Introduction
Colorectal cancer (CRC) is one of the leading causes
of cancer-associated death worldwide (1). While multidisciplinary treatment for
CRC, including surgery, neoadjuvant chemoradiotherapy,
postoperative chemoradiotherapy, chemotherapy (including molecular
targeted therapy), and immunotherapy, has decreased the risk of
recurrence and improved survival and outcomes (2–4), the
5-year survival rates of CRC patients are still not satisfactory,
in part because of multidrug resistance and cancer progression
(5–8).
The high mortality rate of CRC is mainly from late
disease diagnosis and lack of adequate prognostic biomarkers. While
treatment recommendations and prognosis prediction of CRC patients
are determined using the tumor-node-metastasis (TNM) classification
system (9), CRC exhibits
heterogeneity, and even same-stage individuals can show different
clinical outcomes and response to treatment (10). Thus, the identification of robust
prognostic markers is critical. Moreover, such biomarkers possess
adequate prognostic significance for specific subgroups decided by
the TNM staging system (11).
Ferroptosis, which was first discovered in 2012, is
a newly defined form of programmed cell death distinct from
apoptosis, necrosis, and autophagy (12). Ferroptosis is characterized by iron
accumulation, lipid peroxidation, and accumulation of lipid
reactive oxygen species (ROS) within cells. Ferroptosis is distinct
from other types of regulated cell death in various aspects
(12,13). For example, ferroptotic cells show
unique biochemical, morphological, and genetic features, such as
ruptured cellular membrane, lack of chromatin condensation,
shrunken mitochondria, and increased density of the mitochondrial
membrane (14). Several studies
have shown that ferroptosis is involved in various diseases and
processes, including neurodegeneration, neurotoxicity, drug-induced
hepatotoxicity, acute renal failure, tissue ischemia/reperfusion,
immunological abnormality, and carcinogenesis (15–18).
Reports have also shown that therapy-resistant
cancer cells are sensitive to ferroptosis, suggesting that
targeting ferroptosis could be a promising strategy for cancer
(19). We previously demonstrated
that some botanical compounds that possess anti-tumorigenic
potential altered the expression of ferroptosis-related genes; the
compounds induced suppression of cancer progression and restored
chemosensitivity in gastrointestinal cancer (20–22).
We thus hypothesized that negative regulators of ferroptosis may
also have a role in CRC development.
Ferroptosis is negatively regulated by limiting ROS
production and reducing cellular iron uptake. Negative regulation
of ferroptosis is mediated by glutathione peroxidase 4 (GPX4), heat
shock protein β-1 (HSPB1), nuclear factor erythroid 2-related
factor 2 (NRF2), and specific light-chain subunit of the
cystine/glutamate antiporter (SLC7A11) (23). The association between the
expression of negative regulators of ferroptosis and the
oncological outcome of CRC patients has not been examined.
In this study, we conducted a systematic
investigation to first identify candidate ferroptosis negative
regulator genes that may be associated with the prognosis of CRC
patients by in silico analysis. We then confirmed the
clinical importance of candidate genes as prognostic biomarkers by
validation using clinical specimens from CRC patients.
Materials and methods
Study design
The present study consisted of a search for negative
regulators of ferroptosis through in silico testing and
validation in clinical samples of CRC patients. The study flow is
shown in Fig. 1A. Details on the
discovery phase and gene identification are described below.
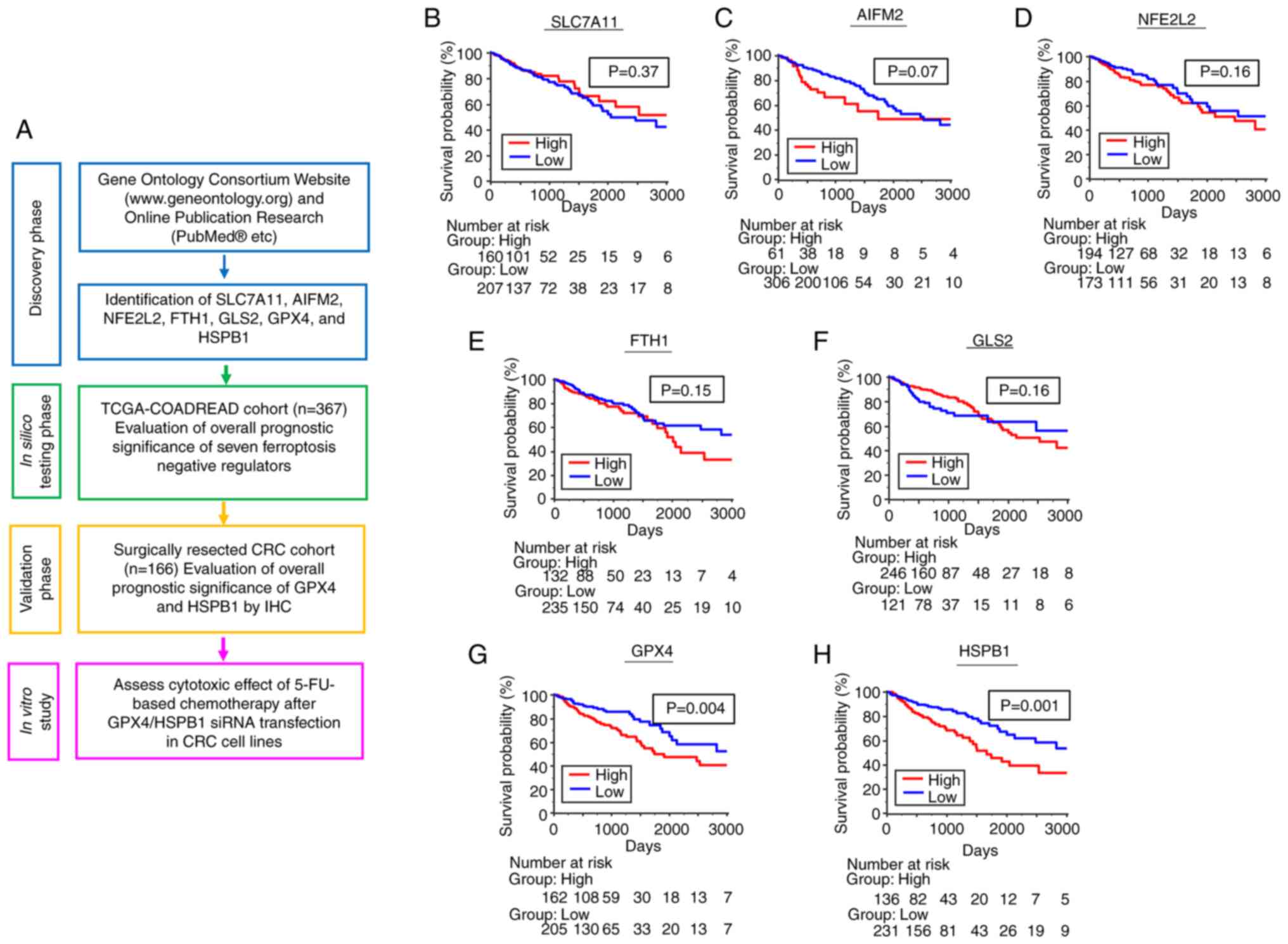 | Figure 1.Identification and in silico
evaluation of candidate ferroptosis negative regulators for
prognostic biomarkers in CRC patients. (A) Schematic of the project
flow. Kaplan-Meier analysis of OS in patients with CRC from TCGA
cohort according to the following individual candidate ferroptosis
negative regulators: (B) SLC7A11, (C) AIFM2, (D) NFE2L2, (E) FTH1,
(F) GLS2, (G) GPX4 and (H) HSPB1. CRC, colorectal cancer; OS,
overall survival; TCGA-COADREAD, The Cancer Genome Atlas-Colon and
Rectal Cancer; 5FU, 5-fluorouracil; siRNA, small interfering
RNA. |
Patient characteristics
Stage 1 to stage 4 CRC patients (n=166) who
underwent surgical resection at the Department of Gastrointestinal
and Pediatric Surgery of the Mie University Graduate School of
Medicine from January 2012 to December 2015 were enrolled in this
study. Patients with incomplete clinical data or inadequate
immunohistochemistry (IHC) results were excluded. Patient
clinicopathological characteristics are shown in Table SI. Staging was performed on the
basis of clinical and histopathological assessment following the
International Union Against Cancer TNM staging system (24). All patients were followed up after
initial hospital discharge, with physical examination and tumor
marker assays (CEA and CA19-9) performed every 1–3 months and
computed tomography every 6 months; endoscopic examinations were
performed when necessary. Written informed consent was obtained
from all patients following the local ethics guidelines, and the
Institutional Review Board (IRB) of Medical Ethics Committee of Mie
University Graduate School of Medicine approved this study (IRB
number: H2023-172).
Comprehensive in silico analysis of
negative regulators of ferroptosis
Several studies have discussed genes associated with
negative regulation of ferroptosis (23,25–29).
In this study, we confirmed the candidate ferroptosis negative
regulator genes using the Gene Ontology Consortium Website
(www.geneontology.org, and http://www.informatics.jax.org/vocab/gene_ontology/).
We downloaded gene expression from the RNAseq (IlluminaHiSeq)
dataset of ‘The Cancer Genome Atlas (TCGA) Colon and Rectal Cancer
(COADREAD)’ patients (https://xenabrowser.net/datapages/). We investigated
the association between the expression profile of seven ferroptosis
negative regulator genes, SLC7A11, AIFM2, NFE2L2 (NRF2), FTH1,
GLS2, GPX4, and HSPB1, and overall prognosis in the 367 CRC
patients in the in silico cohort with available survival
data.
IHC
Formalin-fixed, paraffin-embedded sections (5 µm
thickness) of specimens from the 166 CRC patients were subjected to
immunohistochemical analysis. After deparaffinization by xylene and
rehydration in graded concentrations of ethanol, specimens were
heated at 121°C for 10 min in citrate buffer (pH 6.0) to unmask
antigens. Endogenous peroxidase activity was blocked by incubation
with 3% hydrogen peroxide in distilled water. Nonspecific binding
sites were blocked with 10% normal goat serum (Vector Laboratories,
Burlingame, CA, USA) in Tris-buffered saline with 0.05% Tween 20
(TBST), and samples were incubated with primary antibody overnight
at 4°C. Primary GPX4 antibody (Abcam, Waltham, MA, USA) was used at
1:1,000, and primary HSPB1 antibody (same as HSP27, Santa Cruz
Biotechnology, Dallas TX, USA) was used at 1:1,000. After washing
with TBST, sections were incubated with secondary antibody coupled
with peroxidase-conjugated polymers [Universal Immuno-peroxidase
Polymer method, Histofine SAB-PO(M) Kit, Nichirei Biosciences,
Inc., Tokyo, Japan] for 30 min and detected using the Histofine DAB
substrate kit (Nichirei Biosciences, Inc.). Slides were
counterstained with hematoxylin, as previously described (30–33).
Evaluation of immunohistochemical
staining
GPX4 and HSPB1 expression of CRC specimens were
analyzed three times separately by two investigators who were not
familiar with the clinical or survival data of patients. The
investigators first evaluated the entire tissue specimen at
low-power magnification (×40) and then focused on tumor hotspots at
high-power magnification (×200 and ×400). As previously described
(30–33), an immunohistochemical score was
determined for each case as follows: Staining intensity score: 0,
colorless; 1, weak; 2, moderate; and 3, strong; and staining
percentage score: 1, 1–25; 2, 26–50; 3, 51–75 and 4, >75%
positivity. The IHC score of each patient was obtained by
multiplying the scores for staining intensity and staining
percentage. If the difference between the scores obtained from each
investigator was >3, the stained slide was reassessed.
Cell culture and materials
CRC cell lines (DLD-1, RKO, SW480, and LOVO) were
acquired from the Cell Resource Center of Biomedical Research,
Institute of Development, Aging and Cancer (Tohoku University;
Sendai, Japan). The CRC cells were maintained in RPMI 1640 (Nacalai
Tesque Inc., Kyoto, Japan), supplemented with 10% fetal bovine
serum (FBS, Biowest, Nouille, France) and Antibiotic-Antimycotic
(Nacalai Tesque Inc.) and maintained at 37°C in a humidified
incubator at 5% CO2. All cell lines were checked and
authenticated using a panel of genetic and epigenetic markers and
tested for mycoplasma on a regular basis. 5-Fluorouracil (5FU,
Sigma-Aldrich, MA, USA) was dissolved in dimethyl sulfoxide and
diluted to the appropriate experimental concentrations in cell
culture medium before use.
RNA interference
Specific predesigned small interfering RNA (siRNA)
for GPX4 and HSPB1 and Negative Control siRNA were purchased from
Ambion (Thermo Fisher Scientific, Inc., USA). The siRNA sequences
of GPX4 and HSPB1 were as follows: GPX4: sense,
5′-GGCAAGACCGAAGUAAACUtt-3′ and antisense,
5′-AGUUUACUUCGGUCUUGCCtc-3′; and HSPB1: sense,
5′-CGAGAUCACCAUCCCAGUCtt-3′ and antisense,
5′GACUGGGAUGGUGAUCUCGtt-3′ (SiRNA ID number for GPX4 is 10848, and
for HSPB1 is 121323. Catalogue number for Negative Control siRNA is
4390843). We used two CRC cell lines (DLD-1 and SW480) with high
gene expression of GPX4 and HSPB1 for experiments. DLD-1 and SW480
cells were seeded in six-well culture plates at 2×105
cells per well in 2 ml RPMI 1640. Cells were cultured for 24 h and
then incubated with siRNA oligonucleotides using Lipofectamine
RNAiMAX Reagent and OptiMEM I (both Invitrogen, Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The final
concentration of siRNA oligonucleotide of GPX4/HSPB1 was 50 nM.
After 48 h, transfected cells were harvested and examined by
RT-qPCR and western blotting to check siRNA efficiency.
Total RNA extraction and cDNA
synthesis
Total RNA was extracted from CRC cell lines (DLD1,
RKO, SW480, and LOVO) using miRNeasy (Qiagen, Germany) following
the manufacturer's instructions. RNA quality and concentration were
determined using a Denovix DS-11+ spectrophotometer (DeNovix, Inc.,
USA). cDNA was synthesized from 5 µg of total RNA with random
hexamer primers, dNTPs, 5X buffer, RNase inhibitor, and ReverTra
Ace (Toyobo Co., LTD., Japan).
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR analyses were conducted using the SYBR Green
PCR Master Mix and QuantStudio3 Real-Time PCR System (Applied
Biosystems, Thermo Fisher Scientific, Inc., USA) following the
manufacturer's protocol and as previously described (22). The qPCR cycling conditions were as
follows: 95°C for 10 min, followed by 45 cycles of 15 sec at 95°C
and 60 sec at 60°C. Primers for GPX4, HSPB1 and GAPDH mRNAs were as
follows; GPX4: forward, 5′-GAGGCAAGACCGAAGTAAACTAC-3′ and reverse,
5′-CCGAACTGGTTACACGGGAA-3′; HSPB1: forward,
5′-ACGGTCAAGACCAAGGATGG-3′ and reverse, 5′-AGCGTGTATTTCCGCGTGA-3′;
and GAPDH: forward, 5′-GGAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-AATGAAGGGGTCATTCATGG-3′. Relative expression levels of GPX4 and
HSPB1 mRNA were calculated by normalization to the levels of
endogenous GAPDH mRNA using the 2−ΔΔCq method (22). RT-qPCR assays were performed in
triplicate for each sample and the mean value was calculated.
Western blotting
At 48 h after transfection of GPX4 or HSPB1 siRNA,
cells were lysed in RIPA buffer (BioDynamics Laboratory, Inc.,
Tokyo, Japan) with protease inhibitors, and lysates were
centrifuged for 5 min at 15,000 × g at 4°C. The protein
concentration was measured using the BCA protein assay kit (Thermo
Fisher Scientific, Inc.). Protein samples (20 µg) were separated on
AnykD™ Mini-PROTEAN® TGX™ Precast
Protein Gels and then transferred onto a polyvinylidene difluoride
membrane (Bio-Rad Laboratories, CA, USA). The blots were first
blocked with 5% skimmed milk for 1 h at room temperature and then
incubated overnight with primary antibodies against GPX4 (1:20,000;
Abcam, Cambridge, UK), HSPB1 (1:10,000; Santa Cruz, CA, USA), and
β-actin (1:40,000; MP Biomedicals, Illkirch, France). The blots
were incubated with secondary antibody (Promega, Madison, WI, USA)
for 60 min at room temperature. The protein bands were visualized
by chemiluminescent reaction (Immobilon™ Western;
Millipore, MA, USA) coupled with a WSE-6100 LuminoGraph imaging
system (ATTO Corporation, Tokyo, Japan).
Cell viability assay
After transfection of GPX4 or HSPB1 siRNA, DLD-1 and
SW480 cells were plated in 96-well tissue culture plates (TPP
Techno Plastic Products AG, Switzerland) at a density of 3,000
cells/well in RPMI1640 medium supplemented with 10% FBS and
antibiotics. The cells were allowed to adhere to the plate for 24 h
and then treated with a series of two-fold dilution of 5FU (0,
6.25, 12.5, 25, 50, 100, 200, 400 µM). After 72 h of treatment,
cell proliferation was measured using the WST8 assay (Dojindo
Laboratories, Kumamoto, Japan) following the manufacturer's
instructions. The absorbance in each well was measured at a
wavelength of 450 nm with a Multiskan FC plate reader (Thermo
Fisher Scientific, Inc., USA). Each experiment was performed in
triplicate. The cytotoxic effect of 5FU was assessed by the IC50
concept (34), and the IC50 value
for 5FU was calculated using CompuSyn software (Chou and Martin,
2005, Compusyn, Inc., USA).
Statistical analysis
Statistical analyses were performed using Medcalc
statistical software V.16.2.0 (Medcalc Software bvba, Ostend,
Belgium), JMP software 10.0.2 (SAS Institute, Cary, NC, USA), and
GraphPad Prism software ver.8.2.0 (GraphPad Software Inc., San
Diego, CA). Comparison of IHC score between high- and
low-expression groups and various clinicopathological factors in
the clinical cohort were performed using Fisher's exact test.
Comparisons of differential gene expression between two groups from
in vitro experiments were performed using two-tailed
unpaired Student's t-test. On the other hand, comparisons of
differential gene expression between multiple groups from in
vitro experiments were performed using one-way ANOVA with
Tukey's multiple comparisons test. Comparisons of IHC scores
between various stages were performed using Kruskal-Wallis test and
Dunn's multiple comparisons test. Youden's index for dead/alive or
recurrence/non-recurrence within the observation period was used to
determine the optimal cutoff thresholds to dichotomize patients
into high- and low-expression groups of GPX4 and HSPB1 using
Medcalc statistical software. OS was defined as the period from the
date of CRC diagnosis to the date of last known follow-up;
recurrence-free survival (RFS) was defined as the period from the
date of CRC diagnosis to the date of recurrence. Kaplan-Meier
analyses for OS and RFS were performed by log-rank test. For OS and
RFS survival analyses, we dichotomized patients into GPX4 or HSPB1
high- and low-expression groups as determined by receiver operating
characteristic analysis (same as Youden's index) for dead/alive or
recurrence/non-recurrence. Univariate and multivariate analyses for
OS and RFS were performed using the Cox proportional hazards model
to determine the factors affecting OS and RFS. P<0.05 was
considered to indicate a statistically significant difference.
Results
Comprehensive in silico analysis
identified GPX4 and HSPB1 as candidate prognostic biomarkers in
colorectal cancer patients
As described in the Materials and Methods section,
we determined SLC7A11, AIFM2, NFE2L2 (NRF2), FTH1, GLS2, GPX4, and
HSPB1 genes as candidate negative regulators of ferroptosis. We
then evaluated their prognostic importance by analyzing 367 CRC
patients with available information on survival outcome in
TCGA-COADREAD datasets. We dichotomized patients into high and low
gene expression groups as determined by Youden's index for
dead/alive. High expression of SLC7A11, AIFM2, NFE2L2, FTH1, and
GLS2 genes did not stratify survival outcome of patients (Fig. 1B-F). In contrast, patients with high
expression of GPX4 and HSPB1 genes showed significantly worse OS
(P<0.01 for both factors) than those with low expression
(Fig. 1G and H). Therefore, we
selected GPX4 and HSPB1 as candidate targets for further validation
study.
High GPX4 protein expression was
associated with aggressive cancer phenotype in CRC patients
To evaluate the prognostic potential of GPX4 and
HSPB1 in CRC patients, we first investigated GPX4 expression in a
cohort of stage 1–4 CRC patients by IHC analysis. While GPX4
protein was not expressed or weakly expressed in the adjacent
normal mucosa (Fig. 2A), it was
expressed mainly in the cytoplasm of CRC cells (Fig. 2A). There was varying expression of
GPX4 among CRC cases (Fig. 2B). The
GPX4 IHC score for 151 CRC patients was 3.5±2.1 (mean ± SD), and
median value for GPX4 IHC score was 4; 15 CRC patients with
GPX4-negative staining were excluded from the analysis. The GPX4
IHC score of stage 1 CRC patients was significantly lower than
those of stage 2–4 patients (Fig.
2C). We analyzed the association between GPX4 expression and
clinicopathological parameters and found that high GPX4 expression
(GPX4 IHC score ≥4) was significantly associated with larger tumor
size (P=0.002), advanced T factor (P=0.003), lymph node metastasis
positivity (P=0.002), lymphatic and venous invasion positivity
(P=0.02 and P<0.001, respectively), and stage 4 cases (P=0.02)
(Table I). These results indicate
that high GPX4 protein expression was significantly associated with
an aggressive cancer phenotype in CRC patients in the clinical
cohort.
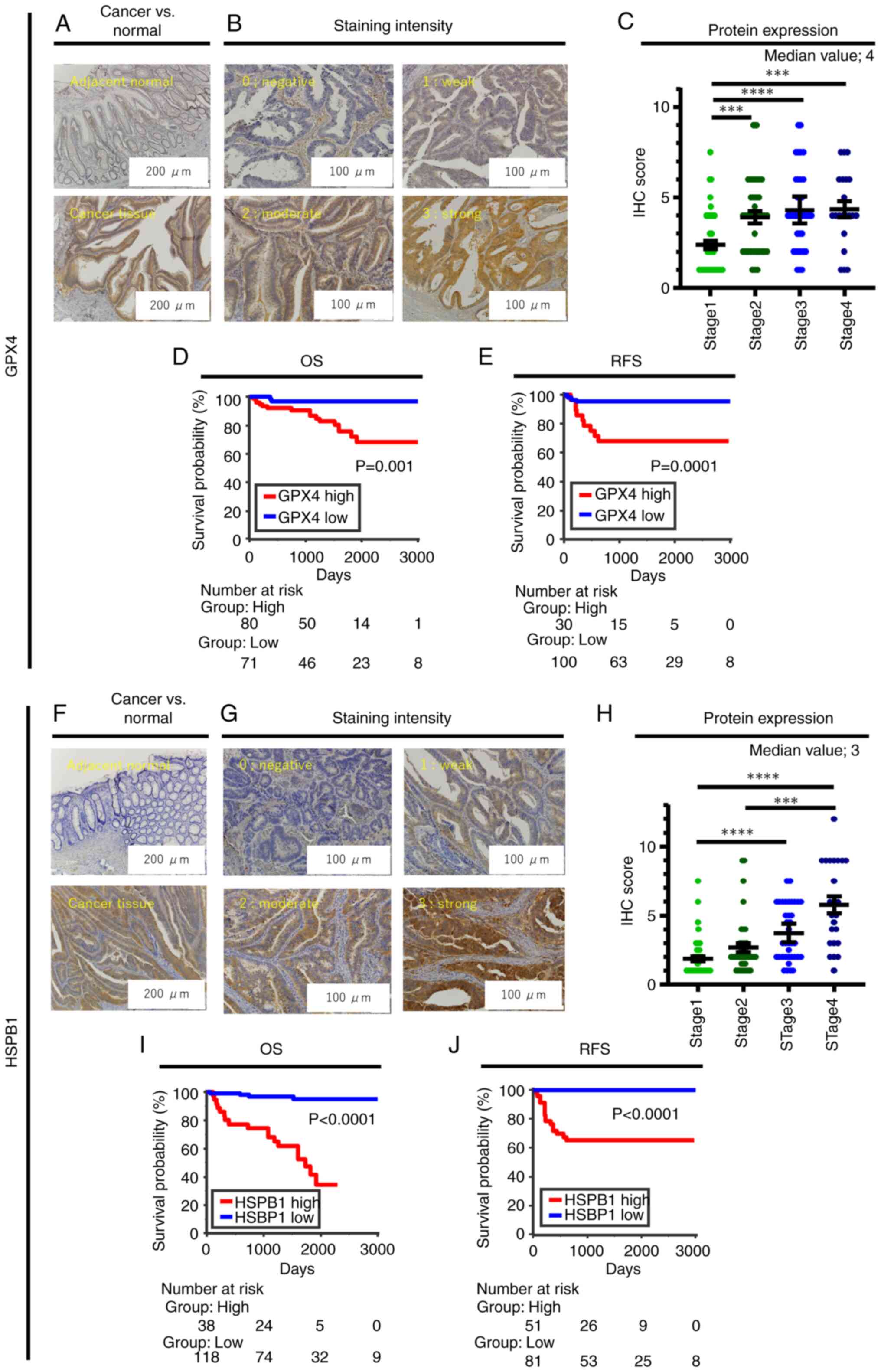 | Figure 2.Representative images of
immunohistochemistry of GPX4 and HSPB1 in CRC specimens and
association between IHC score profiling, UICC stage, and survival
outcome in CRC patients dichotomized by GPX4 and HSPB1 expression.
(A) Representative image of GPX4 expression in adjacent colon
normal mucosa and CRC tissue. (B) Representative staining intensity
of GPX4 scored as 0 (negative), 1 (weak), 2 (moderate), and 3
(strong). (C) Dot plot showing the IHC score of GPX4 for stage 1 to
stage 4 CRC patients. Kaplan-Meier analysis for (D) OS and (E) RFS
in CRC patients classified into high/low GPX4 expression groups.
(F) Representative image of HSPB1 expression in adjacent colon
normal mucosa and CRC tissue. (G) Representative staining intensity
of HSPB1 scored as 0 (negative), 1 (weak), 2 (moderate), and 3
(strong). (H) Dot plot showing the IHC score of HSPB1 for stage 1
to stage 4 CRC patients. Kaplan-Meier analysis for (I) OS and (J)
RFS in CRC patients classified into high/low HSPB1 expression
groups. ***P<0.001, and ****P<0.0001. CRC, colorectal cancer;
OS, overall survival; RFS, recurrence-free survival; IHC,
immunohistochemistry. |
 | Table I.Association between
clinicopathological factors and expression of GPX4 and HSPB1 in the
in-house clinical cohort. |
Table I.
Association between
clinicopathological factors and expression of GPX4 and HSPB1 in the
in-house clinical cohort.
|
| GPX4 | HSPB1 |
|---|
|
|
|
|
|---|
| Variable | Number | Low (n=71) | High (n=80) | P-value | Number | Low (n=118) | High (n=38) | P-value |
|---|
| Age at operation,
years |
|
|
|
|
|
|
|
|
|
>69 | 86 | 39 | 47 |
| 88 | 70 | 18 |
|
|
<69 | 65 | 32 | 33 | 0.74 | 68 | 48 | 20 | 0.26 |
| Sex |
|
|
|
|
|
|
|
|
|
Male | 86 | 39 | 47 |
| 85 | 67 | 18 |
|
|
Female | 65 | 32 | 33 | 0.74 | 71 | 51 | 20 | 0.35 |
| Tumor location |
|
|
|
|
|
|
|
|
|
Colon | 98 | 49 | 49 |
| 101 | 81 | 20 |
|
|
Rectum | 53 | 22 | 31 | 0.39 | 55 | 37 | 18 | 0.08 |
| Macroscopic
type |
|
|
|
|
|
|
|
|
| Type 1,
2 | 137 | 68 | 69 |
| 138 | 107 | 31 |
|
| Type 3,
4 | 14 | 3 | 11 | 0.06 | 18 | 11 | 7 | 0.15 |
| Tumor size, mm |
|
|
|
|
|
|
|
|
|
>40 | 102 | 57 | 45 |
| 51 | 35 | 16 |
|
|
<40 | 49 | 14 | 35 | 0.002a | 105 | 83 | 22 | 0.17 |
| Histological
type |
|
|
|
|
|
|
|
|
|
Intestinal | 140 | 67 | 73 |
| 145 | 113 | 32 |
|
|
Diffuse | 11 | 4 | 7 | 0.54 | 11 | 5 | 6 | 0.03a |
| T factor |
|
|
|
|
|
|
|
|
| T1, T2,
T3 | 128 | 67 | 61 |
| 131 | 105 | 26 |
|
| T4 | 23 | 4 | 19 | 0.003a | 25 | 13 | 12 | 0.005a |
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
|
Negative | 97 | 55 | 42 |
| 97 | 89 | 8 |
|
|
Positive | 54 | 16 | 38 | 0.002a | 59 | 29 | 30 |
<0.0001a |
| Lymphatic
invasion |
|
|
|
|
|
|
|
|
|
Negative | 65 | 38 | 27 |
| 65 | 60 | 5 |
|
|
Positive | 86 | 33 | 53 | 0.02a | 91 | 58 | 33 |
<0.0001a |
| Venous
invasion |
|
|
|
|
|
|
|
|
|
Negative | 73 | 45 | 28 |
| 74 | 67 | 7 |
|
|
Positive | 78 | 26 | 52 | 0.0006a | 82 | 51 | 31 |
<0.0001a |
| UICC stage |
|
|
|
|
|
|
|
|
| 1, 2,
3 | 130 | 66 | 64 |
| 132 | 109 | 23 |
|
| 4 | 21 | 5 | 16 | 0.02a | 24 | 9 | 15 |
<0.0001a |
High GPX4 protein expression was
associated with poor survival outcome in CRC patients
Next, we evaluated the prognostic value of GPX4
protein expression by examining OS using Kaplan-Meier analysis.
Patients with high expression of GPX4 (GPX4 IHC score ≥4) exhibited
significantly worse OS compared with patients with low expression
(P=0.001, Fig. 2D). To elucidate
whether GPX4 protein expression can be used for recurrence
prediction, we investigated the association between GPX4 protein
expression and postoperative recurrence by analyzing RFS of 130
stage 1–3 CRC patients receiving treatment with curative intent. In
line with the OS data, patients with high expression of GPX4 (GPX4
IHC score ≥5; dichotomized by Youden's index for positive/negative
recurrence) showed significantly worse RFS than those with low
expression (P=0.0001, Fig. 2E).
To determine whether high GPX4 protein expression
was a risk factor for OS or RFS, the Cox proportional hazard model
was used to perform univariate and multivariate analysis. In
univariate analysis for OS, along with high GPX4 protein expression
(P=0.0005), several clinicopathological factors such as rectal
cancer (P=0.02), invasive endoscopic type (P=0.04), larger tumor
size (P=0.008), diffuse histological type (P=0.009), advanced T
stage (P<0.0001), lymph node metastasis (P=0.0002), lymphatic
and venous invasion positive cases (P=0.001 for both factors), and
stage 4 cases (P<0.0001) were risk factors for poorer OS
(Table II). Multivariate analysis
showed that diffuse histological type [hazard ratio (HR)=6.48,
P=0.02] and stage 4 cases (HR=5.61, P=0.009) were independent risk
factors for poorer OS; high GPX4 expression did not show
statistical significance (P=0.14, Table II).
 | Table II.Multivariate analysis for overall
survival and recurrence-free survival and GPX4 expression in
clinical cohort. |
Table II.
Multivariate analysis for overall
survival and recurrence-free survival and GPX4 expression in
clinical cohort.
| A, Overall survival
(n=151) |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|---|
| Age >70
years | 0.95 | 0.37–2.44 | 0.92 |
|
|
|
| Male | 1.9 | 0.72–5.92 | 0.2 |
|
|
|
| Tumor location
rectum | 3.07 | 1.21–8.36 | 0.02a | 2.41 | 0.83–7.47 | 0.11 |
| Macroscopic type
3/4/5 | 3.75 | 1.06–10.50 | 0.04a | 3.02 | 0.65–13.20 | 0.15 |
| Tumor size >40
mm | 3.53 | 1.39–9.60 | 0.008a | 0.67 | 0.20–2.31 | 0.52 |
| Poorly
differentiated histology | 5.98 | 1.68–16.95 | 0.009a | 6.48 | 1.30–28.66 | 0.02a |
| T stage greater
than T4 | 11.74 | 4.59–32.12 |
<0.0001a | 2.31 | 0.61–8.92 | 0.22 |
| Lymph node
metastasis positive | 6.39 | 2.29–22.57 | 0.0002a | 1.64 | 0.44–7.98 | 0.48 |
| Lymphatic invasion
positive | 6.76 | 1.92–42.77 | 0.001a | 2.29 | 0.38–19.39 | 0.38 |
| Venous invasion
positive | 7.04 | 2.00–44.53 | 0.001a | 1.79 | 0.38–13.10 | 0.48 |
| Metastasis
positive | 20.55 | 7.91–59.41 |
<0.0001a | 5.61 | 1.55–20.82 | 0.009a |
| GPX4 protein
high | 7.92 | 2.25–50.12 | 0.0005a | 3.29 | 0.70–24.64 | 0.14 |
|
| B,
Recurrence-free survival (n=130) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|
| Age >70
years | 0.99 | 0.33–3.10 | 0.99 |
|
|
|
| Male | 1.1 | 0.37–3.65 | 0.86 |
|
|
|
| Tumor location
rectum | 7.02 | 2.15–31.34 | 0.0009a | 7.07 | 2.12–32.02 | 0.001a |
| Macroscopic type
3/4/5 | 1.32 | 0.07–6.70 | 0.8 |
|
|
|
| Tumor size >40
mm | 1.05 | 0.28–3.21 | 0.94 |
|
|
|
| Poorly
differentiated histology | 5.13E-09 | 2.78–2.78 | 0.23 |
|
|
|
| T Stage greater
than T4 | 1.88 | 0.29–7.01 | 0.45 |
|
|
|
| Lymph node
metastasis positive | 3.77 | 1.26–12.48 | 0.02a | 1.56 | 0.48–5.72 | 0.46 |
| Lymphatic invasion
positive | 13.4 | 2.64–243.96 | 0.0005a | 7.71 | 1.27–150.89 | 0.02a |
| Venous invasion
positive | 6.53 | 1.75–42.20 | 0.004a | 1.35 | 0.23–11.06 | 0.75 |
| GPX4 protein
high | 7.68 | 2.50–28.35 | 0.0004a | 4.11 | 1.18–17.83 | 0.03a |
With regard to univariate analysis for RFS, primary
lesion in rectum (P=0.0007), invasive endoscopic type (P=0.049),
advanced T stage and N stage (P=0.01, P=0.002, respectively),
lymphatic and venous invasion positive cases (P=0.0007, P=0.002,
respectively), and high GPX4 expression (P=0.018) were risk factors
for poor RFS (Table II).
Multivariate analysis showed that primary lesion in rectum
(HR=6.83, P<0.0001), invasive endoscopic type (HR=5.91,
P=0.008), lymphatic invasion positivity (HR=4.66, P=0.02), and high
GPX4 expression (HR=4.11, P=0.03) were independent risk factors for
poor RFS (Table II). Thus, high
GPX4 expression may predict poor RFS in CRC patients.
High HSPB1 protein expression was
associated with aggressive phenotype in CRC patients
We also investigated the prognostic potential of
HSPB1 in the stage 1–4 CRC patients by IHC. While HSPB1 protein was
absent or weakly expressed in the adjacent normal mucosa (Fig. 2F), it was expressed mainly in the
cytoplasm of CRC cells (Fig. 2F),
similar to the expression of GPX4. HSPB1 expression also varied
among CRC cases (Fig. 2G). The
HSPB1 IHC score for 156 patients was 3.1±2.4, and median value for
HSPB1 IHC score was 3; 10 CRC patients with HSPB1-negative staining
were eliminated from analysis. Notably, the IHC score trend of
HSPB1 showed a more prominent ‘stage-dependent elevated pattern’ in
CRC patients (Fig. 2H). Similar to
the results with GPX4 expression, high HSPB1 expression (HSPB1 IHC
score >4; dichotomized by Youden's index for OS) was
significantly associated with aggressive cancer phenotypes such as
diffuse histological type (P=0.03), advanced T factor (P=0.005),
lymph node metastasis positivity (P<0.0001), lymphatic and
venous invasion positivity (P<0.0001 for both factors), and
stage 4 cases (P<0.0001) (Table
I). Thus, similar to GPX4, high HSPB1 protein expression was
significantly associated with aggressive cancer phenotype in CRC
patients.
High HSPB1 protein expression showed
strong robustness as a prognostic and recurrence-predictive
biomarker in CRC patients
We further assessed the overall prognostic
significance of HSPB1 protein expression by analyzing OS using the
Kaplan-Meier method. Patients with high expression of HSPB1 (HSPB1
IHC score ≥5) showed significantly worse OS than patients with low
expression (P<0.0001, Fig. 2I).
Kaplan-Meier analysis for RFS in 132 stage 1–3 CRC patients treated
with curative intent revealed that patients with high expression of
HSPB1 (HSPB1 IHC score ≥3; dichotomized by Youden's index for
positive/negative recurrence) showed significantly worse RFS than
patients with low expression (P<0.0001, Fig. 2J).
We performed Cox proportional univariate and
multivariate analysis for OS and RFS in the CRC patients. In
univariate analysis for OS, along with high HSPB1 expression
(P<0.0001), several aggressive phenotypes such as invasive
endoscopic type (P=0.002), larger tumor size (P=0.002), diffuse
histological type (P=0.002), advanced T stage and N stage
(P<0.0001 for both factors), lymphatic and venous invasion
positive cases (P=0.002, P=0.001, respectively), and stage 4 cases
(P<0.0001) were risk factors for poor OS (Table III). Multivariate analysis
identified that diffuse histological type (HR=5.84, P=0.01), stage
4 patients (HR=3.99, P=0.02), and high HSPB1 expression (HR=6.35,
P=0.006) were independent risk factors for poor OS (Table III). Regarding univariate analysis
for RFS, primary lesion in rectum (P=0.0007), invasive endoscopic
type (P=0.049), advanced T stage and N stage (P=0.01, P=0.002,
respectively), lymphatic and venous invasion positivity (P=0.0007,
P=0.002, respectively), and high HSPB1 expression (P<0.0001)
were risk factors for poor RFS (Table
III). Multivariate analysis demonstrated that primary lesion in
rectum (P=0.0003), advanced T stage (P=0.009), and high HSPB1
expression (P<0.0001) were independent risk factors for poor RFS
(Table III). These results
indicate that high HSPB1 expression may indicate poor prognosis and
high risk of recurrence in CRC patients, and its biomarker
potential may be superior to that of GPX4.
 | Table III.Multivariate analysis for overall
survival and recurrence-free survival and HSPB1 expression in
clinical cohort. |
Table III.
Multivariate analysis for overall
survival and recurrence-free survival and HSPB1 expression in
clinical cohort.
| A, Overall survival
(n=156) |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|---|
| Age >70
years | 0.94 | 0.40–2.21 | 0.89 |
|
|
|
| Male | 1.33 | 0.57–3.34 | 0.51 |
|
|
|
| Tumor location
rectum | 2.28 | 0.98–5.41 | 0.06 |
|
|
|
| Macroscopic type
3/4/5 | 4.92 | 1.88–11.69 | 0.002a | 3.19 | 0.99–10.26 | 0.06 |
| Tumor size >40
mm | 3.96 | 1.69–9.91 | 0.002a | 1.04 | 0.32–3.25 | 0.95 |
| Poorly
differentiated histology | 6.61 | 2.15–16.96 | 0.002a | 5.84 | 1.56–20.05 | 0.01a |
| T stage greater
than T4 | 10.64 | 4.56–25.96 |
<0.0001a | 1.79 | 0.46–6.83 | 0.4 |
| Lymph node
metastasis positive | 10.87 | 3.70–46.29 |
<0.0001a | 2.1 | 0.55–11.37 | 0.3 |
| Lymphatic invasion
positive | 5.21 | 1.78–22.19 | 0.002a | 1.28 | 0.33–6.70 | 0.74 |
| Venous invasion
positive | 5.41 | 1.84–23.04 | 0.001a | 0.95 | 0.23–5.00 | 0.95 |
| Metastasis
positive | 15.31 | 6.51–38.55 |
<0.0001a | 3.99 | 1.23–13.40 | 0.02a |
| HSPB1 protein
high | 16.69 | 6.20–57.96 |
<0.0001a | 6.35 | 1.67–31.59 | 0.006a |
|
| B,
Recurrence-free survival (n=132) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|
| Age >70
years | 1.09 | 0.40–30.8 | 0.87 |
|
|
|
| Male | 0.96 | 0.36–2.68 | 0.93 |
|
|
|
| Tumor location
rectum | 4.82 | 1.75–15.31 | 0.002a | 5.96 | 1.83–22.97 | 0.003a |
| Macroscopic type
3/4/5 | 3.67 | 1.03–10.56 | 0.047a | 1.85 | 0.42–7.28 | 0.39 |
| Tumor size >40
mm | 1.83 | 0.65–4.91 | 0.24 |
|
|
|
| Poorly
differentiated histology | 1.31 | 0.07–6.48 | 0.8 |
|
|
|
| T stage greater
than T4 | 3.58 | 1.00–10.30 | 0.05a | 6.02 | 1.20–28.53 | 0.03a |
| Lymph node
metastasis positive | 3.9 | 1.45–11.45 | 0.007a | 0.9 | 0.24–3.80 | 0.88 |
| Lymphatic invasion
positive | 15.73 | 3.19–284.38 |
<0.001a | 11.87 | 1.45–276.49 | 0.02a |
| Venous invasion
positive | 8.05 | 2.5–51.28 | 0.0006a | 0.9 | 0.15–7.49 | 0.91 |
| HSPB1 protein
high | 1.01E+10 | 14.76–14.76 |
<0.001a | 8.57E+09 | 12.06–1.05e+55 |
<0.001a |
GPX4 and HSPB1 may be biomarkers in
advanced CRC patients treated with adjuvant therapy with curative
intent
We next investigated whether GPX4 and HSPB1
expressions were biomarkers for adjuvant chemotherapy in advanced
(stage 2 and 3) CRC patients. We first investigated the association
between clinicopathological factors and GPX4 and HSPB1 expressions
by Fisher's exact test. We dichotomized cases into high- and
low-expression groups by Youden index for RFS. While there was no
significant correlation between the clinicopathological factors and
GPX4 expression, the frequency of patients with high expression of
HSPB1 was significantly higher in cases with rectal cancer and
cases with positive lymph node metastasis (Table SII).
We then evaluated the recurrence prediction
potential of GPX4 and HSPB1 by Kaplan-Meier analysis in stage 2 CRC
patients who were not administered adjuvant chemotherapy. While
there was no significant difference in RFS between patients with
high and low GPX4 expression (P=0.18), the recurrence rate of
patients with high GPX4 expression was worse than those with low
expression (HR=4.52) (Fig. S1A).
Patients with HSPB1 high expression showed significantly worse RFS
than those with low expression (P=0.0002). HR could not be
calculated because no patients with HSPB1 low expression showed
recurrence (Fig. S1B).
We next evaluated RFS in stage 2 and 3 CRC patients
who were treated with capecitabine. While there was no significant
difference for RFS between patients with high and low GPX4
expression (P=0.26), the recurrence rate of patients with high
expression of GPX4 was worse than that of patients with low
expression (HR=3.61, Fig. S2A).
Similarly, for HSPB1, while there was no significant difference of
RFS between high- and low-expression groups (P=0.25), patients with
high expression of HSPB1 showed worse recurrence rates than those
with low expression (HR=3.52, Fig.
S2B).
Collectively, our findings showed that HSPB1
expression indicated recurrence in stage 2 and 3 CRC patients
treated with curative intent. In patients with high expression of
HSPB1, we may recommend administration or reinforcement of adjuvant
chemotherapy. However, further studies with more patients are
required to strengthen our findings.
GPX4 and HSPB1 may be associated with
5-fluorouracil resistance in CRC cells in vitro
On the basis of the results of subgroup analysis for
GPX4 and HSPB1 in advanced CRC patients treated with adjuvant
therapy with curative intent, we further investigated the
functional role of GPX4 and HSPB1 in the response to 5-fluorouracil
(5FU)-based chemotherapy using CRC cell lines. First, we evaluated
the mRNA and protein expression of GPX4 and HSPB1 in four CRC cell
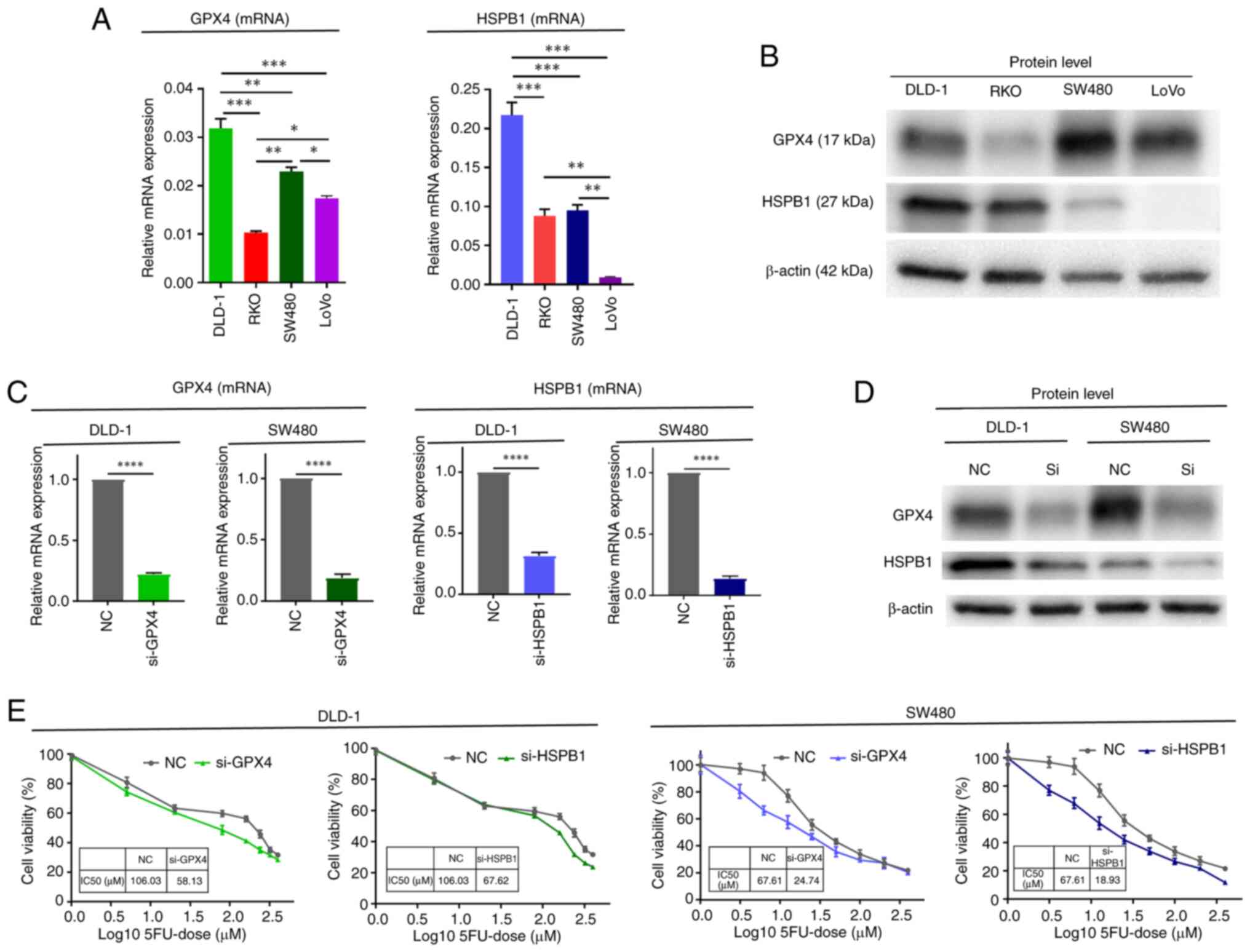
lines (DLD-1, RKO, SW480, and LOVO) (Fig. 3A and B). We selected DLD-1 and SW480
for further experiments, as both GPX4 and HSPB1 mRNA and protein
were detected at higher levels in these two cell lines. Next, we
transfected GPX4 and HSPB1 siRNA in these two CRC cell lines and
confirmed effective knockdown of mRNA (Fig. 3C) and protein levels (Fig. 3D) of both factors. We then compared
the cytotoxic effect of 5FU between cells transfected with negative
control siRNA and cells transfected with siRNA against GPX4 and
HSPB1 using WST8 assays. The IC50 values of 5FU were decreased in
cells transfected with GPX4 siRNA (DLD-1, from 106.03 to 58.13 µM;
SW480, 67.61 to 24.73 µM) and HSPB1 siRNA (DLD-1, from 106.03 to
67.62 µM; SW480, 67.61 to 18.93 µM) (Fig. 3E). Collectively, these in
vitro experiments demonstrated that GPX4 and HSPB1 may play
crucial roles in attenuating the cytotoxic effect of 5FU-based
conventional chemotherapy.
Discussion
Accumulating evidence has revealed the association
between ferroptosis regulators and cancer development in CRC
(35,36). In this study, we performed
comprehensive in silico dataset analysis and evaluated the
association between the expression of negative regulators of
ferroptosis and CRC prognosis; we also performed validation in
clinical specimens from CRC patients by IHC. Our findings indicate
that high expressions of HSPB1 and GPX4 may serve as prognostic
biomarkers for poor outcomes in CRC patients.
Ferroptosis is a newly defined iron-dependent
non-apoptotic cell death form characterized by lipid peroxidation,
iron accumulation, and accumulation of lipid ROS (12). Ferroptosis is distinguished from
other regulated cell death by differences in morphology, genetics,
and biochemistry (12). Multiple
studies have shown that ferroptosis is involved in multiple
pathological processes including tumorigenesis and cancer
development (15–17). Several studies have shown that the
initiation of ferroptosis in CRC cells successfully eliminates
cancer cells that are resistant to other forms of regulated cell
death (35,36). Moreover, GSH, RSL3, ACSL4, LCN2,
SRSF9, GCH1, TMEM16F, SLC7A11, NRF2, and p53 may play a pivotal
role in CRC by ferroptosis-related pathways (37–51).
Hence, we hypothesized that negative regulators of ferroptosis may
also have an important role in CRC progression. In this study, we
performed a comprehensive analysis of public datasets to explore
candidate negative regulators of ferroptosis in CRC.
After the publication of TCGA project of multiple
malignancies including alimentary tract cancer (52–54),
comprehensive expression profiling information is more easily
available and biomarker studies using bioinformatic analysis are
ongoing each year. One bioinformatic study examined the
relationship between ferroptosis-associated gene expression and
prognosis of CRC patients to establish a predictive model and
explored the potential value of ferroptosis as a therapeutic target
(55). Shao et al identified
ferroptosis-related differentially expressed genes between tumor
and normal colon tissues from the GeneCards and FerrDb websites and
analyzed the prognostic information of CRC patients from TCGA
dataset and other public datasets (55). The authors identified a 10-gene
prognostic signature consisting of TFAP2C, SLC39A8, NOS2, HAMP,
GDF15, FDFT1, CDKN2A, ALOX12, AKR1C1, and ATP6V1G2 genes; they
demonstrated that the signature score could effectively predict the
prognosis of CRC patients and the signature score of
cetuximab-resistant CRC patients was significantly higher than that
of sensitive patients (55).
Although the study indicated the importance of this 10-gene
signature, their in-house investigation included a small amount of
evidence such as tissue microarray of FDFT1, GDF15, HAMP, and
TFAP2C to evaluate their differential expression in 75-paired
normal/tumor CRC specimens (55).
In our study, we examined negative regulators of ferroptosis in
cancer progression and demonstrated that HSPB1 and GPX4 were
prognostic biomarkers of CRC patients.
Heat shock proteins increase the migration ability,
decrease apoptosis, and are involved in chemoresistance in cancer
cells (56,57). HSPB1, a chaperone protein also known
as heat shock 27kD protein 1 (HSP27), is stimulated by
transcriptional factor heat shock factor-1 (HSF-1) after elastin
treatment and stabilizes proteins under stress (58). Phosphorylation of HSPB1 by protein
kinase C regulates iron uptake, reduces iron-mediated production of
ROS and inhibits elastin-induced ferroptosis (58). Inhibition of the HSF1-HSPB1 pathway
increased the elastin-mediated anticancer activity in human
cervical carcinoma xenograft mouse models, and an essential role
for HSPB1 on ferroptosis-mediated cancer therapy was indicated
(58).
HSPB1 has also been reported to influence both
cancer progression and chemoresistance (59,60).
Several studies have shown that upregulation of HSPB1 induces
chemoresistance or resistance to radiotherapy in lung (61), breast (62), and colon cancers (63,64).
Several studies showed that HSPB1 suppression increased the
sensitivity of colon cancer cells to 5FU (63,65)
and irinotecan (66). Furthermore,
Liu et al conducted in vitro and in vivo
studies and found that suppression of HSPB1 induced inhibition of
tumor progression and enhancement of sensitivity to 5FU and
vincristine via suppression of the NOTCH1/Akt/mTOR signaling
pathway in colon cancer cells (67). Thus, HSPB1 may be a promising target
with a critical role in CRC development and drug resistance.
Further studies on the mechanism of HSPB1 as a negative regulator
of ferroptosis in CRC should be performed.
GPX4 converts reduced GSH to oxidized glutathione
and reduces lipid hydroperoxides. The function of GPX4 is mainly
inhibited by Ras-selective lethal small molecule 3 (RSL3) or
ferroptosis-inducing agents such as DPI7 (68–70).
Knockdown of GPX4 induces ferroptosis in a MAPK/ERK kinase-, iron-,
and ROS-dependent manner (69).
Zhang et al established 5FU- and AZ628-resistant CRC cells,
revealed the characteristics of the resistant cell lines by in
vitro assays, and evaluated the efficacy and mechanism of GPX4
inhibitor by in vitro and in vivo experiments
(71). While resistant cell lines
exhibited drug sensitivity, GPX4 expression was significantly
upregulated in resistant cells, and persister cells were more
sensitive and underwent ferroptosis induced by the GPX4 inhibitor.
Studies in a mouse model revealed that GPX4 inhibition restrained
tumor regrowth after discontinuation of anti-CRC drug treatment
(71). The authors concluded that
upregulated GPX4 in drug-resistant cells was a potential
therapeutic target and GPX4 inhibition by combination chemotherapy
with molecular targeted therapy may be a promising anti-CRC
treatment (71). From this
evidence, GPX4 may be a therapeutic target for CRC development and
chemoresistance. The mechanisms and downstream targets of GPX4
should be explored in future studies.
In this study, subgroup analysis of stage 2–3 CRC
patients showed that HSPB1 expression could clearly stratify the
recurrence prognosis of this patient group. We recommend
administration or reinforcement of adjuvant chemotherapy in
patients with high expression of HSPB1. While National
Comprehensive Cancer Network and Japanese Society for Cancer of the
Colon and Rectum have provided definitions for high-risk stage 2
CRC patients (72,73), there are still controversial issues
regarding the detailed regimen or administration period of adjuvant
chemotherapy. A recent prospective randomized phase III ACHIEVE-2
trial revealed that three months of adjuvant chemotherapy of
mFOLFOX6/CAPOX showed significantly less adverse effects and 3-year
RFS did not differ compared with the six-month regimen in high-risk
stage 2 colon cancer patients (74). In future studies, we will analyze a
larger group of patients and investigate whether HSPB1 may be a
biomarker to help provide detailed directions for adjuvant
chemotherapy treatment.
This study has several limitations. First, this was
a retrospective single institutional study with a relatively small
sample size. There were some patients (approximately 6.6%, data not
shown) whose postoperative follow-up period was short (less than 30
days) because of lost follow-up. Additionally, while we found that
GPX4 and HSPB1 may exhibit important roles in attenuating the
cytotoxic effect of conventional cytotoxic chemotherapy through
in vitro experiments, we could not directly confirm
induction of ferroptosis in response to siRNA transfection of GPX4
or HSPB1 because of technical limitations. Therefore, in the
future, multi-center large sample number studies are warranted and
in-depth mechanistic experiments are required to reveal the
detailed biological function of GPX4 and HSPB1, especially in the
negative regulation of ferroptosis in CRC.
In conclusion, our study provides novel insights
into the prognostic biomarker potential of negative regulators of
ferroptosis in CRC patients using in silico identification
and evaluation and validation in clinical samples. Moreover,
attenuation of the cytotoxic effect of chemotherapy induced by GPX4
and HSPB1 may be one of the mechanisms to shorten the overall
and/or recurrence-free prognosis of CRC patients with high GPX4 and
HSPB1 expression. Collectively, our results demonstrated that GPX4
and HSPB1 may be effective biomarkers to predict the survival
outcome and postoperative recurrence in CRC patients.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to acknowledge Ms. Yuki Orito
and Ms. Amphone Okada (Department of Gastrointestinal and Pediatric
Surgery, Mie University Graduate School of Medicine) for
experimental support, and Dr Gabrielle White Wolf for editing a
draft of this manuscript.
Funding
This work was partially supported by a Grant in Aid for
Scientific Research (grant nos. 22K08868 and 23K08210) from the
Ministry of Education, Culture, Sports, Science, and Technology,
Japan.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
TS, CY, YOku and YT contributed to conceptualization
and design of the study. TS, CY, RM, AZ, YN, AS, HO, XZ, YOku and
YT acquired, analyzed or interpretated the data. TS, CY, YOku and
YT drafted the manuscript. TS, CY, RM, AZ, SYa, KH, YS, HImao, TK,
MK, YK, YOki, SYo, MO, AH, HImai, YOku and YT performed statistical
analysis. CY, RM, AZ, YN, AS, HO, AH, HImai and YOku provided
administrative, technical or material support. YT supervised the
study. TS and CY confirm the authenticity of all the raw data. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All study-related procedures were performed as per
The Declaration of Helsinki, wherein a written informed consent was
obtained from each patient, and the Institutional Review Board
(IRB) of Medical Ethics Committee of Mie University Graduate School
of Medicine approved this study (IRB number: H2023-172).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
GPX4
|
glutathione peroxidase 4
|
|
HSPB1
|
heat shock protein β-1
|
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gill S, Loprinzi CL, Sargent DJ, Thomé SD,
Alberts SR, Haller DG, Benedetti J, Francini G, Shepherd LE,
Francois Seitz J, et al: Pooled analysis of fluorouracil-based
adjuvant therapy for stage II and III colon cancer: Who benefits
and by how much? J Clin Oncol. 22:1797–1806. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyerhardt JA: Adjuvant therapy for stage
II and III colon cancer. Clin Adv Hematol Oncol. 8:772–774.
2010.PubMed/NCBI
|
|
4
|
Wu C: Systemic therapy for colon cancer.
Surg Oncol Clin N Am. 27:235–242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dallas NA, Xia L, Fan F, Gray MJ, Gaur P,
van Buren G II, Samuel S, Kim MP, Lim SJ and Ellis LM:
Chemoresistant colorectal cancer cells, the cancer stem cell
phenotype, and increased sensitivity to insulin-like growth
factor-I receptor inhibition. Cancer Res. 69:1951–1957. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu T, Li Z, Gao CY and Cho CH: Mechanisms
of drug resistance in colon cancer and its therapeutic strategies.
World J Gastroenterol. 22:6876–6889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ganesh K, Stadler ZK, Cercek A, Mendelsohn
RB, Shia J, Segal NH and Diaz LA Jr: Immunotherapy in colorectal
cancer: Rationale, challenges and potential. Nat Rev Gastroenterol
Hepatol. 16:361–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Guo BC, Sun LR, Wang JW, Fu XH,
Zhang SZ, Poston G and Ding KF: TNM staging of colorectal cancer
should be reconsidered by T stage weighting. World J Gastroenterol.
20:5104–5112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gallois C, Pernot S, Zaanan A and Taieb J:
Colorectal cancer: Why does side matter? Drugs. 78:789–798. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duffy MJ: Carcinoembryonic antigen as a
marker for colorectal cancer: Is it clinically useful? Clin Chem.
47:624–630. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: an iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Wei Z, Pan K, Li J and Chen Q: The
function and mechanism of ferroptosis in cancer. Apoptosis.
25:786–798. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedmann Angeli JP, Schneider M, Proneth
B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch
A, Eggenhofer E, et al: Inactivation of the ferroptosis regulator
Gpx4 triggers acute renal failure in mice. Nat Cell Biol.
16:1180–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsushita M, Freigang S, Schneider C,
Conrad M, Bornkamm GW and Kopf M: T cell lipid peroxidation induces
ferroptosis and prevents immunity to infection. J Exp Med.
212:555–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen L, Hambright WS, Na R and Ran Q:
Ablation of the ferroptosis inhibitor glutathione peroxidase 4 in
neurons results in rapid motor neuron degeneration and paralysis. J
Biol Chem. 290:28097–28106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi ZZ, Fan ZW, Chen YX, Xie XF, Jiang W,
Wang WJ, Qiu YT and Bai J: Ferroptosis in carcinoma: Regulatory
mechanisms and new method for cancer therapy. Onco Targets Ther.
12:11291–11304. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen P, Li X, Zhang R, Liu S, Xiang Y,
Zhang M, Chen X, Pan T, Yan L, Feng J, et al: Combinative treatment
of β-elemene and cetuximab is sensitive to KRAS mutant colorectal
cancer cells by inducing ferroptosis and inhibiting
epithelial-mesenchymal transformation. Theranostics. 10:5107–5119.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharma P, Shimura T, Banwait JK and Goel
A: Andrographis-mediated chemosensitization through activation of
ferroptosis and suppression of β-catenin/Wnt-signaling pathways in
colorectal cancer. Carcinogenesis. 41:1385–1394. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shimura T, Sharma P, Sharma GG, Banwait JK
and Goel A: Enhanced anti-cancer activity of andrographis with
oligomeric proanthocyanidins through activation of metabolic and
ferroptosis pathways in colorectal cancer. Sci Rep. 11:75482021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma R, Shimura T, Yin C, Okugawa Y,
Kitajima T, Koike Y, Okita Y, Ohi M, Uchida K, Goel A, et al:
Antitumor effects of andrographis via ferroptosis-associated genes
in gastric cancer. Oncol Lett. 22:5232021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie Y, Hou W, Song X, Yu Y, Huang J, Sun
X, Kang R and Tang D: Ferroptosis: Process and function. Cell Death
Differ. 23:369–379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sharma A and Flora SJS: Positive and
negative regulation of ferroptosis and its role in maintaining
metabolic and redox homeostasis. Oxid Med Cell Longev.
2021:90742062021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Li J, Kang R, Klionsky DJ and Tang
D: Ferroptosis: Machinery and regulation. Autophagy. 17:2054–2081.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ouyang S, Li H, Lou L, Huang Q, Zhang Z,
Mo J, Li M, Lu J, Zhu K, Chu Y, et al: Inhibition of
STAT3-ferroptosis negative regulatory axis suppresses tumor growth
and alleviates chemoresistance in gastric cancer. Redox Biol.
52:1023172022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu W, Zhang C, Wu R, Sun Y, Levine A and
Feng Z: Glutaminase 2, a novel p53 target gene regulating energy
metabolism and antioxidant function. Proc Natl Acad Sci USA.
107:7455–7460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu S, Li T, Liu W and Huang Y: Ferroptosis
and cancer: Complex relationship and potential application of
exosomes. Front Cell Dev Biol. 9:7337512021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shimura T, Toiyama Y, Tanaka K, Saigusa S,
Kitajima T, Kondo S, Okigami M, Yasuda H, Ohi M, Araki T, et al:
Angiopoietin-like protein 2 as a predictor of early recurrence in
patients after curative surgery for gastric cancer. Anticancer Res.
35:4633–4639. 2015.PubMed/NCBI
|
|
31
|
Ichikawa T, Okugawa Y, Toiyama Y, Tanaka
K, Yin C, Kitajima T, Kondo S, Shimura T, Ohi M, Araki T and
Kusunoki M: Clinical significance and biological role of L1 cell
adhesion molecule in gastric cancer. Br J Cancer. 121:1058–1068.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mori K, Toiyama Y, Otake K, Ide S, Imaoka
H, Okigami M, Okugawa Y, Fujikawa H, Saigusa S, Hiro J, et al:
Successful identification of a predictive biomarker for lymph node
metastasis in colorectal cancer using a proteomic approach.
Oncotarget. 8:106935–106947. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shigemori T, Toiyama Y, Okugawa Y,
Yamamoto A, Yin C, Narumi A, Ichikawa T, Ide S, Shimura T, Fujikawa
H, et al: Soluble PD-L1 expression in circulation as a predictive
marker for recurrence and prognosis in gastric cancer: Direct
comparison of the clinical burde between tissue and serum PD-L1
expression. Ann Surg Oncol. 26:876–883. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stewart MJ and Watson ID: Standard units
for expressing drug concentrations in biological fluids. Br J Clin
Pharmacol. 16:3–7. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Zhang Z, Sun W, Zhang J, Xu Q,
Zhou X and Mao L: Ferroptosis in colorectal cancer: Potential
mechanisms and effective therapeutic targets. Biomed Pharmacother.
153:1135242022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang X, You Z, Chen X and Li J: Targeting
ferroptosis in colorectal cancer. Metabolites. 12:7452022.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen HHW and Kuo MT: Role of glutathione
in the regulation of Cisplatin resistance in cancer chemotherapy.
Met Based Drugs. 2010:4309392010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sui X, Zhang R, Liu S, Duan T, Zhai L,
Zhang M, Han X, Xiang Y, Huang X, Lin H and Xie T: RSL3 drives
ferroptosis through GPX4 inactivation and ROS production in
colorectal cancer. Front Pharmacol. 9:13712018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tian X, Li S and Ge G: Apatinib promotes
ferroptosis in colorectal cancer cells by targeting ELOVL6/ACSL4
signaling. Cancer Manag Res. 13:1333–1342. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chaudhary N, Choudhary BS, Shah SG,
Khapare N, Dwivedi N, Gaikwad A, Joshi N, Raichanna J, Basu S,
Gurjar M, et al: Lipocalin 2 expression promotes tumor progression
and therapy resistance by inhibiting ferroptosis in colorectal
cancer. Int J Cancer. 149:1495–1511. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang R, Su Q, Yin H, Wu D, Lv C and Yan Z:
Inhibition of SRSF9 enhances the sensitivity of colorectal cancer
to erastin-induced ferroptosis by reducing glutathione peroxidase 4
expression. Int J Biochem Cell Biol. 134:1059482021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xiang R, Fu J, Ge Y, Ren J, Song W and Fu
T: Identification of subtypes and a prognostic gene signature in
colon cancer using cell differentiation trajectories. Front Cell
Dev Biol. 9:7055372021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ousingsawat J, Schreiber R and Kunzelmann
K: TMEM16F/anoctamin 6 in ferroptotic cell death. Cancers (Basel).
11:6252019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo C, Liu P, Deng G, Han Y, Chen Y, Cai
C, Shen H, Deng G and Zeng S: Honokiol induces ferroptosis in colon
cancer cells by regulating GPX4 activity. Am J Cancer Res.
11:3039–3054. 2021.PubMed/NCBI
|
|
46
|
Chen Y, Zhang F, Du Z, Xie J, Xia L, Hou
X, Hao E and Deng J: Proteome analysis of Camellia nitidissima Chi
revealed its role in colon cancer through the apoptosis and
ferroptosis pathway. Front Oncol. 11:7271302021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He J, Ding H, Li H, Pan Z and Chen Q:
Intra-tumoral expression of SLC7A11 is associated with immune
microenvironment, drug resistance, and prognosis in cancers: A
pan-cancer analysis. Front Genet. 12:7708572021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gnanapradeepan K, Basu S, Barnoud T,
Budina-Kolomets A, Kung CP and Murphy ME: The p53 tumor suppressor
in the control of metabolism and ferroptosis. Front Endocrinol
(Lausanne). 9:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wei G, Sun J, Hou Z, Luan W, Wang S, Cui
S, Cheng M and Liu Y: Novel antitumor compound optimized from
natural saponin Albiziabioside A induced caspase-dependent
apoptosis and ferroptosis as a p53 activator through the
mitochondrial pathway. Eur J Med Chem. 157:759–772. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wei R, Zhao Y, Wang J, Yang X, Li S, Wang
Y, Yang X, Fei J, Hao X, Zhao Y, et al: Tagitinin C induces
ferroptosis through PERK-Nrf2-HO-1 signaling pathway in colorectal
cancer cells. Int J Biol Sci. 17:2703–2717. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang L, WenTao T, ZhiYuan Z, Qi L, YuXiang
L, Peng Z, Ke L, XiaoNa J, YuZhi P, MeiLing J, et al: Cullin-9/p53
mediates HNRNPC degradation to inhibit erastin-induced ferroptosis
and is blocked by MDM2 inhibition in colorectal cancer. Oncogene.
41:3210–3221. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cancer Genome Atlas Research Network;
Analysis Working Group; Asan University; BC Cancer Agency; Brigham
and Women's Hospital; Broad Institute; Brown University; Case
Western Reserve University; Dana-Farber Cancer Institute; Duke
University; Greater Poland Cancer Centre, et al, . Integrated
genomic characterization of oesophageal carcinoma. Nature.
541:169–175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shao Y, Jia H, Huang L, Li S, Wang C,
Aikemu B, Yang G, Hong H, Yang X, Zhang S, et al: An original
ferroptosis-related gene signature effectively predicts the
prognosis and clinical status for colorectal cancer patients. Front
Oncol. 11:7117762021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chatterjee S and Burns TF: Targeting heat
shock proteins in cancer: A promising therapeutic approach. Int J
Mol Sci. 18:19782017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wu J, Liu T, Rios Z, Mei Q, Lin X and Cao
S: Heat shock proteins and cancer. Trends Pharmacol Sci.
38:226–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X,
Wang H, Cao L and Tang D: HSPB1 as a novel regulator of ferroptotic
cancer cell death. Oncogene. 34:5617–5625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Garrido C, Brunet M, Didelot C, Zermati Y,
Schmitt E and Kroemer G: Heat shock proteins 27 and 70:
Anti-apoptotic proteins with tumorigenic properties. Cell Cycle.
5:2592–2601. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cheng J, Lv Z, Weng X, Ye S, Shen K, Li M,
Qin Y, Hu C, Zhang C, Wu J and Zheng S: Hsp27 acts as a master
molecular chaperone and plays an essential role in hepatocellular
carcinoma progression. Digestion. 92:192–202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu CL, Chen SF, Wu MZ, Jao SW, Lin YS,
Yang CY, Lee TY, Wen LW, Lan GL and Nieh S: The molecular and
clinical verification of therapeutic resistance via the p38
MAPK-Hsp27 axis in lung cancer. Oncotarget. 7:14279–14290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Oesterreich S, Weng CN, Qiu M, Hilsenbeck
SG, Osborne CK and Fuqua SA: The small heat shock protein hsp27 is
correlated with growth and drug resistance in human breast cancer
cell lines. Cancer Res. 53:4443–4448. 1993.PubMed/NCBI
|
|
63
|
Shimada T, Tsuruta M, Hasegawa H,
Okabayashi K, Shigeta K, Ishida T, Asada Y, Suzumura H, Koishikawa
K, Akimoto S and Kitagawa Y: Heat shock protein 27 knockdown using
nucleotide-based therapies enhances sensitivity to 5-FU
chemotherapy in SW480 human colon cancer cells. Oncol Rep.
39:1119–1124. 2018.PubMed/NCBI
|
|
64
|
Liang HH, Huang CY, Chou CW, Makondi PT,
Huang MT, Wei PL and Chang YJ: Heat shock protein 27 influences the
anti-cancer effect of curcumin in colon cancer cells through ROS
production and autophagy activation. Life Sci. 209:43–51. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hayashi R, Ishii Y, Ochiai H, Matsunaga A,
Endo T, Hasegawa H and Kitagawa Y: Suppression of heat shock
protein 27 expression promotes 5-fluorouracil sensitivity in colon
cancer cells in a xenograft model. Oncol Rep. 28:1269–1274. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ishida T, Ishii Y, Tsuruta M, Okabayashi
K, Akimoto S, Koishikawa K, Hasegawa H and Kitagawa Y: Cetuximab
promotes SN38 sensitivity via suppression of heat shock protein 27
in colorectal cancer cells with wild-type RAS. Oncol Rep.
38:926–932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu Z, Liu Y, Long Y, Liu B and Wang X:
Role of HSP27 in the multidrug sensitivity and resistance of colon
cancer cells. Oncol Lett. 19:2021–2027. 2020.PubMed/NCBI
|
|
68
|
Yang WS and Stockwell BR: Synthetic lethal
screening identifies compounds activating iron-dependent,
nonapoptotic cell death in oncogenic-RAS-harboring cancer cells.
Chem Biol. 15:234–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dixon SJ, Winter GE, Musavi LS, Lee ED,
Snijder B, Rebsamen M, Superti-Furga G and Stockwell BR: Human
haploid cell genetics reveals roles for lipid metabolism genes in
nonapoptotic cell death. ACS Chem Biol. 10:1604–1609. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang X, Ma Y, Ma J, Yang L, Song Q, Wang
H and Lv G: Glutathione peroxidase 4 as a therapeutic target for
anti-colorectal cancer drug-tolerant persister cells. Front Oncol.
12:9136692022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Benson AB, Venook AP, Al-Hawary MM, Arain
MA, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Farkas L, et
al: Colon cancer, version 2.2021, NCCN clinical practice guidelines
in oncology. J Natl Compr Canc Netw. 19:329–359. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hashiguchi Y, Muro K, Saito Y, Ito Y,
Ajioka Y, Hamaguchi T, Hasegawa K, Hotta K, Ishida H, Ishiguro M,
et al: Japanese Society for Cancer of the Colon and Rectum (JSCCR)
guidelines 2019 for the treatment of colorectal cancer. Int J Clin
Oncol. 25:1–42. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yamazaki K, Yamanaka T, Shiozawa M, Manaka
D, Kotaka M, Gamoh M, Shiomi A, Makiyama A, Munemoto Y, Rikiyama T,
et al: Oxaliplatin-based adjuvant chemotherapy duration (3 vs. 6
months) for high-risk stage II colon cancer: The randomized phase
III ACHIEVE-2 trial. Ann Oncol. 32:77–84. 2021. View Article : Google Scholar : PubMed/NCBI
|