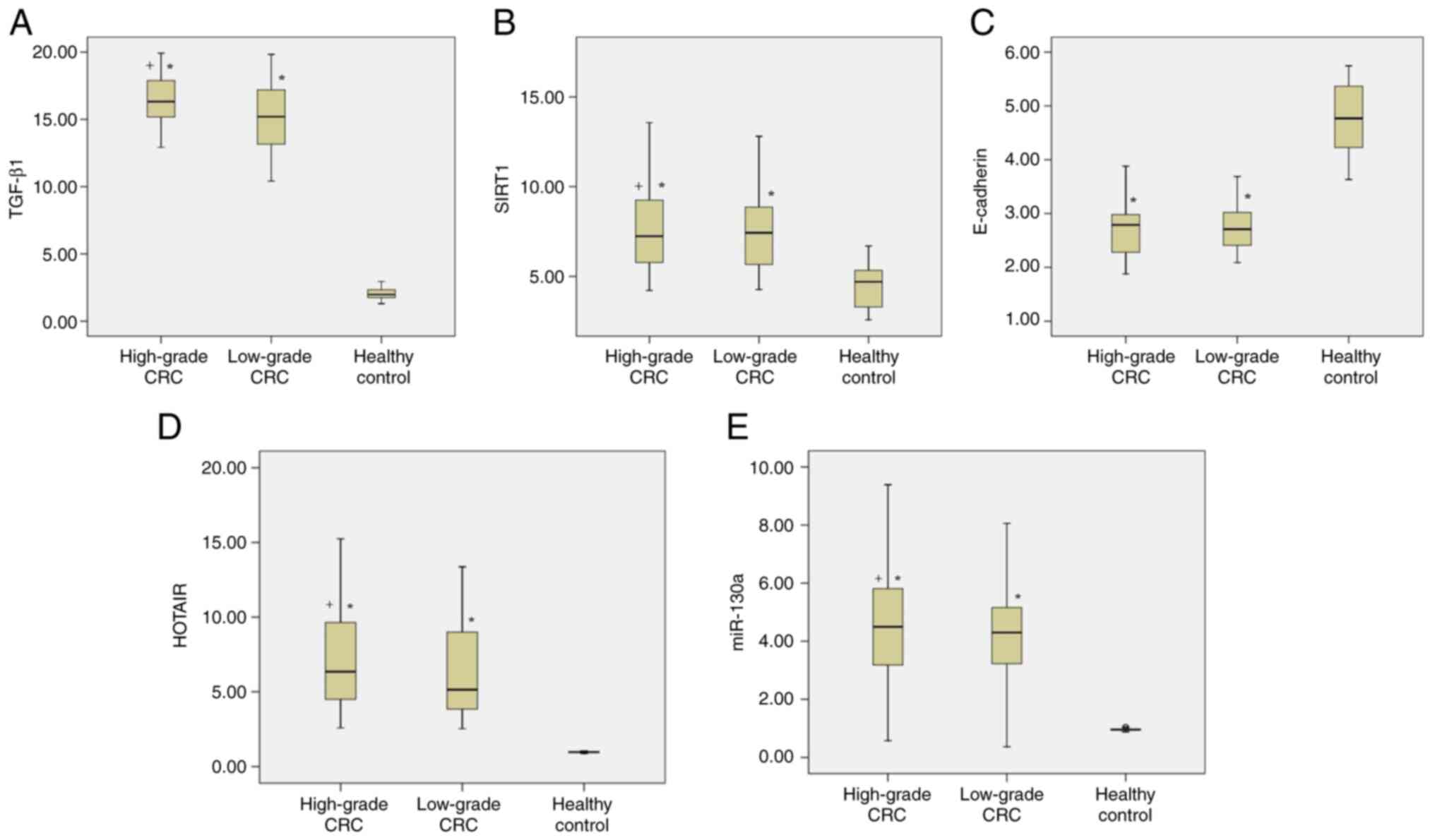 | Figure 1.Comparison of serum levels of TGF-β1,
SIRT1, E-cadherin, HOTAIR and miR-130a among the study
participants. Serum levels of (A) TGF-β1, (B) SIRT1, (C)
E-cadherin, (D) HOTAIR and (E) miR-130a in patients with high-grade
and low-grade CRC and healthy controls. *P<0.05 vs. healthy
controls; +P<0.05 vs. low-grade CRC. TGF-β1,
transforming growth factor-β1; SIRT1, sirtuin 1; HOTAIR, HOX
transcript antisense intergenic RNA; miR, microRNA; CRC, colorectal
cancer. |
 | Table II.Comparisons of serum levels of
TGF-β1, SIRT1, E-cadherin, HOTAIR and miR-130a among the study
participants. |
Table II.
Comparisons of serum levels of
TGF-β1, SIRT1, E-cadherin, HOTAIR and miR-130a among the study
participants.
| Variables | TGF-β1, ng/ml | SIRT1, ng/ml | E-cadherin,
ng/ml | HOTAIR | miR-130a |
|---|
| Healthy
control | 2.10±0.10 | 4.43±0.23 | 4.75±0.11 | 0.98±0.01 | 0.97±0.01 |
| Low-grade CRC |
15.11±0.42a |
7.39±0.40a |
2.77±0.08a |
6.38±0.82a |
4.14±0.38a |
| High-grade CRC |
16.30±0.36a,b |
7.87±0.47a,b |
2.65±0.07a |
7.06±0.53a,b |
4.54±0.34a,b |
| F-ratio | 528.47 | 20.80 | 182.546 | 52.136 | 38.99 |
| P-value | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 |
A clear and significant reduction in the serum
levels of E-cadherin was observed in patients with CRC compared
with those in healthy individuals (P<0.05). However, no
significant difference in serum E-cadherin levels was detected
between patients with low-grade and high-grade CRC (Table II; Fig.
1C).
To determine whether the serum levels of HOTAIR and
miR-130a differ between patients with CRC and control individuals,
RT-qPCR analysis was performed. The serum levels of HOTAIR and
miR-130a in patients with CRC were significantly higher than those
in healthy controls (P<0.05). Furthermore, patients with
high-grade CRC had significantly higher serum HOTAIR and miR-130a
levels compared with those of patients with low-grade CRC
(P<0.05; Table II; Fig. 1D and E).
Association of serum analyte with
lesion location and inter-analyte correlations
No significant association was detected between the
serum levels of TGF-β1, SIRT1, E-cadherin and HOTAIR and the lesion
location. However, a significant association between lesion site
and the serum level of miR-130a was detected. In this regard, a
statistically significant increase in the expression level of
miR-130a was observed in patients with CRC located in the sigmoid,
ascending colon, rectum and cecum compared with that in the
patients with colon tumors located in the transverse colon and
rectosigmoid (P<0.05; Table
III).
 | Table III.Associations between lesion location
and serum levels of TGF-β1, SIRT1, E-cadherin, HOTAIR and miR-130a
in patients with colorectal cancer. |
Table III.
Associations between lesion location
and serum levels of TGF-β1, SIRT1, E-cadherin, HOTAIR and miR-130a
in patients with colorectal cancer.
| Lesion
location | TGF-β1, ng/ml | SIRT1, ng/ml | E-cadherin,
ng/ml | HOTAIR | miR-130a |
|---|
| Sigmoid colon | 16.137±0.536 | 7.493±0.797 | 2.585±0.100 | 5.967±0.521 |
4.39±0.411a,b |
| Ascending
colon | 14.922±0.804 | 7.262±0.694 | 2.652±0.087 | 6.585±1.134 |
6.158±0.984b |
| Transverse
colon | 15.137±0.650 | 7.059±0.536 | 2.866±0.131 | 6.374±0.688 |
3.596±0.534a |
| Rectosigmoid
region | 16.105±0.415 | 7.941±0.773 | 2.798±0.142 | 7.145±1.151 |
3.789±0.767a |
| Rectum | 16.033±0.736 | 8.625±0.588 | 2.676±0.110 | 7.788±1.031 |
4.603±0.534a,b |
| Cecum | 15.557±1.512 | 6.750±0.664 | 2.582±0.162 | 7.480±1.790 |
4.827±0.568a,b |
The correlations among the serum levels of HOTAIR,
miR-130a, TGF-β1, SIRT1 and E-cadherin were evaluated using
Pearson's correlation analysis. Positive correlations were
identified between HOTAIR and miR-130a, TGF-β1 and SIRT1 (r=0.478,
0.738 and 0.455, respectively). However, negative correlations were
observed between E-cadherin and HOTAIR, miR-130a, TGF-β1 and SIRT1
(r=−0.621, −0.592, −0.838 and −0.515, respectively). In addition,
miR-130a was positively correlated with TGF-β1 and SIRT1 (r=0.662
and 0.366, respectively) (Table
IV; Fig. S1, Fig. S2, Fig.
S3).
 | Table IV.Pearson's correlation coefficients
between miR-130a, HOTAIR, TGF-β1, SIRT1 and E-cadherin among CRC
participants. |
Table IV.
Pearson's correlation coefficients
between miR-130a, HOTAIR, TGF-β1, SIRT1 and E-cadherin among CRC
participants.
| Parameters | miR-130a | HOTAIR | TGF-β1 | SIRT1 | E-cadherin |
|---|
| miR-130a | - | 0.478 | 0.662 | 0.366 | −0.592 |
| HOTAIR | 0.478 | - | 0.738 | 0.455 | −0.621 |
| TGF-β1 | 0.662 | 0.738 | - | 0.529 | −0.838 |
| SIRT1 | 0.366 | 0.455 | 0.529 | - | −0.515 |
| E-cadherin | −0.592 | −0.621 | −0.838 | −0.515 | - |
Diagnostic performance
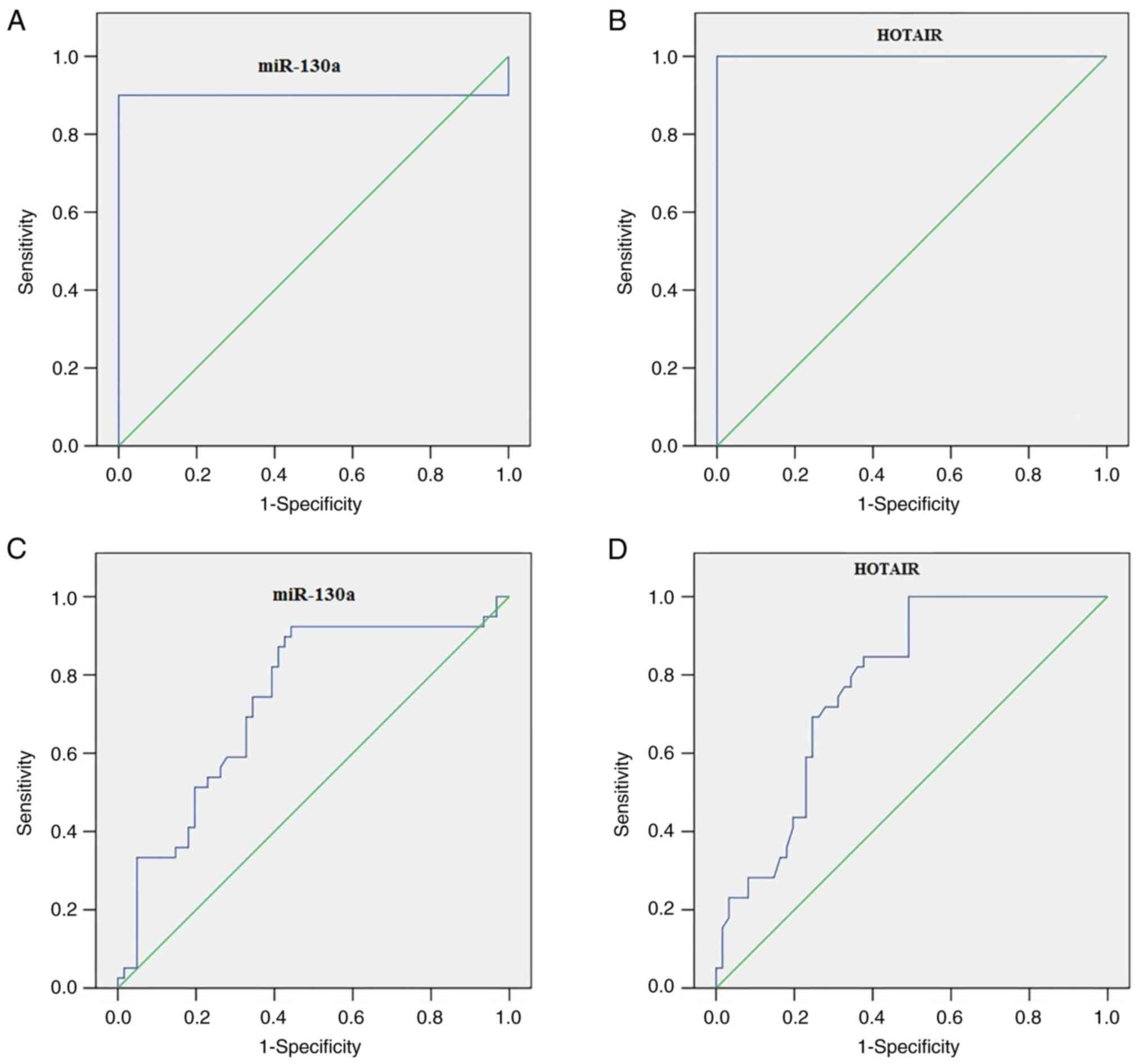
ROC curve analysis revealed that serum miR-130a
differentiated patients with CRC from healthy controls with an
optimum cutoff value of 1.195 [area under the ROC curve (AUC),
0.90; 95% confidence interval (CI), 0.830–0.970; P<0.001], 90%
sensitivity and 100% specificity (Fig.
2A). Furthermore, serum HOTAIR differentiated patients with CRC
from healthy controls with an optimum cutoff value of 1.79 (AUC,
1.00; P<0.001), sensitivity of 100% and specificity of 100%
(Fig. 2B). Serum miR-130a
distinguished patients with high-grade CRC from all other
participants with an optimum cutoff value of 2.41 (AUC, 0.735;
P<0.001), sensitivity of 89.74% and specificity of 57.38%
(Fig. 2C). In addition, serum
HOTAIR distinguished patients with high-grade CRC from all other
participants, with an optimum cutoff value of 2.56 (AUC, 0.682;
P<0.004), sensitivity of 90.77% and specificity of 53.08%
(Fig. 2D).
Discussion
Numerous biological factors have been shown to
contribute to the growth and progression of CRC, both directly and
indirectly (13,23). In our previous study, the potential
of miR-146a and miR-215 as reliable biological markers for
detecting CRC and predicting associated complications was
identified, suggesting that using these miRNAs to target TGF-β and
IL-6 could provide a promising approach for the treatment of CRC
(23).
An intricate mRNA-miRNA-lncRNA network critically
regulates various biological processes and molecular mechanisms in
tumors, with lncRNAs acting as sponges that sequester miRNAs,
thereby modulating miRNA levels and affecting mRNA modulation
(24,25). An intricate mechanism, involving
dysregulation of the mRNA-miRNA-lncRNA network, plays a crucial
role in the regulation of gene transcription and
post-transcriptional translation. This network has prognostic
utility and can be used to identify therapeutic targets (26,27).
The present study aimed to explore the levels of
HOTAIR and miR-130a in the serum of patients with CRC and evaluate
their correlation with the serum levels of TGF-β1, SIRT1 and
E-cadherin. Using RT-qPCR, it was identified that patients with CRC
had significantly increased serum levels of HOTAIR and miR-130a, in
addition to significantly elevated serum levels of TGF-β1 and
SIRT1, and significantly reduced serum levels of E-cadherin
compared with those of healthy individuals. Additionally, the
levels of miR-130a and HOTAIR increased with the grade of CRC.
These findings suggest that miR-130a and HOTAIR could potentially
serve as reliable biomarkers for detecting and predicting the
outcomes of CRC. Previous studies have consistently reported that
miR-130 and HOTAIR have oncogenic effects in CRC (26–30).
Zhang et al (31) detected a substantial association
between the prognosis of patients with breast cancer after systemic
treatment and the differential expression of miR-130a/HOTAIR in
these patients. Furthermore, the oncogenic nature of HOTAIR
promotes cell migration and invasion while suppressing apoptosis;
HOTAIR has also been identified as a strong predictor of metastasis
and mortality for numerous types of cancers, including prostate
cancer (32), lung cancer (33) and breast cancer (16).
Regarding miR-130, a number of studies have shown
that it is involved in the pathogenesis of various tumors,
including ovarian (34), esophageal
(35), liver (36) and stomach (37) cancer. Additionally, Wang et
al (38) demonstrated that
circulating miR-130a levels are upregulated in patients with
high-grade bladder cancer and significantly correlated with tumor
stage.
In CRC, persistent TGF-β expression is primarily
associated with the advanced stages of the disease (39). The present research revealed that
the plasma levels of SIRT1 and TGF-β1 were increased in patients
with higher grades of CRC compared with those with lower grades.
This increase in plasma level is likely to be related to tumor
progression and the associated oncogenic activity. The upregulation
of SIRT1 and TGF-β accelerates tumor growth and metastasis while
preventing immune surveillance (40,41).
Previous studies have shown that the expression of SIRT1 and TGF-β
is significantly increased in CRC, suggesting the oncogenic roles
of these factors in CRC progression (23,42).
The present study revealed that the serum levels of
E-cadherin are lower in patients with CRC than in healthy
individuals. This finding aligns with a study by Hydru and Das
(43), who reported a
downregulation in cadherin expression levels in the tumor tissues
of patients with CRC, and suggested that this reduction could be
used as a diagnostic biomarker to track the progression of the
disease and predict the invasiveness and migration the tumor cells.
The downregulation of E-cadherin is a key factor in EMT, which has
been linked to invasiveness in various types of cancer, such as
pancreatic cancer (44).
In the present study, the relationships between the
serum levels of TGF-β1, SIRT1 and E-cadherin and those of the
HOTAIR/miR-130a were investigated. Pearson's correlation analysis
revealed significant positive correlations between serum HOTAIR
levels and those of miR-130a and TGF-β1. Notable positive
correlations were also observed among the serum levels of TGF-β1,
SIRT and miR-130. Therefore, it is hypothesized that TGF-β
upregulates SIRT1, which then induces changes in E-cadherin
expression. This is supported by previous studies by Carafa et
al (45) and Palmirotta et
al (46) in which it is
reported that TGF-β upregulates SIRT1, which interacts with other
transcription factors, thereby leading to the downregulation of
E-cadherin, and promoting the migration, invasion and death
resistance of cancer cells. In addition, the present study found a
negative correlation between HOTAIR and E-cadherin levels,
suggesting a potential contribution of HOTAIR to EMT, due to the
downregulation of E-cadherin and associated promotion of CRC cell
migration and invasiveness.
The ROC curve analysis performed in the present
study indicate that miR-130a and HOTAIR have good sensitivity and
specificity as biomarkers for the discrimination of patients with
CRC from healthy study participants. Notably, these results are
consistent with a previous study by Wang et al (38), in which serum levels of miR-130 were
identified as a potential biomarker for distinguishing patients
with bladder cancer from healthy individuals.
To the best of our knowledge, the present study is
the first to suggest correlations of HOTAIR/miR-130 with TGF-β1,
SIRT1 and E-cadherin. The miR-130a/HOTAIR and
TGF-β1/SIRT1/E-cadherin axis may serve as a novel biomarker for the
early diagnosis of CRC. However, the main limitation of the study
is the small sample size, which is due to the lack of financial
support and funding. Therefore, future large-scale studies and
clinical trials are necessary to establish the relationships of
HOTAIR, miR-130a, TGF-β1, SIRT1 and E-cadherin with their
therapeutic effects in clinical settings.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Zarqa University, Jordan provided partial funding for this
study.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
OGS, GA and NAH contributed to the conception and
design of the study. Material preparation, data collection and
analysis were performed by BMB, KD, TIA, EAH, RAN, SG and NAH. OGS
and NAH confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The study was conducted in compliance with the
Declaration of Helsinki, and was approved by Medical Ethics and
Human Clinical Trial Committee of the Faculty of Medicine, Fayoum
University (approval no. R492; date of approval, September 17,
2023), following the ethical principles of the Declaration of
Helsinki. Written informed consent was obtained from all subjects
prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sharma R, Abbasi-Kangevari M, Abd-Rabu R,
Abidi H, Abu-Gharbieh E, Acuna JM, Adhikari S, Advani SM, Afzal MS,
Aghaie Meybodi M, et al: Global, regional, and national burden of
colorectal cancer and its risk factors, 1990–2019: A systematic
analysis for the global burden of disease study 2019. Lancet
Gastroenterol Hepatol. 7:627–647. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goïta AA and Guenot D: Colorectal cancer:
The contribution of CXCL12 and its receptors CXCR4 and CXCR7.
Cancers (Basel). 14:18102022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kumar A, Gautam V, Sandhu A, Rawat K,
Sharma A and Saha L: Current and emerging therapeutic approaches
for colorectal cancer: A comprehensive review. World J Gastrointest
Surg. 15:495–519. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Q, Geng S, Luo H, Wang W, Mo YQ, Luo Q,
Wang L, Song GB, Sheng JP and Xu B: Signaling pathways involved in
colorectal cancer: Pathogenesis and targeted therapy. Signal
Transduct Target Ther. 9:2662024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jayanthi P, Varun BR and Selvaraj J:
Epithelial-mesenchymal transition in oral squamous cell carcinoma:
An insight into molecular mechanisms and clinical implications. J
Oral Maxillofac Pathol. 24:1892020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xue VW, Chung JYF, Córdoba CAG, Cheung AH,
Kang W, Lam EW, Leung KT, To KF, Lan HY and Tang PM: Transforming
growth factor-β: A multifunctional regulator of cancer immunity.
Cancers (Basel). 12:30992020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baba AB, Rah B, Bhat GR, Mushtaq I,
Parveen S, Hassan R, Hameed Zargar M and Afroze D: Transforming
growth factor-beta (TGF-β) signaling in cancer-a betrayal within.
Front Pharmacol. 13:7912722022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jaiganesh A, Narui Y, Araya-Secchi R and
Sotomayor M: Beyond cell-cell adhesion: Sensational cadherins for
hearing and balance. Cold Spring Harb Perspect Biol.
10:a0292802018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaszak I, Witkowska-Piłaszewicz O,
Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F and Jurka P: Role
of cadherins in cancer-a review. Int J Mol Sci. 21:76242020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu QJ, Zhang TN, Chen HH, Yu XF, Lv JL,
Liu YY, Liu YS, Zheng G, Zhao JQ, Wei YF, et al: The sirtuin family
in health and disease. Signal Transduct Target Ther. 7:4022022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao L, Cao J, Hu K, He X, Yun D, Tong T
and Han L: Sirtuins and their biological relevance in aging and
age-related diseases. Aging Dis. 11:927–945. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee GJ, Jung YH, Kim TJ, Chong Y, Jeong
SW, Lee IK and Woo IS: Surtuin 1 as a potential prognostic
biomarker in very elderly patients with colorectal cancer. Korean J
Intern Med. 36 (Suppl 1):S235–S244. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khalil EH, Shaker OG and Hasona NA: Impact
of rs2107425 polymorphism and expression of lncH19 and miR-200a on
the susceptibility of colorectal cancer. Indian J Clin Biochem.
38:331–337. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khalil EH, Shaker OG and Hasona NA: lncRNA
H-19 and miR-200a implication and frequency of lncRNA H-19
rs2170425 SNP in ulcerative colitis and Crohn's disease. Comp Clin
Pathol. 32:565–571. 2023. View Article : Google Scholar
|
|
15
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1086–1099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khaliefa AK, Desouky EM, Hozayen WG,
Shaaban SM and Hasona NA: miRNA-1246, HOTAIR, and IL-39 signature
as potential diagnostic biomarkers in breast cancer. Noncoding RNA
Res. 8:205–210. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
DiStefano JK and Gerhard GS: Long
noncoding RNAs and human liver disease. Annu Rev Pathol. 17:1–21.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao Y, Wang JW, Ren JY, Guo M, Guo CW,
Ning SW and Yu S: Long noncoding RNAs in gastric cancer: From
molecular dissection to clinical application. World J
Gastroenterol. 26:3401–3412. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen S, Zhang C and Feng M: Prognostic
Value of LncRNA HOTAIR in colorectal cancer: A meta-analysis. Open
Med (Wars). 15:76–83. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Qiu WQ, Zhu H, Liu H, Sun JH,
Chen Y, Shen H, Qian CL and Shen ZY: HOTAIR contributes to the
carcinogenesis of gastric cancer via modulating cellular and
exosomal miRNAs level. Cell Death Dis. 11:7802020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jass JR, Sobin LH and Watanabe H: The
World Health Organization's histologic classification of
gastrointestinal tumors. A commentary on the second edition.
Cancer. 66:2162–2167. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ayeldeen G, Shaker OG, Khairy AM, Elfert
AY and Hasona NA: Signature of micro RNA 146a/215 and IL-6/TGF-β
levels in a cross-link axis between obesity and colorectal cancer.
Noncoding RNA Res. 8:187–191. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abdel Hameed NA, Shaker OG and Hasona NA:
Significance of LINC00641 and miR-378 as a potential biomarker for
colorectal cancer. Comp Clin Pathol. 31:807–814. 2022. View Article : Google Scholar
|
|
25
|
Ratti M, Lampis A, Ghidini M, Salati M,
Mirchev MB, Valeri N and Hahne JC: MicroRNAs (miRNAs) and long
non-coding RNAs (lncRNAs) as new tools for cancer therapy: First
steps from bench to bedside. Target Oncol. 15:261–278. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu
J, Yang H, Chen Q, Chen M, Ye L, et al: Potential regulatory role
of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother.
121:1096272020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shengnan J, Dafei X, Hua J, Sunfu F,
Xiaowei W and Liang X: Long non-coding RNA HOTAIR as a competitive
endogenous RNA to sponge miR-206 to promote colorectal cancer
progression by activating CCL2. J Cancer. 11:4431–4441. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen W, Tong K and Yu J: MicroRNA-130a is
upregulated in colorectal cancer and promotes cell growth and
motility by directly targeting forkhead box F2. Mol Med Rep.
16:5241–5248. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu L, Nie J, Chen L, Dong G, Du X, Wu X,
Tang Y and Han W: The oncogenic role of microRNA-130a/301a/454 in
human colorectal cancer via targeting Smad4 expression. PLoS One.
8:e555322013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang M, Wu K, Zhang P, Qiu Y, Bai F and
Chen H: HOTAIR Facilitates endocrine resistance in breast cancer
through ESR1/miR-130b-3p axis: Comprehensive analysis of
mRNA-miRNA-lncRNA network. Int J Gen Med. 14:4653–4663. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Liu N, Gao Y, Quan Z, Hao Y, Yu C,
Li L, Yuan M, Niu L, Luo C and Wu X: Long noncoding RNA HOTAIR
regulates the invasion and metastasis of prostate cancer by
targeting hepaCAM. Br J Cancer. 124:247–258. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ren MM, Xu S, Wei YB, Yang JJ, Yang YN,
Sun SS, Li YJ, Wang PY and Xie SY: Roles of HOTAIR in lung cancer
susceptibility and prognosis. Mol Genet Genomic Med. 8:e12992020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Huang L, Zhao Y and Tan W:
Downregulation of miR-130a contributes to cisplatin resistance in
ovarian cancer cells by targeting X-linked inhibitor of apoptosis
(XIAP) directly. Acta Biochim Biophys Sin (Shanghai). 45:995–1001.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Ji F, Liu G, Wang W, Li Z, Yue Y
and Wang Z: Upregulation of circulating miR130a is correlated with
development of Barrett's esophagus and esophageal adenocarcinoma.
Onco Targets Ther. 12:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
El-Naidany SS, Zid E, Reda FM, Nada A and
Fouda EAM: Clinical significance of MiR-130b and MiR-125b as
biomarkers in hepatocellular carcinoma. Asian Pac J Cancer Prev.
23:2687–2693. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu W, Zheng X, Liu J, Zhang M, Liang Y and
Song M: MicroRNA MiR-130a-3p promotes gastric cancer by targeting
Glucosaminyl N-acetyl transferase 4 (GCNT4) to regulate the
TGF-β1/SMAD3 pathway. Bioengineered. 12:11634–11647. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, Zhao L, Peng X, Liu K, Zhang C,
Chen X, Han Y and Lai Y: Evaluation of miR-130 family members as
circulating biomarkers for the diagnosis of bladder cancer. J Clin
Lab Anal. 34:e235172020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Waldner MJ and Neurath MF: TGFβ and the
tumor microenvironment in colorectal cancer. Cells. 12:11392023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chan MKK, Chan ELY, Ji ZZ, Chan ASW, Li C,
Leung KT, To KF and Tang PMK: Transforming growth factor-β
signaling: From tumor microenvironment to anticancer therapy.
Explor Target Antitumor Ther. 4:316–343. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khalil M, Desouky EM, Khaliefa AK, Hozyen
WG, Mohamed SS and Hasona NA: Insights into the crosstalk between
miR-200a/lncRNA H-19 and IL-6/SIRT-1 axis in breast cancer. J
Interferon Cytokine Res. 44:191–197. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu DF, Jiang SJ, Pan ZP, Cheng WD, Zhang
WJ, Yao XK, Li YC and Lun YZ: Expression and clinical significance
of Sirt1 in colorectal cancer. Oncol Lett. 11:1167–1172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hydru SP and Das NM: Expression of
E-cadherin in colorectal cancer and its association with
morphological features. J Evolution Med Dent Sci. 11:163–168. 2022.
View Article : Google Scholar
|
|
44
|
Nagathihalli NS and Merchant NB:
Src-mediated regulation of E-cadherin and EMT in pancreatic cancer.
Front Biosci (Landmark Ed). 17:2059–2069. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Carafa V, Altucci L and Nebbioso A: Dual
tumor suppressor and tumor promoter action of sirtuins in
determining malignant phenotype. Front Pharmacol. 9:4166872019.
|
|
46
|
Palmirotta R, Cives M, Della-Morte D,
Capuani B, Lauro D, Guadagni F and Silvestris F: Sirtuins and
cancer: Role in the epithelial-mesenchymal transition. Oxid Med
Cell Longev. 2016:30314592016. View Article : Google Scholar : PubMed/NCBI
|