Introduction
Non-small cell lung cancer is the most common
histological type of lung cancer, and the main histological
classification of non-small cell lung cancer is lung adenocarcinoma
(1). According to Global Cancer
Statistics data published in 2020, lung cancer continues to pose a
significant threat to human health as one of the most perilous
malignant tumors, with an estimated 2.2 million new cases, and one
of the leading causes of cancer-related mortality, with an
estimated 1.8 million associated deaths (2). Typically, lung cancer has a poor
prognosis. With the diverse treatment strategies now available, the
survival prospects of individuals with advanced lung cancer have
improved. However, despite these advancements, the overall 5-year
survival rate remains at <20% (3–5). The
primary factors contributing to this low survival rate include the
fact that early symptoms that often go unnoticed, leading to a
postponed diagnosis (6,7), and that, following surgery, lymphatic
and distant metastases are common, with 34–45% of patients
experiencing recurrence (8). In
addition, although targeted therapy and immunotherapy have shown
significant effectiveness and made some progress in lung cancer
therapeutics, the available options remain limited (9). Therefore, the identification of
molecular targets for lung cancer is vital.
Various bioinformatics analyses have been conducted
to identify novel and potential markers that can detect cancers in
their early stages, predict prognosis or serve as therapeutic
targets (10,11). For example, Ma et al
(11) reported that RUNX family
transcription factor 1 is pivotal to non-small cell lung cancer by
using The Cancer Genome Atlas (https://portal.gdc.cancer.gov/), Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/), Gene Expression
Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) and Kaplan-Meier (KM)
plotter databases (https://kmplot.com/analysis/). Jiang et al
(12) utilized bioinformatic
approaches and found that chromatin licensing and DNA replication
factor 1 could predict the prognosis of human lung adenocarcinoma.
Furthermore, Gong et al (13) identified the therapeutic targets of
aspirin in small cell lung cancer using bioinformatics
analysis.
Pulmonary nodules are the main manifestations of
early lung cancer (14). In the
present study, the National Center for Biotechnology Information
Gene Expression Omnibus (NCBI GEO) GSE135304 dataset was
downloaded, which contains sequencing data of whole blood from
patients with malignant nodules and benign nodules, and nodule-free
patients, and analyzed differentially expressed genes (DEGs) to
identify key genes involved in lung cancer. The associations
between key genes and lung cancer prognosis were verified using
online KM Plotter. Functions and mechanisms of lung
cancer-associated genes were also investigated using molecular
experiments.
Materials and methods
Bioinformatics analysis
Setting ‘Homo sapiens’ and ‘pulmonary nodules’ as
keywords, the GSE135304 dataset (15) was downloaded from the NCBI GEO
database (https://www.ncbi.nlm.nih.gov/). The dataset consisted
of three main types of samples: Whole-blood samples from patients
with malignant nodules, patients with benign nodules and
nodule-free patients. After removing duplicate and treated samples,
587 samples remained in the following groups: Malignant (n=303),
benign (n=196) and no nodules (n=88). Association between RNASE2
expression and nodule size was also analyzed.
The identification of statistically different genes
was completed using the ‘Limma’ package (R Core Team) with false
discovery rate <0.05 and log(fold-change) >0.05.
Overlapping DEGs from malignant/benign and
malignant/(benign + no nodules) nodules were identified using VENNY
2.0 (https://bioinfogp.cnb.csic.es/tools/venny/index2.0.2.html).
The KM Plotter (http://kmplot.com/analysis/index.php?p=service) was
used to assess the association between the expression of all genes
and survival in lung cancer, and all analysis of results has been
taken directly from Kaplan-Meier Plotter without reanalysis.
The GSE68465 dataset, which contained expression
data for 462 samples [lung adenocarcinoma (n=443) and normal
(n=19)] and survival information, was also downloaded from NCBI GEO
in order to verify the expression of RNASE2.
The online databases miRDB (https://mirdb.org/) and TargetScanHuman (http://www.targetscan.org/) were used to predict the
microRNA (miR/miRNA) targets.
Cell culture and transfection
Human lung adenocarcinoma cell lines (A-549 and
ABC-1) were purchased from Nanjing Cobioer Gene Technology Co.,
Ltd. The A-549 cells were cultured in F12K + 10% FBS (Nanjing
Cobioer Gene Technology Co., Ltd.) and the ABC-1 cells were
cultured in MEM + 1% non-essential amino acids + 10% FBS + 1 mM
sodium pyruvate (Nanjing Cobioer Gene Technology Co., Ltd.), both
in a 37°C incubator with 5% CO2.
The ribonuclease A family member 2 (RNASE2) pVITRO2
overexpression vector (RNASE2 OE), small interfering RNA (siRNA)
against RNASE2 (RNASE2 siRNA), miR-185-5p mimic and negative
controls were designed, synthesized and obtained from GenePharma
Co., Ltd. Empty vector and miR-NC were used as negative
controls.
A-549 and ABC-1 cells were infected with RNASE2
siRNA, RNASE2 OE, RNASE2 OE, miR-185-5p mimic using Lipofectamine
3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
for 24 h according to the manufacturer's instructions. The cells
were collected after 48 h of culture for subsequent
experiments.
The sequence of the miR-185-5p mimic was as follows:
Sense, 5′-UGGAGAGAAAGGCAGUUCCUGA-3′ and antisense,
5′-AGGAACUGCCUUUCUCUCCAUU-3′. The sequence of the mimic-NC was as
follows: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
3′-TTAAGAGGCUUGCACAGUGCA-5′.
RNA immunoprecipitation (RIP)
assay
An EZ Magna RNA immunoprecipitation Kit (cat. no.
17-701; MilliporeSigma) was used according to the manufacturer's
guidelines. Briefly, A-549 and ABC-1 cells were lysed in 500 µl RIP
lysis buffer. Centrifugation was performed at 21,367 × g at 4°C for
15 min to extract the supernatant. Magnetic beads (100 µl) were
preincubated with 5 µg anti-protein argonaute-2 (AGO2) or 5 µg
immunoglobulin G (IgG) antibodies for 30 min at room temperature.
Cell lysates (100 µl) were immunoprecipitated with beads for 6 h at
4°C. After incubation and brief centrifugation, the EP tube was
placed on a magnetic rack, waiting for the solution to clear, and
the supernatant discarded. A total of 500 µl RIP Wash Buffer was
added, and then the contents of the EP tube was vortexed and placed
back on the magnetic rack. The supernatant was discarded again and
this step was repeated five times for a total of six washes. qPCR
was used to confirm the target protein.
Cell counting Kit-8 (CCK-8) assay
The proliferation of the two cell lines (A-549 and
ABC-1) was measured using the CCK-8 assay (MilliporeSigma). The
cells were routinely digested using trypsin and seeded onto 96-well
plates. After 24 h, 10 µl CCK-8 solution was added per well and
incubated for 2 h at 37°C. Finally, absorbance was measured at 450
nm using a microplate reader (Molecular Devices, LLC).
Transwell cell migration and invasion
assay
After coating the upper chamber with 50 mg/l
Matrigel (BD Biosciences) and leaving to air dry at 4°C,
1×105 A-549 or ABC-1 cells with 100 µl serum-free medium
(Thermo Fisher Scientific, Inc.) were seeded into Transwell
chambers (8.0-µm pore size; MilliporeSigma). The complete medium
was added to the lower chamber. After 48 h of incubation at 37°C,
the upper chamber was cleaned using cotton swabs, whereas the cells
in the lower chamber were fixed with 70% ethanol for 20 min and
stained with 0.1% crystal violet for 15 min, both at 25°C. The
number of invasive cells was counted using a light microscope
(Olympus Corporation). The steps for the cell migration assay were
the same as those for the invasion assay, except that the upper
chamber was not pre-coated.
Luciferase reporter gene analysis
pmirGLO vectors (Promega Corporation) were inserted
with binding site-amplified RNASE2 3′UTR, through which RNASE2
3′UTR wild-type reporter (RNASE2-Wt) and a mutated isoform
(RNASE2-Mut) with altered binding sites were constructed.
Transfection of RNASE2-Wt and RNASE2-Mut with Lipofectamine was
conducted along with the aforementioned miR-185-5p mimic. Next, the
fluorescence intensity indicating luciferase activity was measured
using a Reporter Assay Kit (cat. no. 16186; Thermo Fisher
Scientific, Inc.) at 36 h post-transfection, while normalization
was performed with reference to Renilla luciferase activity,
according to the manufacturer's protocol.
Statistical analysis
Using GraphPad Prism (version 8.0.1; Dotmatics),
data from three independent experiments (with the exception of
clinical data) were analyzed and exhibited as the mean ± SD. The
statistically significant differences between tumor tissues and
adjacent normal tissues were determined using unpaired Student's
t-test. The statistically significant differences between other two
groups comparisons were determined using Mann-Whitney U-test or
unpaired Student's t-test, where appropriate. ANOVA with Tukey's
post hoc test was used in datasets containing three or more groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Gene profiling in whole blood samples
from patients with malignant nodules and benign nodules, and from
nodule-free patients
Pulmonary nodules are the main manifestations of
early lung cancer (14). The
GSE135304 dataset, which contains the sequencing data of whole
blood from patients with malignant nodules and benign nodules, and
from nodule-free patients, was downloaded from the GEO database.
Duplicates and treated samples were removed, leaving 587 samples
(303 samples with malignant nodules, 196 with benign nodules and 88
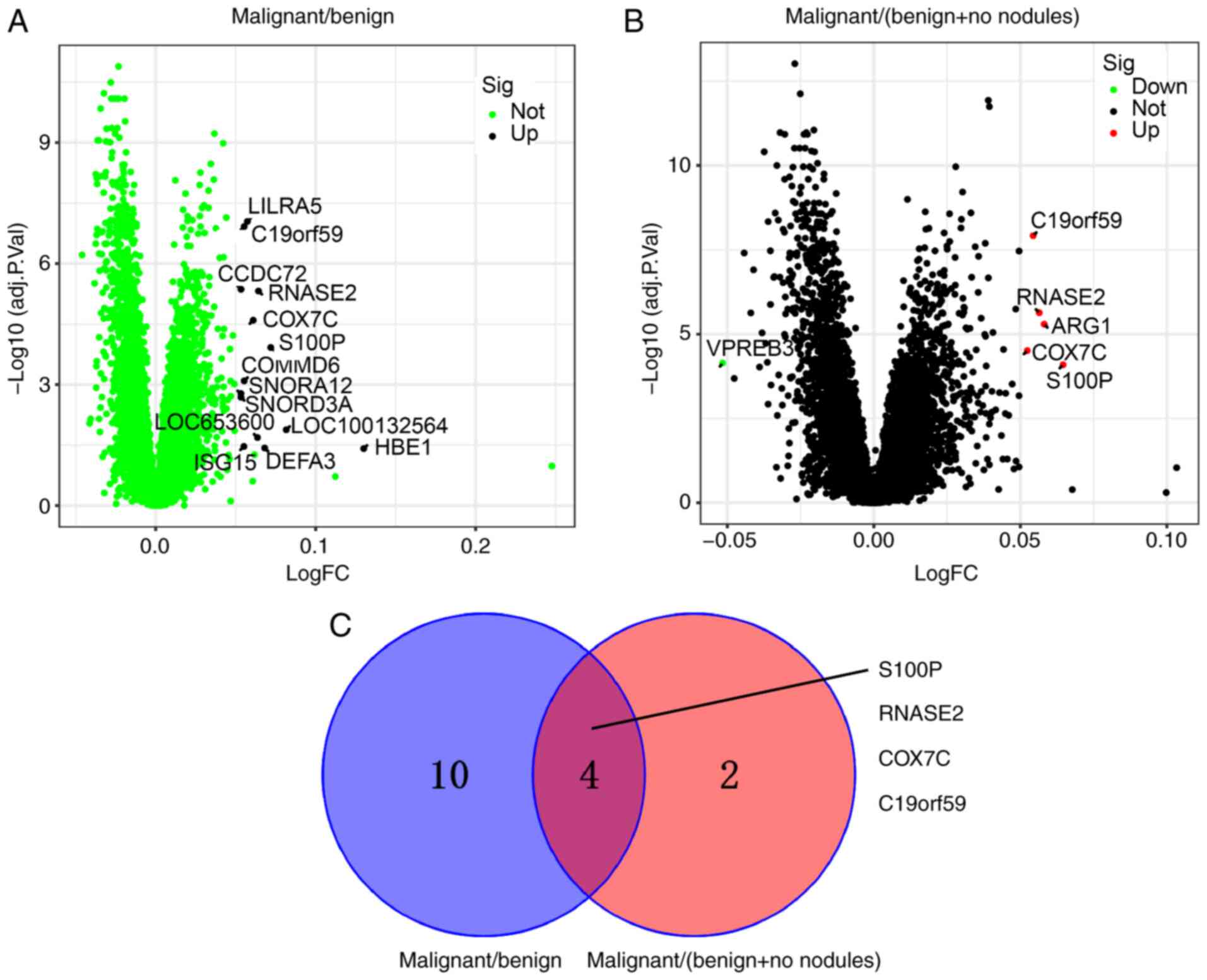
with no nodules). Fig. 1A shows
that 14 DEGs were upregulated in the malignant nodule blood samples
compared with the benign nodule blood samples. Fig. 1B shows that 5 DEGs were upregulated
and one was downregulated in the malignant nodule blood samples
compared with the benign nodule and no nodules blood samples. Four
overlapping DEGs [S100 calcium binding protein P (S100P), RNASE2,
cytochrome c oxidase subunit 7C (COX7C) and mast cell
expressed membrane protein 1 (C19orf59)] from malignant/benign and
malignant/(benign + no nodules) nodules were obtained using VENNY
(Fig. 1C).
Expression characteristics of the four
overlapped DEGs
The four overlapping DEGs (S100P, RNASE2, COX7C and
C19orf59) were upregulated in the malignant nodule blood samples
compared with those in the benign nodule samples and no nodules
samples, and their RNA expression levels did not differ between the
benign and no nodules blood samples (Fig. 2A). Using the KM Plotter, the
connection between these DEGs and the overall survival of patients
with lung cancer was analyzed. Patients with high S100P and RNASE2
expression exhibited a significantly poorer prognosis compared with
those patients with low expression, whereas patients with high
C19orf59 expression exhibited a better prognosis compared with the
patients with low expression (Fig.
2B). The survival of patients with lung cancer was not affected
by COX7C expression.
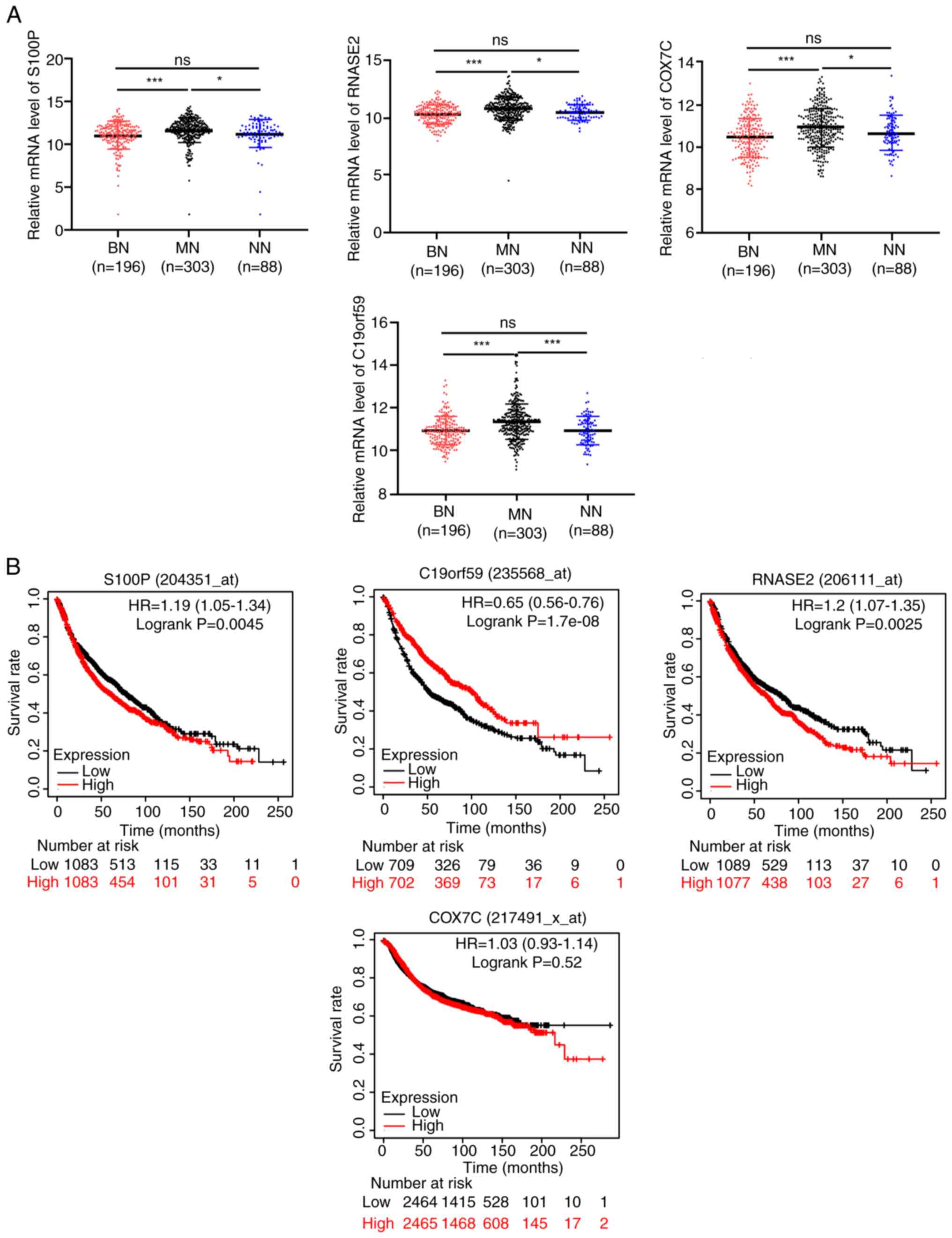 | Figure 2.Expression characteristics of the four
overlapped DEGs. (A) The expression of S100P, RNASE2, COX7C and
C19orf59 in malignant and benign blood samples, and those with no
nodules. (B) The connection between these DEGs and the overall
survival of lung cancer patients was analyzed though Kaplan-Meier
Plotter. *P<0.05 and ***P<0.001. ns, not significant; BN,
benign nodules; MN, malignant nodules; NN, no nodules; S100, S100
calcium binding protein P; RNASE2, ribonuclease A family member 2;
COX7C, cytochrome c oxidase subunit 7C; C19orf59, mast cell
expressed membrane protein 1; DEG, differentially expressed
gene. |
RNASE2 promotes cell proliferation,
migration and invasion
A literature search revealed that, to the best of
our knowledge, no study has yet reported the association between
RNASE2 and lung cancer. Therefore, RNASE2 was selected as a novel
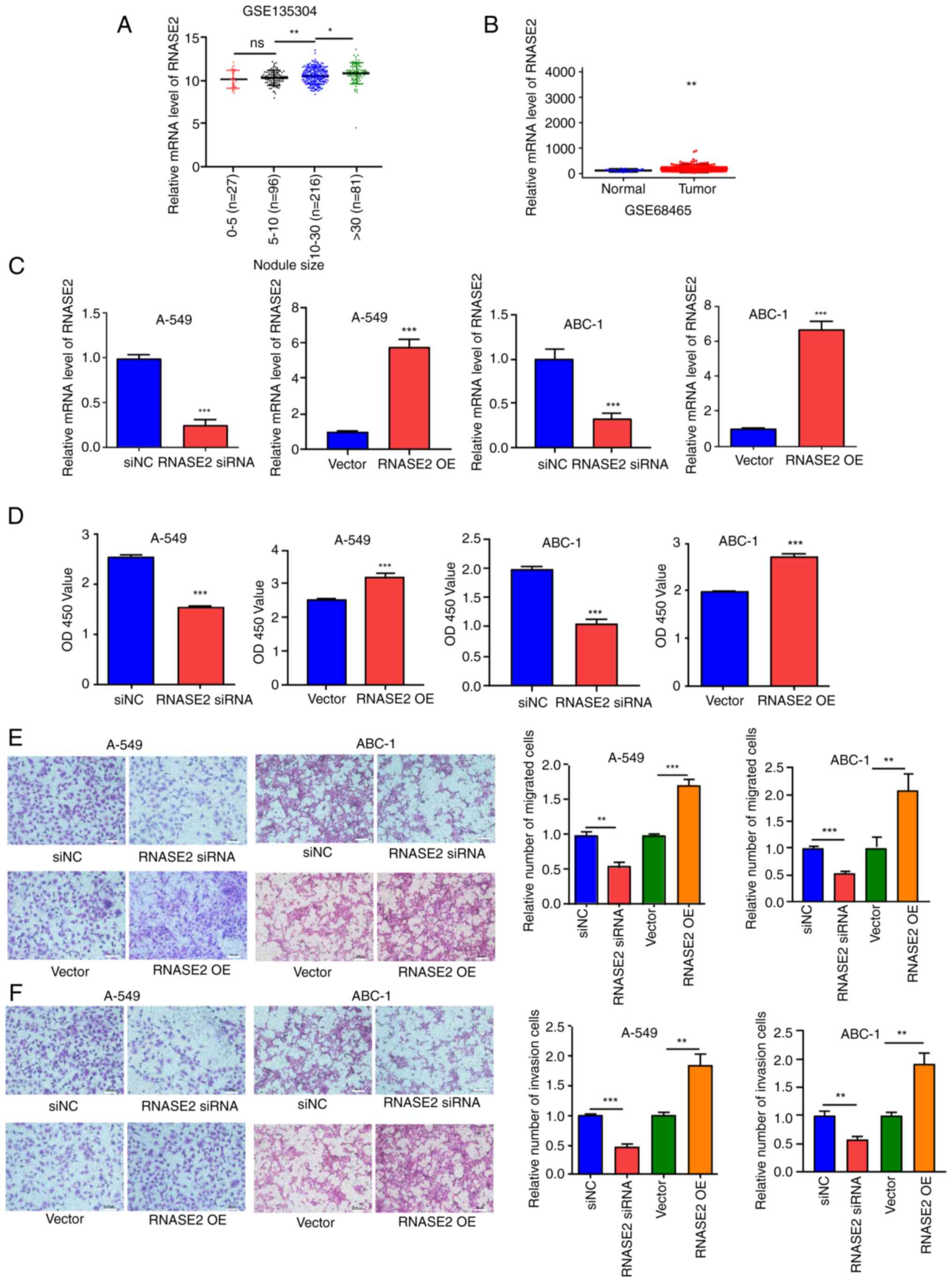
target to explore its role in lung cancer. The analysis of the
GSE135304 dataset revealed an association between RNASE2 expression
and nodule size; in general, the larger the nodule, the higher the
gene expression level (Fig. 3A).
Using the GSE68465 dataset, it was found that the expression of
RNASE2 was significantly higher in lung adenocarcinoma tissues than
that in normal tissues (Fig. 3B).
RNASE2 was knocked down and RNASE2 was overexpressed in A-549 and
ABC-1 cells (Fig. 3C), and the
proliferation of transfected A-549 and ABC-1 cells was measured
using a CCK-8 assay. Proliferation was inhibited by RNASE2 siRNA
and promoted by RNASE2 overexpression (Fig. 3D). The ability of the cells to
migrate and invade was also tested, which similarly showed
inhibition after RNASE2 siRNA treatment and promotion by RNASE2
overexpression (Fig. 3E and F).
RNASE2 is a target gene of
miR-185-5p
The upstream regulatory mechanism of RNASE2 was also
explored. As a number of mRNAs are regulated by miRNAs that play a
role in the occurrence and development of tumors (16–18),
28 miRNAs were predicted to target RNASE2 in miRDB (https://mirdb.org/) (Table SI). The 18 miRNAs with target
scores of >60 were analyzed. Additionally, the miRNA targets
were predicted using TargetScanHuman (http://www.targetscan.org/) (Table SII). In TargetScanHuman, the
context++ score is an indicator used to assess the reliability and
strength of the binding between a miRNA and its target genes.
Specifically, the context++ score is calculated based on various
features within the target mRNA sequence, including the position of
binding sites, sequence specificity, and whether there are
evolutionary conservation features such as immunohistochemistry
staining. A lower context++ score indicates a stronger predicted
binding and a more likely miRNA-target relationship. In total, 17
miRNAs with a context ++ score percentile of 99 were selected.
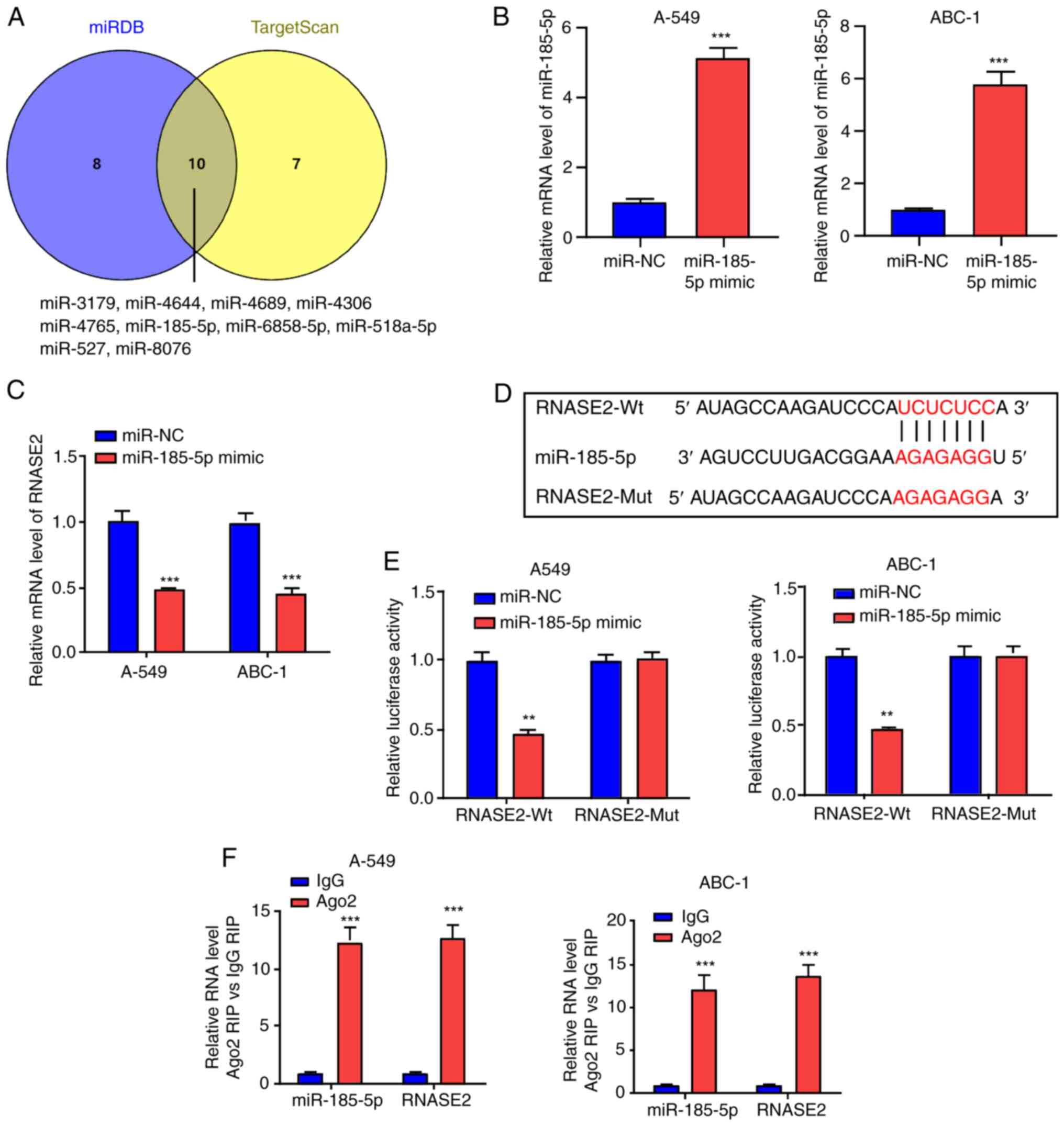
Using VENNY, 10 overlapping miRNAs were identified between the
miRDB and TargetScan miRNAs (Fig.
4A). Among these, miR-185-5p is closely associated with lung
cancer (19–21). The miR-185-5p mimic was therefore
used to transfect A-549 and ABC-1 cells to upregulate miR-185-5p
level (Fig. 4B), and RNASE2 was
found to be inhibited by the miR-185-5p mimic (Fig. 4C). Next, RNASE2-Wt and RNASE2-Mut
were constructed (Fig. 4D). The
results of a luciferase reporter assay showed that the luciferase
activity of RNASE2-Wt was inhibited by the miR-185-5p mimic,
whereas no such effect was shown for RNASE2-Mut (Fig. 4E). The RNA-induced silencing complex
(RISC) is a complex protein complex in cells that regulates gene
expression. This complex can target a specific mRNA through miRNA
guidance, inhibiting its translation or promoting its degradation,
thus affecting gene expression levels. RISC contains miRNA, mRNA
and Ago2 protein. To confirm whether miR-185-5p and RNASE2 form a
RISC, RIP experiments were conducted, which found that both
miR-185-5p and RNASE2 were enriched in the Ago2 group (Fig. 4F). This indicates that RNASE2 can
bind to miR-185-5p.
miR-185-5p inhibits the proliferation,
migration and invasion of lung adenocarcinoma cells by targeting
RNASE2
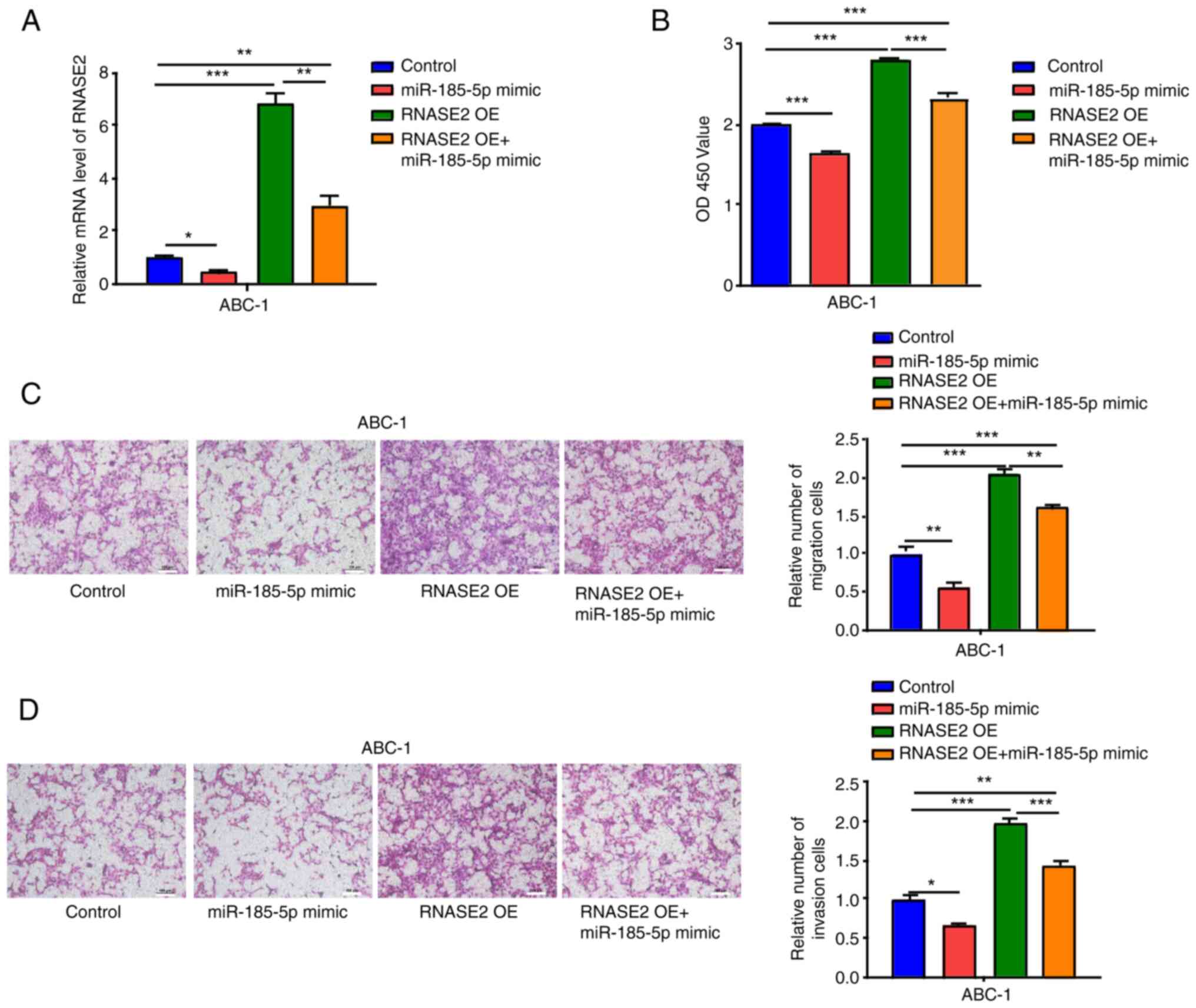
Using the miR-185-5p mimic alone or in combination
with RNASE2 OE, transfection was conducted in the ABC-1 cells.
RNASE2 expression was significantly inhibited by the miR-185-5p
mimic in comparison with the control (Fig. 5A). Moreover, significantly
suppressed cell proliferation was observed in the ABC-1 cells
transfected with miR-185-5p mimic compared with the control, which
partially reversed the enhanced cell proliferation induced by
RNASE2 OE (Fig. 5B). The migration
and invasion of ABC-1 cells were also inhibited by the miR-185-5p
mimic, which also partially reversed the enhanced cell migration
and invasion induced by RNASE2 OE (Fig.
5C and D).
Discussion
In the present study, DEGs in blood samples with or
without malignant nodules were analyzed to identify potential
biomarkers for the treatment of lung cancer. Four key genes (S100P,
C19orf59, COX7C and RNASE2) were identified that may play important
roles in lung cancer. Using KM Plotter datasets, it was found that
S100P, C19orf59 and RNASE2 were associated with the prognosis of
patients with lung cancer. High S100P and RNASE2 expression was
associated with a poor prognosis, whereas high C19orf59 levels
indicated a better prognosis. As S100P, C19orf59 and RNASE2 are
upregulated in malignant nodule samples, a focus was placed on
S100P and RNASE2. C19orf59 was not a focus as its expression was
inconsistent with the prognosis trend.
The role of S100P in the development and occurrence
of lung cancer was established through a literature search. Rehbein
et al (22) reported that
S100P induction is vital to early lung adenocarcinomas, whereas in
advanced stages, tumor progression partially relies on its
downregulation. Tan et al (23) reported that the lncRNA NORAD,
through the sequestering of S100P, inhibits the metastasis of lung
and breast cancer (23). Hsu et
al (24) revealed that in lung
cancer, S100P interacts with integrin α7, leading to heightened
migration and invasion activities (24). Chien et al (25) reported that, in lung
adenocarcinomas, through the control of S100P, the Keap1-Nrf2
interaction can suppress cell motility. However, to the best of our
knowledge, no studies have reported the association between RNASE2
and lung diseases.
Although no study has found an association between
RNASE2 and lung cancer, RNASE2 was previously found to promote the
progression of gliomas (26). In
the present study, GSE135304 analysis revealed a significant
association between RNASE2 expression and nodule size. Using the
GSE68465 dataset, it was found that the expression levels of RNASE2
were higher in lung adenocarcinoma tissues than those in normal
tissues. In A-549 and ABC-1 cells, the ability to proliferate,
migrate and invade was promoted by the overexpression of RNASE2 and
inhibited by RNASE2 siRNA. In addition, RNASE2 was regulated by
miR-185-5p in lung adenocarcinoma cells. miR-185-5p was previously
reported to target tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein ζ to regulate non-small cell
lung cancer progression (27).
miR-185-5p has anticancer effects via the regulation of transgelin
2 in lung adenocarcinoma (21).
Ras-associated binding protein 35 acts as a target of miR-185-5p,
which subsequently regulates the proliferation, migration and
invasion of non-small cell lung cancer cells, particularly through
tumor cell-derived exosomes (28).
In the present study, RNASE2 was highly expressed in lung
adenocarcinoma tissues compared with normal tissues. Through the
inhibition of RNASE2, miR-185-5p mimic could suppress the
proliferation, migration and invasion of lung adenocarcinoma
cells.
The present study has certain limitations. First,
in vivo experiments were not performed, and these are
required to elucidate the mechanism of action of RNASE2. Second,
the analysis was based on public data, and clinical specimens
should be collected and sequenced to verify the expression of
RNASE2. Third, other candidate targets should be studied carefully
to elucidate their roles in lung cancer. Fourth, in Fig. 2B, there is late-stage crossover of
curves in the graph for RNASE2, and it would be better to
re-analyse this dataset, either by restricting the analyzed period
of time to exclude this late crossover event, or by using a
weighted test, such as Renyi or Cramer-von Mises (29). Finally, genes other than miR-185-5p
that may regulate RNASE2 and other miRNAs involved in lung cancer
pathogenesis should be further explored.
In summary, the present study showed that miR-185-5p
regulates RNASE2 to promote the proliferation, migration and
invasion of lung adenocarcinoma cells. RNASE2 is a potential
therapeutic target for the treatment of lung adenocarcinoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
FL conceived the study idea and designed the
experiments. GL, YH, LW, DL and SX performed the experiments. QL,
NZ and NG performed the bioinformatics analysis. GL, LZ and YH
collected and analyzed the data. GL was a major contributor in
writing the manuscript. All authors have read and approved the
final manuscript. FL and GL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sharma P, Mahadevia H, Donepudi S, Kujtan
L, Gustafson B, Ponvilawan B, Al-Obaidi A, Subramanian J and Bansal
D: A novel EGFR germline mutation in lung adenocarcinoma: Case
report and literature review. Clin Lung Cancer. 25:479–482. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun R, Hou Z, Zhang Y and Jiang B: Drug
resistance mechanisms and progress in the treatment of EGFR-mutated
lung adenocarcinoma. Oncol Lett. 24:4082022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li N and Zhan X: Identification of
pathology-specific regulators of m(6)A RNA modification to optimize
lung cancer management in the context of predictive, preventive,
and personalized medicine. EPMA J. 11:485–504. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shu J, Jiang J and Zhao G: Identification
of novel gene signature for lung adenocarcinoma by machine learning
to predict immunotherapy and prognosis. Front Immunol.
14:11778472023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu A, Wang Z, Yang Y, Wang J, Dai X, Wang
L, Lu Y and Xue F: Preoperative diagnosis of malignant pulmonary
nodules in lung cancer screening with a radiomics nomogram. Cancer
Commun (Lond). 40:16–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prosper AE, Kammer MN, Maldonado F, Aberle
DR and Hsu W: Expanding role of advanced image analysis in
CT-detected indeterminate pulmonary nodules and early lung cancer
characterization. Radiology. 309:e2229042023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma L, Qiu B, Zhang J, Li QW, Wang B, Zhang
XH, Qiang MY, Chen ZL, Guo SP and Liu H: Survival and prognostic
factors of non-small cell lung cancer patients with postoperative
locoregional recurrence treated with radical radiotherapy. Chin J
Cancer. 36:932017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran Jr WJ, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hajipour S, Hosseini SM, Irani S and
Tavallaie M: Identification of novel potential drugs and miRNAs
biomarkers in lung cancer based on gene co-expression network
analysis. Genomics Inform. 21:e382023. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma H, Jiang S, Yuan Y, Li J, Li Y, Lv Y,
Du T, Guan J, Jiang X, Tian L, et al: RUNX1 promotes proliferation
and migration in non-small cell lung cancer cell lines via the mTOR
pathway. FASEB J. 37:e231952023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang J, Zhang Y, Wang J, Yang X, Ren X,
Huang H, Wang J, Lu J, Zhong Y, Lin Z, et al: Identification of
CDT1 as a prognostic marker in human lung adenocarcinoma using
bioinformatics approaches. Peer J. 11:e161662023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong L, Zhang D, Dong Y, Lei Y, Qian Y,
Tan X, Han S and Wang J: Integrated bioinformatics analysis for
identificating the therapeutic targets of aspirin in small cell
lung cancer. J Biomed Inform. 88:20–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Peng Y and Guo Y: Pulmonary
nodules detection based on multi-scale attention networks. Sci Rep.
12:14662022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kossenkov AV, Qureshi R, Dawany NB,
Wickramasinghe J, Liu Q, Majumdar RS, Chang C, Widura S, Kumar T,
Horng WH, et al: A gene expression classifier from whole blood
distinguishes benign from malignant lung nodules detected by
low-dose CT. Cancer Res. 79:263–273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei L, Wu Y, Cai S, Qin Y, Xing S and Wang
Z: Long non-coding RNA linc01224 regulates hypopharyngeal squamous
cell carcinoma growth through interactions with miR-485-5p and
IGF2BP3. J Cancer. 14:3009–3022. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song S, Xie S, Liu X, Li S, Wang L, Jiang
X and Lu D: miR-3200 accelerates the growth of liver cancer cells
by enhancing Rab7A. Noncoding RNA Res. 8:675–685. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai C, He N, Zeng J, Long C, Shi M, Li J,
Ma S, Xiong Y and Liang X: Highly expressed miR-144-3p promotes the
proliferation, migration and invasion of colon carcinoma cells by
activating the Wnt/beta-catenin signaling pathway through targeting
SFRP1. J Cancer. 14:3117–3129. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang D, Zhang S, Zhao M and Chen F: LncRNA
MALAT1 accelerates non-small cell lung cancer progression via
regulating miR-185-5p/MDM4 axis. Cancer Med. 9:9138–9149. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Hu Y, Wu Y, Zhang D and Huang D:
LINC00205 promotes tumor malignancy of lung adenocarcinoma through
sponging miR-185-5p. Lab Med. 53:39–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu N, Gong H, Chen W and Peng W: CircRNA
ZKSCAN1 promotes lung adenocarcinoma progression by
miR-185-5p/TAGLN2 axis. Thorac Cancer. 14:1467–1476. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rehbein G, Simm A, Hofmann HS, Silber RE
and Bartling B: Molecular regulation of S100P in human lung
adenocarcinomas. Int J Mol Med. 22:69–77. 2008.PubMed/NCBI
|
|
23
|
Tan BS, Yang MC, Singh S, Chou YC, Chen
HY, Wang MY, Wang YC and Chen RH: LncRNA NORAD is repressed by the
YAP pathway and suppresses lung and breast cancer metastasis by
sequestering S100P. Oncogene. 38:5612–5626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsu YL, Hung JY, Liang YY, Lin YS, Tsai
MJ, Chou SH, Lu CY and Kuo PL: S100P interacts with integrin alpha7
and increases cancer cell migration and invasion in lung cancer.
Oncotarget. 6:29585–29598. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chien MH, Lee WJ, Hsieh FK, Li CF, Cheng
TY, Wang MY, Chen JS, Chow JM, Jan YH, Hsiao M, et al: Keap1-Nrf2
interaction suppresses cell motility in lung adenocarcinomas by
targeting the s100p protein. Clin Cancer Res. 21:4719–4732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu T, Chen Y, Yang L, Wang X, Chen K and
Xu D: Ribonuclease a family member 2 promotes the malignant
progression of glioma through the PI3K/Akt signaling pathway. Front
Oncol. 12:9210832022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma J, Bai Y, Chen F, Zhou F, Zhang L, Xue
P and Wang D: MicroRNA-185-5p targets tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
to regulate non-small cell lung cancer progression. J Cardiothorac
Surg. 18:2412023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen H, Liu Z, Tang J and Bu L: MiR-185-5p
targets RAB35 gene to regulate tumor cell-derived exosomes-mediated
proliferation, migration and invasion of non-small cell lung cancer
cells. Aging (Albany NY). 13:21435–21450. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|