Introduction
Lung cancer is the most common malignant tumor,
ranking as the second most prevalent and lethal malignant tumor
globally (1). Lung cancer can be
broadly classified into two main subtypes: i) Non-small cell lung
cancer (NSCLC) and ii) small cell lung cancer, from both
pathological and therapeutic perspectives. Lung adenocarcinoma
(LUAD) is a pathological subtype of NSCLC, accounting for ~45% of
all cases (2). Despite important
advancements in molecular targeting, chemotherapy, radiation
therapy and immunotherapy, the prognosis of LUAD remains
unfavorable, with a 5-year overall survival rate of ~15% (3,4).
Previous studies have demonstrated the pivotal role of genetic
analysis in the diagnosis and treatment of lung cancer (5–7). For
instance, the polymorphisms of the epidermal growth factor receptor
are associated with side effects and outcome of tyrosine kinase
inhibitor therapy administered to patients with NSCLC (5–7).
Therefore, there is an urgent need for novel and effective
screening methods based on genetic analysis to improve diagnostic
accuracy and treatment efficiency, in order to improve the
prognosis of patients with LUAD.
The Kelch like ECH associated protein 1
(KEAP1)/nuclear factor erythroid 2-related factor 2 (NRF2)/heme
oxygenase 1 (HO-1) pathway serves a pivotal role in metabolic
reprogramming, immune remodeling and treatment resistance in LUAD
(8). In total, 20–30% of LUADs
exhibit KEAP1 loss-of-function mutations, leading to aberrant
activation of NRF2 and subsequently resulting in poor prognosis
(9,10). KEAP1 mutations facilitate the
progression of LUAD by augmenting glutamine metabolism and
upregulating solute carrier family 33 member 1 expression (11,12).
Furthermore, KEAP1-mutant tumors diminish dendritic cell and T cell
responses, thus driving immunotherapy resistance in LUAD (13). NRF2, a transcription factor, serves
a crucial role in the antioxidant pathway, promoting tumor cell
growth, proliferation and drug resistance (14). NRF2 is the target of KEAP1 and
abnormal activation of NRF2 is not only associated with KEAP1
mutations but also with gain-of-function mutations in NRF2
(15). Previous studies have found
multiple gain-of-function mutation sites affecting NRF2 in LUAD
(16). NRF2 mutations promote tumor
cell immune escape by inhibiting stimulator of interferon response
CGAMP interactor 1 activation (17). HO-1, which is regulated by NRF2,
serves a crucial role as an antioxidant factor. When HO-1
expression is dysregulated, it can promote the proliferation of
lung cancer cells and make them resistant to radiotherapy (18,19).
However, several crucial and unresolved issues remain, encompassing
the potential reclassification of LUAD based on KEAP1/NRF2/HO-1
mutations, the intricate interplay between KEAP1/NRF2/HO-1
mutations and the tumor microenvironment, as well as the strategic
implementation of therapeutic interventions to effectively control
tumor progression contingent upon KEAP1/NRF2/HO-1 mutations.
Therefore, the present study aimed to identify
specific genes regulated by gene mutations of the KEAP1/NRF2/HO-1
axis and construct a prognostic model using upregulated genes. All
patients were stratified into low- or high-risk groups based on the
risk score, and the diagnostic efficacy was validated in both the
training and test sets. Furthermore, the association between risk
score and immune cell infiltration in the tumor microenvironment
was explored and the individualized sensitivity to chemotherapy and
immunotherapy were simultaneously predicted for each patient with
LUAD.
Materials and methods
Validation of the expression of KEAP1,
NRF2, HO-1 and Ki-67
Postoperative paraffin-embedded tissue blocks of
patients with LUAD who underwent surgical procedures at Xishan
People's Hospital of Wuxi City (Wuxi, China) between January 2020
and June 2023 were included in the present study. The inclusion
criterion was a histologically confirmed diagnosis of LUAD, whilst
the exclusion criteria were prior chemotherapy or radiation
therapy. After selection, a total of 104 tissue wax blocks from
patients with LUAD were included in the study. The patients
consisted of 42 men and 62 women, with an mean age of 66.6 years
(range, 32–92 years). The present study was approved by the Ethics
Committee of Xishan People's Hospital (approval no.
xs2023ky021).
Tissue sections (5 µm) were prepared and the
expression of KEAP1, NRF2 and HO-1 was assessed using
immunohistochemistry as previously described (19), using anti-KEAP1 (cat. no. ab226997;
1:100 dilution; Abcam), anti-NRF2 (cat. no. ab313825; 1:100;
Abcam), anti-HO-1 (cat. no. ab137749; 1:100; Abcam) and anti-Ki-67
(cat. no. ab230460; 1:100; Abcam) antibodies as the primary
antibodies. The Elivision™ plus Polyer HRP (mouse/rabbit) IHC Kit
(cat. no. KIT-9902; Maixin Biotech. Co., Ltd.) served as the
secondary antibody reagent in this study.
Data acquisition
High-throughput sequence-fragments per kilobase of
transcript per million mapped reads data and clinicopathological
information of patients with LUAD were acquired from The Cancer
Genome Atlas (TCGA)-LUAD database (https://portal.gdc.cancer.gov/). Gene expression and
survival data associated with the GSE68465 (20) dataset were obtained from the Gene
Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Gene mutation
information for KEAP1, NRF2 and HO-1 was retrieved from the cBio
Cancer Genomics Portal (cBioPortal) website (http://www.cbioportal.org/). The association between
KEAP1, NRF2 and HO-1 expression levels and patient survival was
analyzed using the Kaplan-Meier Plotter website (https://kmplot.com/analysis/index.php?p=service)
(21). All patients diagnosed with
lung adenocarcinoma across the 17 datasets were included in this
study. All possible cut-off values between the lower and upper
quartiles were computed and the best performing threshold was used
as a cut-off.
Identification of mutation-associated
differentially expressed genes
Based on the mutation information obtained from the
cBioPortal website, 112 tumor samples were classified into the
mutation group, 403 tumor samples into the wild-type group and 59
normal pulmonary tissues into the normal group. The ‘limma’ package
in R (version 4.1.3; http://www.r-project.org) was utilized to identify
differentially expressed genes (log2 fold-change >1;
P<0.05) by comparing gene expression between the mutant group
and both the wild-type and normal groups (22).
Construction of a prognostic
model
Candidate genes in the TCGA-LUAD and GSE68465
datasets were initially screened using univariate Cox analysis.
Subsequently, 101 combinations of algorithms were used to construct
a prognostic model based on the leave-one-out cross-validation
framework (23). The TCGA-LUAD
dataset was utilized as the training group, whilst the GSE68465
dataset served as the validation group. Furthermore, the
consistency index (C-index) of each model was calculated in the
validation group (23).
Survival analysis and nomogram
construction based on clinical characteristics
Univariate and multivariate Cox regression analyses
were used to identify independent prognostic factors in patients
with LUAD. The ‘rms’ package in R (version 4.1.3) was utilized for
constructing a nomogram based on risk score and two characteristic
factors. Calibration curves were employed to evaluate the accuracy
of these predictions. Additionally, decision curves generated using
the ‘rmda’ package in R (version 4.1.3) were used to assess the
predictive utility of the nomogram and to compare different
prediction models.
Immune infiltration assessment and
immunotherapy response prediction
The ‘ESTIMATE’ package in R (version 4.1.3) was used
to evaluate the levels of immune cells and stromal cells in tumor
tissues. The ImmuCellAI website (https://guolab.wchscu.cn/ImmuCellAI/#!/) was utilized
for the analysis of immune cell types and levels within tumor
tissues, facilitating a comparative assessment of immune cell
levels between low- and high-risk groups (24). The effectiveness of immunotherapy in
the aforementioned groups was evaluated through the prediction of
individual response to immunotherapy among patients with LUAD.
Assessment of chemotherapy
effectiveness
The ‘oncoPredict’ package in R (version 4.1.3) was
used to predict chemotherapy drug sensitivity in patients with
LUAD, aiming to compare the sensitivity to chemotherapy drugs
between the high- and low-risk groups (25).
Expression of hub genes at the
single-cell level
The Tumor Immune Single-cell Hub website (http://tisch1.comp-genomics.org/) was utilized to
evaluate the expression of these hub genes in several cell
types.
Statistical analysis
Statistical analyses were performed using R software
(version 4.1.3). The Wilcoxon rank-sum test was used to determine
the significance of differences between two groups. Frequency
counts were used for statistical description of categorical
(qualitative) data, and comparisons between groups were performed
using either the χ2 test or the Fisher's exact test. All
P-values were two-tailed. P<0.05 was considered to indicate a
statistically significant difference.
Results
Association between KEAP, NRF2 and
HO-1 expression and clinicopathology
The expression of KEAP1, NRF2 and HO-1 was assessed
using immunohistochemistry based on 104 cases of LUAD and their
association with pathological characteristics was evaluated. The
immunohistochemical analysis revealed that KEAP1 and HO-1 were
predominantly expressed in the cytoplasm, whereas NRF2 and Ki-67
were primarily localized in the nucleus (Fig. 1). No significant association was
demonstrated between the expression of KEAP1 and clinical features
(Table I). Compared to the
NRF2-negative group, the NRF2-positive group exhibited enhanced
tumor aggressiveness, higher proportions of lymph node and distant
metastases, more advanced TNM stage (26) and increased proliferative activity
(Table I). Compared to the
HO-1-negative group, the HO-1-positive group exhibited
significantly higher grades in T stage, N stage, M stage and the
overall TNM staging system. Additionally, the HO-1 positive group
demonstrated a larger tumor diameter and a markedly increased
expression level of Ki-67 (Table
I).
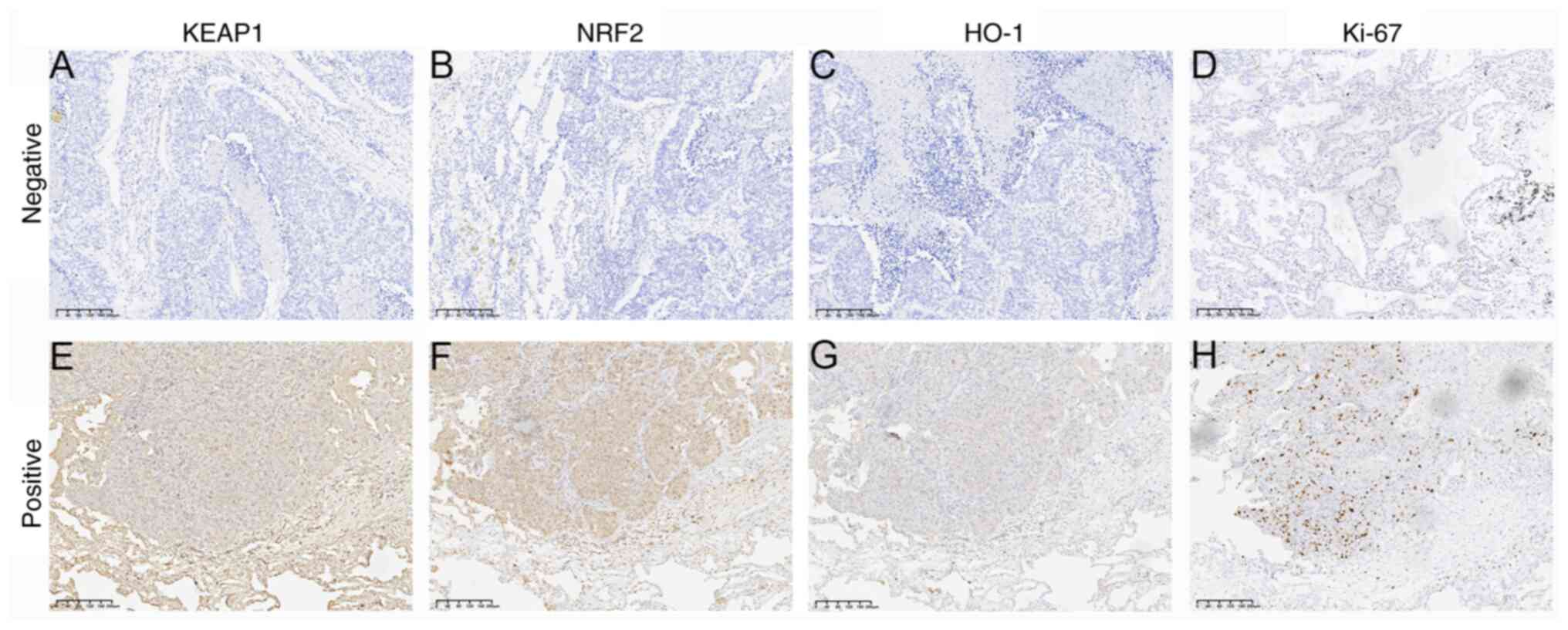 | Figure 1.Immunohistochemical detection of
KEAP1, NRF2, HO-1 and Ki-67 expression in lung adenocarcinoma
tissues. Negative expression of (A) KEAP1, (B) NRF2, (C) HO-1 and
(D) Ki-67. Positive expression of (E) KEAP1, (F) NRF2, (G) HO-1 and
(H) Ki-67 (magnification, ×100). KEAP1, Kelch like ECH associated
protein 1; NRF2, nuclear factor erythroid 2-related factor 2; HO-1,
heme oxygenase 1. |
 | Table I.Association between KEAP1/NRF2/HO-1
expression and clinicopathological features in lung
adenocarcinoma. |
Table I.
Association between KEAP1/NRF2/HO-1
expression and clinicopathological features in lung
adenocarcinoma.
| A, KEAP1
expression |
|---|
|
|---|
| Clinicopathological
feature | Positive
(n=72) | Negative
(n=32) | P-value |
|---|
| Age, years | 66.04±11.58 | 67.78±11.15 | 0.47 |
| Sex |
|
| 0.80 |
|
Female | 44 | 18 |
|
|
Male | 28 | 14 |
|
| T stage |
|
| 0.06 |
|
T1-2 | 62 | 32 |
|
|
T3-4 | 10 | 0 |
|
| N stage |
|
| 0.50 |
| N0 | 50 | 25 |
|
|
N1-3 | 22 | 7 |
|
| M stage |
|
| 0.09 |
| M0 | 63 | 32 |
|
| M1 | 9 | 0 |
|
| TNM stage |
|
| 0.12 |
|
I–II | 54 | 29 |
|
|
III–IV | 18 | 3 |
|
| Tumor diameter,
cm | 2.01±1.44 | 2.10±1.03 | 0.72 |
| Ki-67 expression
ratio, % | 41±15 | 39±13 | 0.37 |
|
| B, NRF2
expression |
|
|
Clinicopathological feature | Positive
(n=58) | Negative
(n=46) | P-value |
|
| Age, years | 68.38±11.57 | 64.30±10.93 | 0.07 |
| Sex |
|
| 0.44 |
|
Female | 37 | 25 |
|
|
Male | 21 | 21 |
|
| T stage |
|
| <0.01 |
|
T1-2 | 48 | 46 |
|
|
T3-4 | 10 | 0 |
|
| N stage |
|
| <0.01 |
| N0 | 34 | 41 |
|
|
N1-3 | 24 | 5 |
|
| M stage |
|
| 0.01 |
| M0 | 49 | 46 |
|
| M1 | 9 | 0 |
|
| TNM stage |
|
| <0.001 |
|
I–II | 39 | 44 |
|
|
III–IV | 19 | 2 |
|
| Tumor diameter,
cm | 2.52±1.50 | 1.43±0.68 | <0.001 |
| Ki-67 expression
ratio, % | 46±15 | 34±10 | <0.001 |
|
| C, HO-1
expression |
|
|
Clinicopathological feature | Positive
(n=63) | Negative
(n=41) | P-value |
|
| Age, years | 68.00±11.24 | 64.39±11.49 | 0.12 |
| Sex |
|
| 0.21 |
|
Female | 34 | 28 |
|
|
Male | 29 | 13 |
|
| T stage |
|
| 0.02 |
|
T1-2 | 53 | 41 |
|
|
T3-4 | 10 | 0 |
|
| N stage |
|
| <0.001 |
| N0 | 36 | 39 |
|
|
N1-3 | 27 | 2 |
|
| M stage |
|
| 0.03 |
| M0 | 54 | 41 |
|
| M1 | 9 | 0 |
|
| TNM stage |
|
| <0.001 |
|
I–II | 42 | 41 |
|
|
III–IV | 21 | 0 |
|
| Tumor diameter,
cm | 2.43±1.50 | 1.42±0.64 | <0.001 |
| Ki-67 expression
ratio, % | 45±15 | 33±9 | <0.001 |
Association between KEAP1, NRF2 and
HO-1 expression and prognosis of patients with LUAD
The association between the expression of KEAP1,
NRF2 and HO-1 and the prognosis of patients with LUAD was analyzed
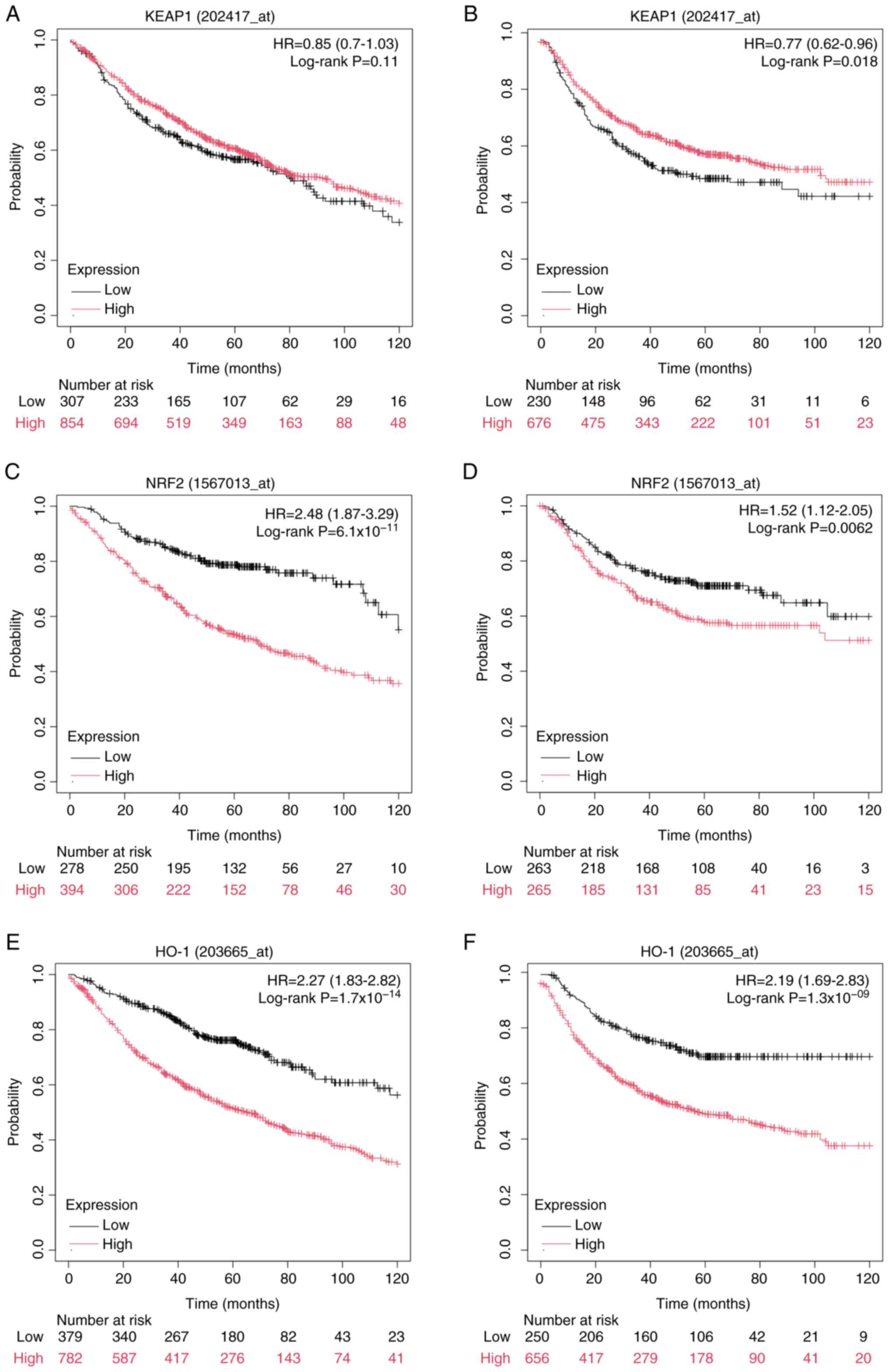
using the Kaplan-Meier Plotter website. The results indicated that,
compared with low KEAP1 expression, high KEAP1 expression was not
associated with overall survival (OS; Fig. 2A), but was significantly associated
with improved first progression survival (FPS; Fig. 2B). Conversely, high expression of
NRF2 and HO-1 was significantly associated with worse OS and FPS,
in comparison with low NRF2 and HO-1 expression (Fig. 2C-F). The association between KEAP1,
NRF2 and HO-1 expression levels and disease-specific survival
(DSS), disease-free interval and progression-free interval (PFI)
were further assessed based on data from the TCGA-LUAD dataset. The
results revealed that only the expression of HO-1 was significantly
associated with poor DSS and PFI (Fig.
S1).
Identification of KEAP1/NRF2/HO-1
mutation-mediated upregulated genes (KNHMUGs)
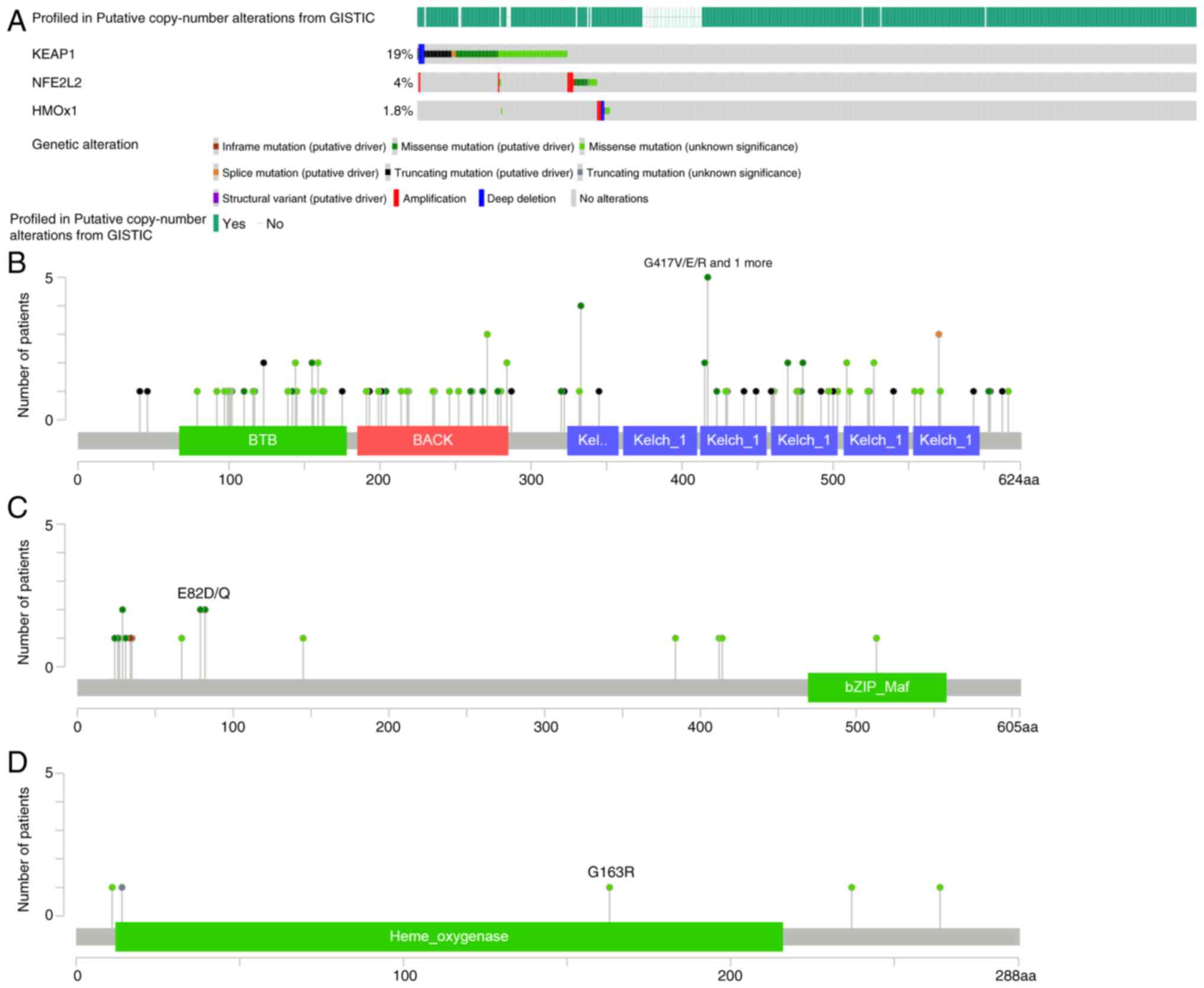
From the cBioPortal database, KEAP1, NRF2 and HO-1
mutations in LUAD from TCGA pan-cancer atlas, which included 566
patients, were analyzed. Genetic alterations, such as in-frame,
missense and truncation mutations of KEAP1 (19%), NRF2 (4%) and
HO-1 (1.8%), were identified in patients with LUAD (Fig. 3). Based on the status of
KEAP1/NRF2/HO-1 mutations, patients were classified into two
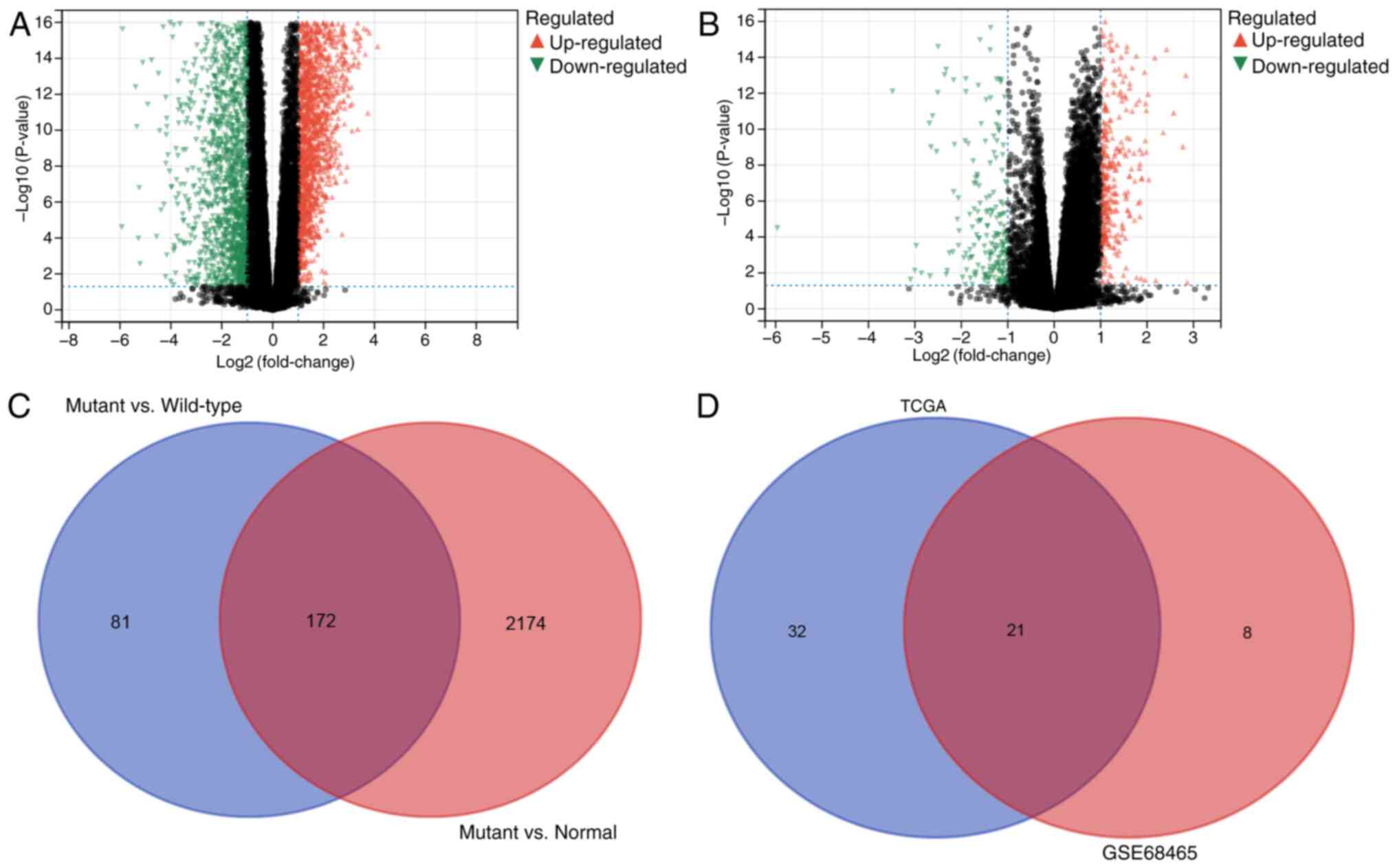
groups: i) Mutant and ii) wild-type. Subsequently, the differential
genes between the mutant, wild-type and normal groups were
compared. A total of 2,346 upregulated genes specific to the mutant
group were identified compared with the normal group (Fig. 4A), with 253 upregulated genes in the
mutant group compared with the wild-type group (Fig. 4B). Furthermore, 172 common
differentially upregulated genes in the mutant group were
identified using a Venn diagram (Fig.
4C).
Prognostic model based on KNHMUGs
Univariate Cox regression analysis was then
performed on 172 candidate genes in the TCGA and GSE68465 datasets.
The TCGA dataset identified a total of 53 genes associated with
prognosis, whereas the GSE68465 dataset revealed 29 genes that were
prognostically relevant. Ultimately, an overlap of 21 genes with
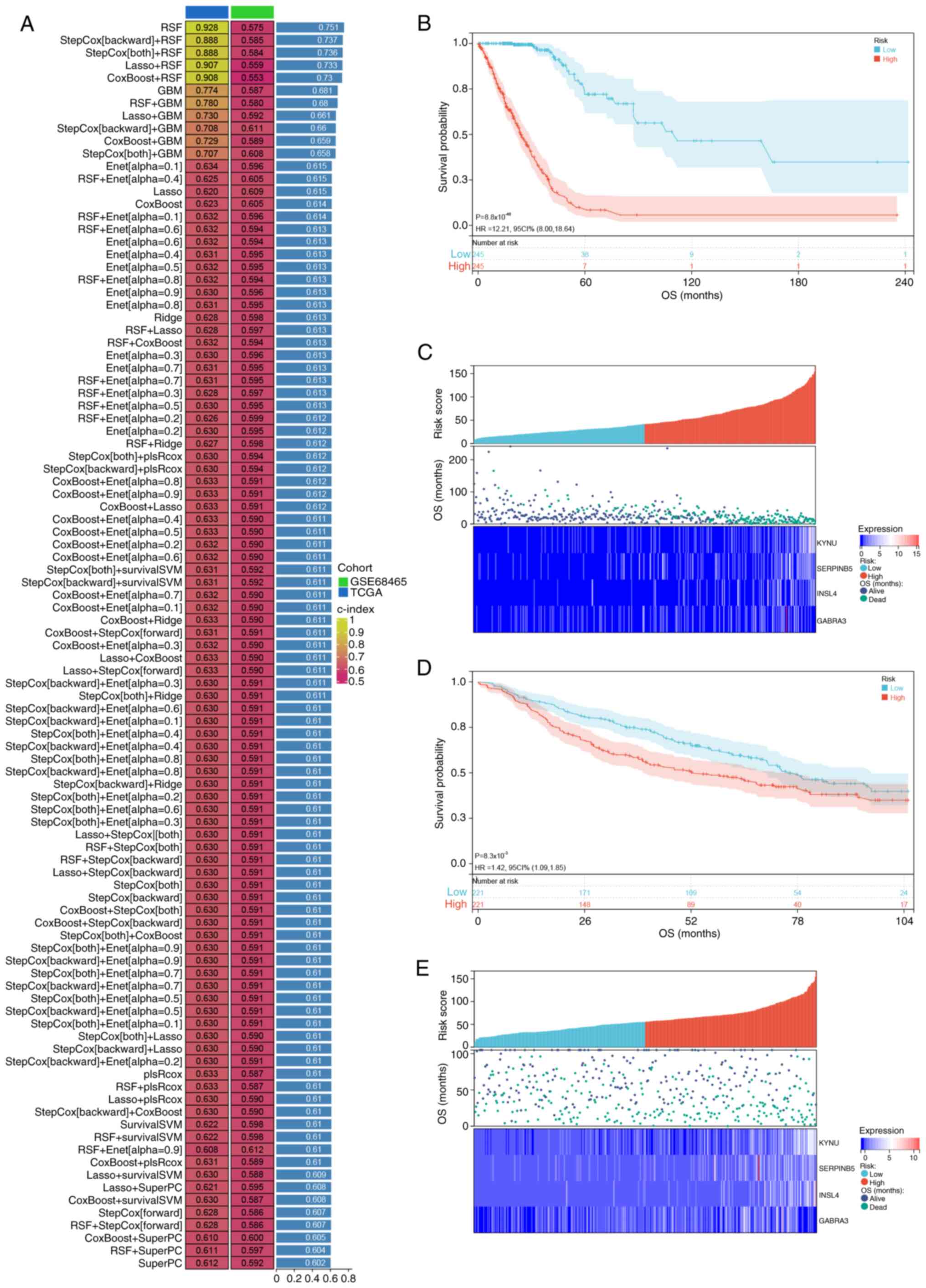
prognostic significance was observed in both datasets (Fig. 4D). Subsequently, these 21 genes were
subjected to analysis using a combination of machine algorithms
(101 combinations) with TCGA as the training set and GSE68465 as
the validation set. By evaluating the C-index of both sets and
considering the number of genes in optimal combinations, StepCox
(backward) + random survival forest (RSF) algorithms emerged as the
most effective prediction model (Fig.
5A), comprising four characterized genes, kynureninase (KYNU),
serpin family B member 5 (SERPINB5), insulin like 4 (INSL4) and
γ-aminobutyric acid type A receptor subunit α3 (GABRA3). Based on
the median risk score, patients were stratified into high- and
low-risk groups. Notably, OS was significantly lower for patients
classified as high-risk compared with that for patients categorized
as low-risk in both the TCGA [hazard ratio (HR), 12.21; 95%
confidence interval (CI), 8.00–18.64; P<0.001] and GSE68465 (HR,
1.36; 95% CI, 1.05–1.75; P=0.012) datasets (Fig. 5B and D). It was also observed that
the survival rate of patients significantly decreased as the risk
score increased. Notably, the KYNU, SERPINB5, INSL4 and GABRA3
genes were identified as risk factors with an increasing trend in
expression as the risk score rose (Fig.
5C and F).
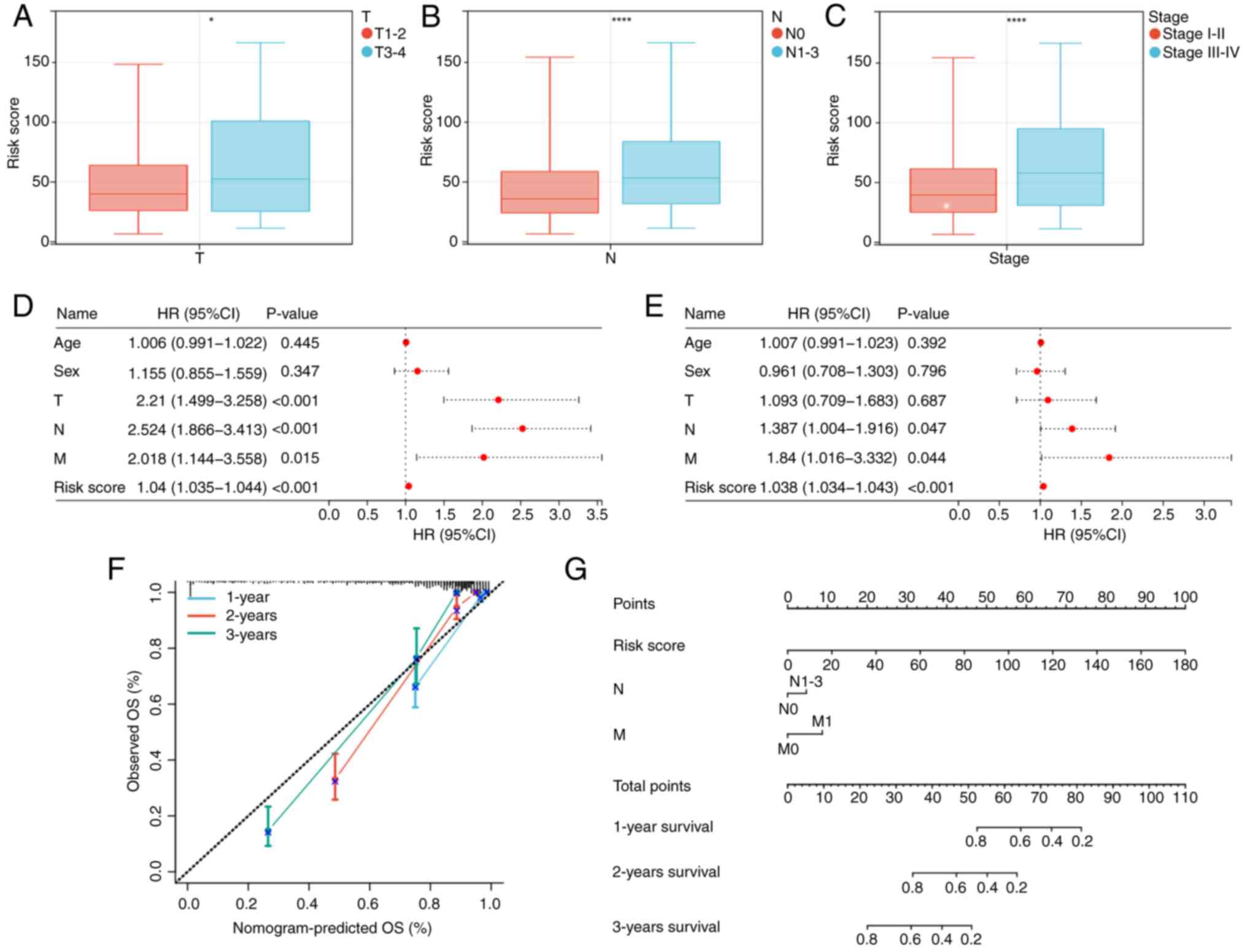
Association between risk score and
clinicopathological features
The association between risk score and
clinicopathological characteristics was assessed. Patients with
advanced-stage tumors [tumor stage (T) 3–4; lymph node stage
(N)1-3; and stage III–IV] demonstrated a significantly higher risk
score compared with those with early-stage tumors (T1-2; N0; and
stage I–II), suggesting that this could be one of the factors
contributing to the unfavorable prognosis observed in the high-risk
group (Fig. 6A-C).
Construction of the nomogram
Univariate and multivariate Cox regression analyses
were performed to assess the clinical characteristics and risk
score of 490 patients (Fig. 6D and
E). The results of the multivariate regression analysis
revealed that risk score, N stage and metastasis (M) stage
independently served as prognostic factors for patients with LUAD.
Consequently, a nomogram was constructed based on these three
independent risk factors (Fig. 6G)
and calibration curves demonstrated a notable agreement between the
predicted and actual values of the prediction model at 1, 2 and 3
years (Fig. 6F).
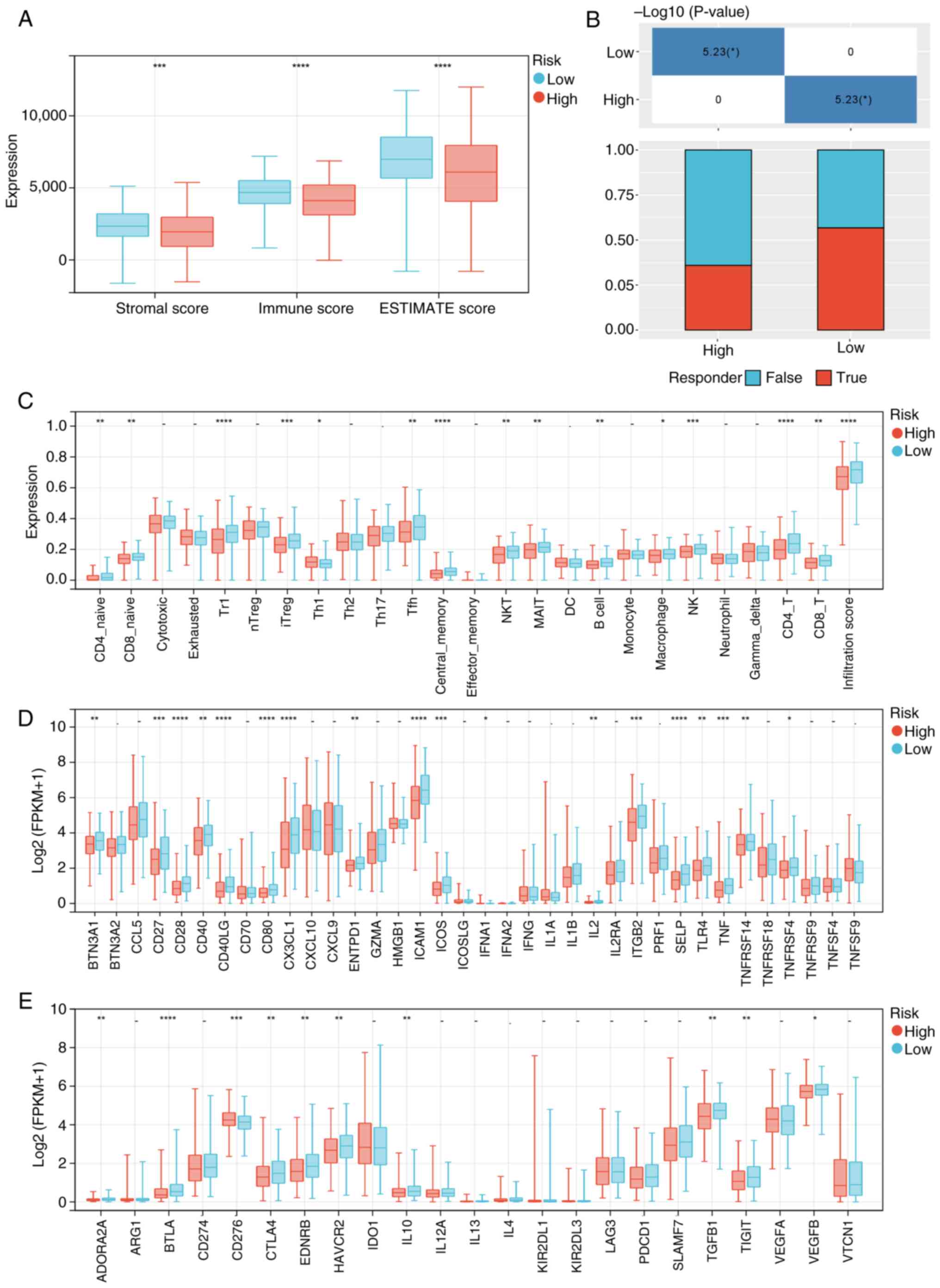
Association of risk score with immune
infiltration and immunotherapy
The ESTIMATE results revealed that the low-risk
group had significantly increased stromal and immune scores
compared with the high-risk group, indicating a higher level of
immune cell infiltration within this group (Fig. 7A). Subsequently, further analysis on
immune cell infiltration in tumor tissues was performed. It was
revealed that the low-risk group exhibited significantly enhanced
infiltration of CD4+ naive, CD8+ naive,
CD4+ T, CD8+ T, natural killer (NK), NKT
cells and other immune cells compared with the high-risk group
(Fig. 7C). Additionally, the
expression levels of immune checkpoint-related genes were assessed.
Notably, the low-risk group demonstrated a significantly increased
expression of immune checkpoint-stimulated genes such as CD27, CD80
and C-X3-C motif chemokine ligand 1, as well as immune
checkpoint-inhibited genes such as cytotoxic T-lymphocyte
associated protein 4 (CTLA4) and hepatitis A virus cellular
receptor 2 (HAVCR2), compared with the high-risk group (Fig. 7D and E). Furthermore, the efficacy
of immunotherapy in different subgroups was predicted, and it was
revealed that immune-checkpoint blockade therapy response
prediction indicated a higher rate of response in the low-risk
group compared with that in the high-risk group (Fig. 7B).
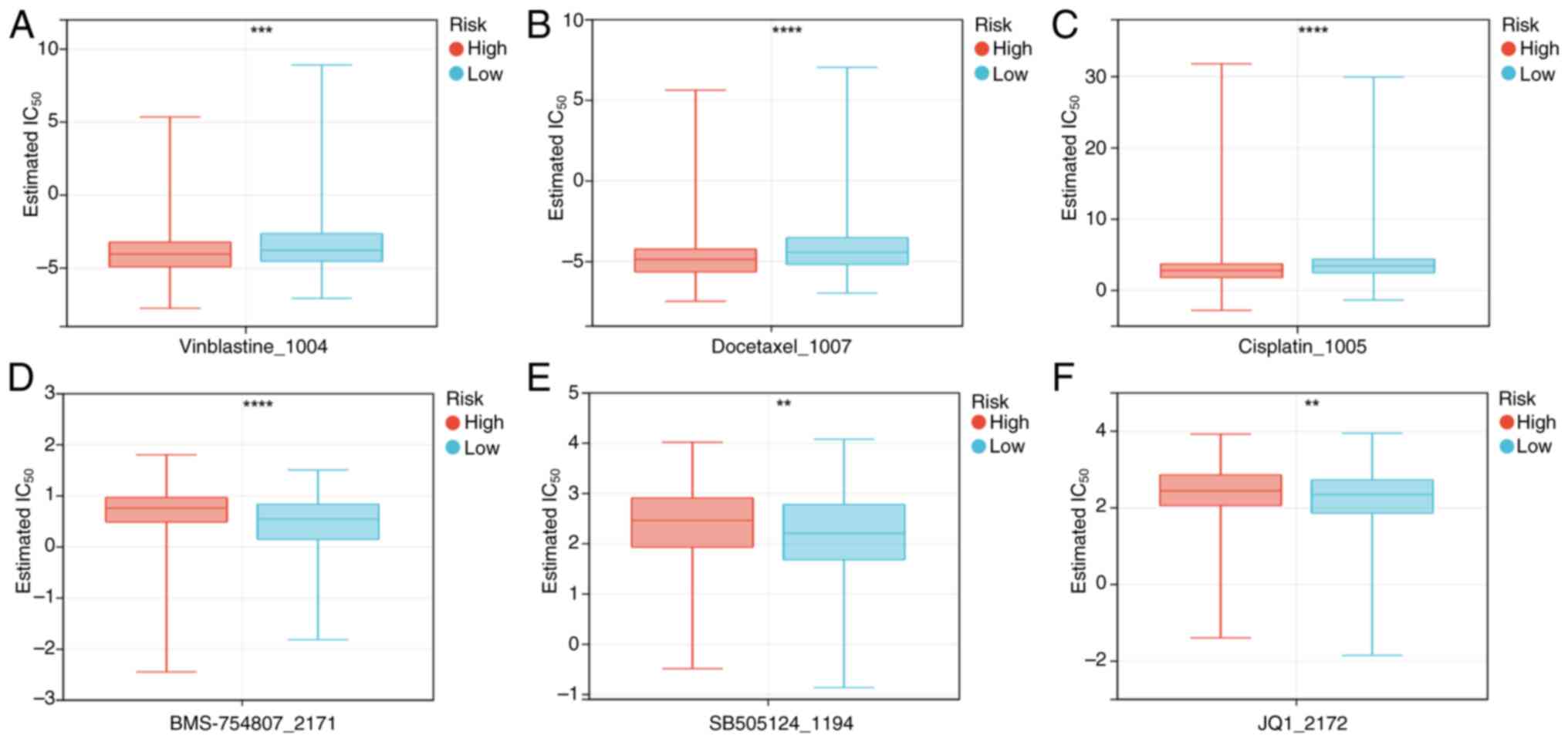
Association of risk score with drug
therapy
The ‘oncoPredict’ R package was used to compare the
differences in chemotherapy sensitivity between the high- and
low-risk groups. The results demonstrated that the high-risk group
had a significantly increased sensitivity to vincristine, docetaxel
and cisplatin, whilst the low-risk group exhibited a significantly
increased sensitivity to BMS_754807 (insulin receptor inhibitor),
SB505124_1194 (TGF-β receptor inhibitor) and JQ1_2172 [bromodomain
and extraterminal (BET) inhibitor] (Fig. 8).
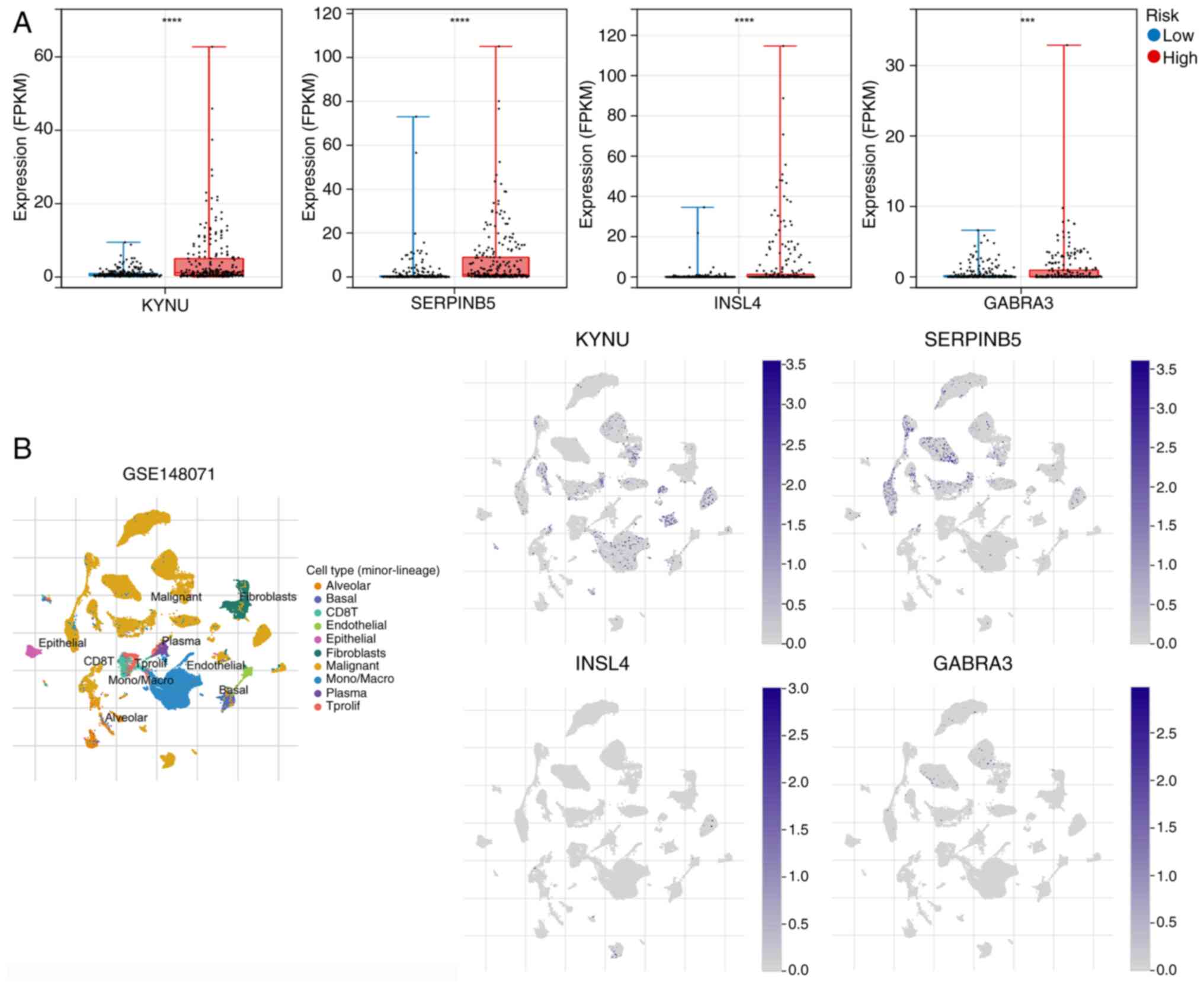
Expression of hub genes in different
risk groups and their localization in LUAD
To analyze the expression of hub genes in different
risk groups, the mRNA expression levels of hub genes were assessed.
The findings revealed that KYNU, SERPINB5, INSL4 and GABRA3 were
significantly upregulated in the high-risk group compared with that
of the low-risk group (P<0.05; Fig.
9A). Additionally, the Tumor Immune Single-cell Hub website was
utilized to evaluate the expression of these hub genes in several
cell types. Annotation analysis of the GSE148071 single-cell
dataset demonstrated that tumor tissue consists of diverse cells,
including malignant cells, epithelial cells and fibroblasts.
Notably, KYNU, SERPINB5, INSL4 and GABRA3 exhibited a predominant
expression in malignant cells (Fig.
9B). These results suggest that these hub genes primarily
contribute to LUAD progression by modulating biological functions,
specifically within malignant cells.
Discussion
A total of 20–30% LUAD tumors carry mutations in
KEAP1 and NRF2. Both loss-of-function mutations in KEAP1 and
gain-of-function mutations in NFE2-like BZIP transcription factor 2
can activate the KEAP1/NRF2 pathway in LUAD (15). Previous studies have demonstrated
that the KEAP1/NRF2/HO-1 axis inhibits cisplatin-induced
ferroptosis by upregulating the iron death-related genes solute
carrier family 7 member 11 and NAD(P)H quinone dehydrogenase 1
(27,28). Furthermore, the NRF2/HO-1 axis could
attenuate resistance to programmed cell death 1 immunotherapy by
inhibiting the transformation of M2 into M1 macrophages (29). Notably, KEAP1/NRF2 mutations are
considered a pivotal driver of immune-suppressive tumor
microenvironment metabolic reprogramming in LUAD, including potent
induction of T-regulatory cells and diminishing the number of
dendritic cells and tissue-resident memory CD8+ T cells, thereby
compromising the efficacy of immunotherapy (13,30,31).
However, combination therapy involving glutaminase inhibition and
immune checkpoint blockade is reported to hold promise for
reversing the immune suppression mediated by KEAP1/NRF2 mutations
(13,30). Mutations in the KEAP1/NRF2/HO-1
genes serve a crucial role in the development of LUAD. However,
there is a lack of established clinical models to assess the
prognosis, tumor microenvironment and response to chemotherapy and
immunotherapy in patients with LUAD based on genes regulated by
KEAP1/NRF2/HO-1 mutations.
The prognostic model in the present study consisted
of four genes: KYNU, SERPINB5, INSL4 and GABRA3. KYNU acts as a key
enzyme in tryptophan metabolism, and its metabolite
3-hydroxyanthranilic acid is notably associated with poor prognosis
in patients with NSCLC (32,33).
Further research reported that NRF2 upregulated KYNU, which was
positively associated with an inhibitory tumor microenvironment
(34). SERPINB2 belongs to the
family of protease inhibitors, which is associated with
tumorigenesis and regulation of several vital biological processes
(35,36). The expression of SERPINB5 has been
reported to be upregulated in LUAD, and its high expression levels
were notably associated with poor OS. Furthermore, overexpression
of SERPINB5 promoted cell proliferation, migration and invasion and
epithelial-mesenchymal transition (EMT) in LUAD cells (37). INSL4 is a member of the insulin
superfamily, which includes genes for insulin, insulin-like growth
factors and relaxin. In LUAD, high levels of INSL4 expression have
been reported to be associated with poor prognosis (38). Increased expression of INSL4 can
promote tumor cell proliferation and invasion (38,39).
GABRA3 acts as a key chloride ion transporter and also functions as
a subunit of γ-aminobutyric acid A receptors (40). In LUAD, increased expression of
GABRA3 has been closely associated with poor prognosis. By
upregulating the expression of MMP-2 and MMP-9 via activation of
the JNK/activator protein-1 signaling pathway, GABRA3 facilitates
lymphatic metastasis in LUAD (41).
Therefore, the aforementioned four genes are associated with the
pathogenesis and progression of LUAD and may be considered
potential therapeutic targets. Currently, there are studies
investigating the regulatory association between KEAP1, NRF2 and
HO-1 and KYNU (34). Subsequent
research should further explore the interaction between KEAP1, NRF2
and HO-1, and SERPINB5, along with INSL4 and GABRA3.
The tumor microenvironment consists of diverse
immune cells, stromal cells, extracellular matrix and several
cytokines (42). These different
components not only impact patient prognosis but also have a close
association with immunotherapy (43). The present study revealed that the
low-risk group exhibited a higher percentage of infiltration of
CD4+ T, CD8+ T, NK and NKT cells compared
with the high-risk group. Previous research has demonstrated that
an increased presence of CD4+ T and CD8+ T
cells is associated with improved prognosis and enhanced
immunotherapeutic efficacy (44).
NK cells are a subset of innate immune cells capable of rapidly
recognizing and eliminating tumor cells whilst also promoting
adaptive T-cell immune responses to suppress tumor aggressiveness
(45). NKT cells represent a class
of lymphocytes expressing both NK and T cell surface markers. They
serve as a link between intrinsic and adaptive immunity and serve a
crucial role in the tumor immune microenvironment (46). Therefore, increased infiltration of
these cells may explain why the low-risk group exhibited improved
prognosis compared with the high-risk group.
At present, research on immune checkpoint inhibitors
is a hot topic. The current study assessed the expression of immune
checkpoint suppressor genes in both the high- and low-risk groups.
The results revealed high expression of immune checkpoint
inhibitors, including CTLA4, T cell immunoreceptor with Ig and ITIM
domains (TIGIT), HAVCR2 and B and T lymphocyte associated protein
(BTLA) in the low-risk group. CTLA4 is a suppressor molecule
located on the surface of T cells that can promote tumor tolerance
and T-cell exhaustion when abnormally expressed (47). TIGIT is an immune checkpoint gene
expressed on several types of T and NK cells. Its binding to CD155
on immune-suppressive cells or tumor cell surfaces inhibits immune
cell function (48). HAVCR2, as a
negatively regulated immune checkpoint, not only regulates the
exhaustion of T and NK cells, but also regulates the intrinsic
immune escape of tumors mediated by macrophages and dendritic cells
(49). BTLA is a suppressor
expressed on the surface of immune cells such as T and B cells.
Upon binding to herpesvirus entry mediator, it sends inhibitory
signals to T cells, leading to a decrease in T cell proliferation
and activation (50). These
findings indicate that there are more immunotherapeutic targets
within the low-risk group with higher potential benefits from
immunotherapy compared with the high-risk group, aligning with the
predicted response rate for immunotherapy.
Notably, chemotherapy is a crucial component of LUAD
treatment. Half-maximal inhibitory concentration analysis was
performed to screen potential compounds and the findings revealed
that the high-risk group exhibited greater sensitivity to
vincristine, docetaxel and cisplatin, whilst the low-risk group
showed increased sensitivity to BMS_754807, SB505124_1194 and
JQ1_2172. Clinically, vincristine, docetaxel and cisplatin are
commonly utilized chemotherapeutic agents for LUAD treatment, which
are often administered in combination to enhance patient prognosis
(51). BMS-754807 is a potent
inhibitor of the insulin-like growth factor 1 receptor/insulin
receptor family of kinases that synergistically interacts with
platinum-based chemotherapeutics to induce apoptosis in lung cancer
cells (52). Furthermore, dasatinib
combined with BMS-754807 could inhibit lung cancer cell
proliferation by inducing autophagy and arresting the cell cycle at
G1 phase (53). SB505124_1194 is a
selective TGF-β1 receptor inhibitor that notably impedes
TGF-β1-mediated EMT in lung cancer cells, thereby reducing
recurrence and metastasis (54,55).
In the present study, higher expression of TGF-β1 in the low-risk
group was associated with drug prediction. JQ1_2172 is a small
molecule inhibitor targeting the BET protein that binds reversibly
to its bromodomain, thus disrupting its interaction with acetylated
lysines on histones or transcription factors. This inhibition leads
to the suppression of oncogene expression, ultimately resulting in
cancer cell proliferation arrest (56). Additionally, JQ1_2172 inhibits the
upregulation of programmed death-ligand 1 during concurrent
radiotherapy for lung cancer, thus exerting antitumor immune escape
effects (57). Despite their
effective eradication of tumor cells, anti-neoplastic drugs exert
certain effects on both patients and hospital workers exposed to
them, including increased DNA damage, elevated oxidative stress
parameters and impairment on complete blood counts (58). Therefore, the implementation of
personalized treatment strategies based on risk score could
facilitate the selection of appropriate drug treatment plans,
thereby enhancing treatment efficacy and minimizing the adverse
effects of anti-neoplastic drugs on patients and healthcare
workers.
Although a robust prognostic model based on KNHMUGs
was constructed in the present study using public data, it is
relied on public databases and further clinical validation is
therefore required to confirm the model efficacy in prognostic
prediction and assessment of immunotherapy and chemotherapy
sensitivity. Secondly, whilst the hub genes screened in the present
study have been investigated in a small number of LUAD cases, their
mechanisms remain unclear and require further exploration.
Subsequent studies should be conducted to address the
aforementioned questions.
In summary, KEAP1, NRF2 and HO-1 expression is
closely associated with clinicopathological features and prognosis
in LUAD. The prognostic model constructed based on KNHMUGs may
serve as an independent prognostic factor for predicting the
prognosis of patients with LUAD. It integrates important clinical
parameters and risk scores of prognoses, demonstrating reliability
and effectiveness. Furthermore, the association between risk score
and tumor immune microenvironment, immunotherapy and chemotherapy
response may offer a potentially powerful new tool for clinical
decision-making, patient prognosis determination and personalized
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was supported by the scientific research
projects of the Top Talent Support Program for Young and
Middle-Aged People of Wuxi Health Committee (grant no. HB2023116)
and the Innovation Cultivation Fund of Xishan People's Hospital
(grant no. 202103).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZX designed the research and drafted the manuscript.
CC and QS utilized software code to perform an in-depth analysis
and visualization of the data and reviewed the present paper. WZ
and YZ analyzed the paraffin sections. LY designed the mathematical
methods, collected and analyzed the data. LC performed the
immunohistochemistry. WZ and ZX confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study complied with the Declaration of
Helsinki and was approved by the Xishan People's Hospital Ethics
Committee (approval no. xs2024ky037). The samples used in the
present study were sourced from Xishan People's Hospital in Wuxi
City, collected from patients with LUAD undergoing surgery between
January 2020 and June 2023. Written informed consent forms were
signed by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death-based treatment of lung adenocarcinoma. Cell Death
Dis. 9:1172018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reck M and Rabe KF: Precision diagnosis
and treatment for advanced non-small-cell lung cancer. N Engl J
Med. 377:849–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Y, Yan B and He S: Advances and
challenges in the treatment of lung cancer. Biomed Pharmacother.
169:1158912023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Obradovic J, Todosijevic J and Jurisic V:
Application of the conventional and novel methods in testing EGFR
variants for NSCLC patients in the last 10 years through different
regions: A systematic review. Mol Biol Rep. 48:3593–3604. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jurisic V, Vukovic V, Obradovic J,
Gulyaeva LF, Kushlinskii NE and Djordjevic N: EGFR polymorphism and
survival of NSCLC patients treated with TKIs: A systematic review
and meta-analysis. J Oncol. 2020:19732412020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Obradovic J, Todosijevic J and Jurisic V:
Side effects of tyrosine kinase inhibitors therapy in patients with
non-small cell lung cancer and associations with EGFR
polymorphisms: A systematic review and meta-analysis. Oncol Lett.
25:622023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu K, Ma J, Hall SRR, Pengs RW, Yang H and
Yao F: Battles against aberrant KEAP1-NRF2 signaling in lung
cancer: Intertwined metabolic and immune networks. Theranostics.
13:704–723. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghareghomi S, Moosavi-Movahedi F, Saso L,
Habibi-Rezaei M, Khatibi A, Hong J and Moosavi-Movahedi AA:
Modulation of Nrf2/HO-1 by natural compounds in lung cancer.
Antioxidants (Basel). 12:7352023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Romero R, Sanchez-Rivera FJ, Westcott PMK,
Mercer KL, Bhutkar A, Muir A, Robles TJ, Rodriguez SL, Liao LZ, Ng
SR, et al: Keap1 mutation renders lung adenocarcinomas dependent on
Slc33a1. Nat Cancer. 1:589–602. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Romero R, Sayin VI, Davidson SM, Bauer MR,
Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A,
Sanchez-Rivera FJ, et al: Keap1 loss promotes Kras-driven lung
cancer and results in dependence on glutaminolysis. Nat Med.
23:1362–1368. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zavitsanou AM, Pillai R, Hao Y, Wu WL,
Bartnicki E, Karakousi T, Rajalingam S, Herrera A, Karatza A,
Rashidfarrokhi A, et al: KEAP1 mutation in lung adenocarcinoma
promotes immune evasion and immunotherapy resistance. Cell Rep.
42:1132952023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ghareghomi S, Habibi-Rezaei M, Arese M,
Saso L and Moosavi-Movahedi AA: Nrf2 modulation in breast cancer.
Biomedicines. 10:26682022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Menegon S, Columbano A and Giordano S: The
dual roles of NRF2 in cancer. Trends Mol Med. 22:578–593. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kerins MJ and Ooi A: A catalogue of
somatic NRF2 gain-of-function mutations in cancer. Sci Rep.
8:128462018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Liang G, Yan K and Wang Y: NRF2
mutation enhances the immune escape of hepatocellular carcinoma by
reducing STING activation. Biochem Biophys Res Commun.
698:1495362024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang H, Liu S, Yan X, Jin Y, He X, Huang
H, Liu L, Hu W and Wu A: Inhibition of LNC EBLN3P enhances
radiation-induced mitochondrial damage in lung cancer cells by
targeting the Keap1/Nrf2/HO-1 axis. Biology (Basel).
12:12082023.PubMed/NCBI
|
|
19
|
Spampinato M, Sferrazzo G, Pittala V, Di
Rosa M, Vanella L, Salerno L, Sorrenti V, Carota G, Parrinello N,
Raffaele M, et al: Non-competitive heme oxygenase-1 activity
inhibitor reduces non-small cell lung cancer glutathione content
and regulates cell proliferation. Mol Biol Rep. 47:1949–1964. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Director's Challenge Consortium for the
Molecular Classification of Lung Adenocarcinoma, Shedden K, Taylor
JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S,
Jurisica I, Giordano TJ, et al: Gene expression-based survival
prediction in lung adenocarcinoma: A multi-site, blinded validation
study. Nat Med. 14:822–827. 2008. View
Article : Google Scholar
|
|
21
|
Lanczky A and Gyorffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H,
Wang L, Lu T, Zhang Y, Sun Z and Han X: Machine learning-based
integration develops an immune-derived lncRNA signature for
improving outcomes in colorectal cancer. Nat Commun. 13:8162022.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miao YR, Zhang Q, Lei Q, Luo M, Xie GY,
Wang H and Guo AY: ImmuCellAI: A unique method for comprehensive
T-cell subsets abundance prediction and its application in cancer
immunotherapy. Adv Sci (Weinh). 7:19028802020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Detterbeck FC, Woodard GA, Bader AS, Dacic
S, Grant MJ, Park HS and Tanoue LT: The proposed ninth edition TNM
classification of lung cancer. Chest. 166:882–895. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lou JS, Zhao LP, Huang ZH, Chen XY, Xu JT,
Tai WC, Tsim KWK, Chen YT and Xie T: Ginkgetin derived from Ginkgo
biloba leaves enhances the therapeutic effect of cisplatin via
ferroptosis-mediated disruption of the Nrf2/HO-1 axis in EGFR
wild-type non-small-cell lung cancer. Phytomedicine. 80:1533702021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bi G, Liang J, Zhao M, Zhang H, Jin X, Lu
T, Zheng Y, Bian Y, Chen Z, Huang Y, et al: miR-6077 promotes
cisplatin/pemetrexed resistance in lung adenocarcinoma via
CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol Ther
Nucleic Acids. 28:366–386. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mei L, Long J, Wu S, Mei M, Mei D and Qiu
H: APOC1 reduced anti-PD-1 immunotherapy of nonsmall cell lung
cancer via the transformation of M2 into M1 macrophages by
ferroptosis by NRF2/HO-1. Anticancer Drugs. 35:333–343. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei XW, Lu C, Zhang YC, Fan X, Xu CR, Chen
ZH, Wang F, Yang XR, Deng JY, Yang M, et al: Redox(high) phenotype
mediated by KEAP1/STK11/SMARCA4/NRF2 mutations diminishes
tissue-resident memory CD8+ T cells and attenuates the efficacy of
immunotherapy in lung adenocarcinoma. Oncoimmunology.
13:23401542024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fahrmann JF, Tanaka I, Irajizad E, Mao X,
Dennison JB, Murage E, Casabar J, Mayo J, Peng Q, Celiktas M, et
al: Mutational activation of the NRF2 pathway upregulates
kynureninase resulting in tumor immunosuppression and poor outcome
in lung adenocarcinoma. Cancers (Basel). 14:25432022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phillips RS: Structure and mechanism of
kynureninase. Arch Biochem Biophys. 544:69–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karayama M, Masuda J, Mori K, Yasui H,
Hozumi H, Suzuki Y, Furuhashi K, Fujisawa T, Enomoto N, Nakamura Y,
et al: Comprehensive assessment of multiple tryptophan metabolites
as potential biomarkers for immune checkpoint inhibitors in
patients with non-small cell lung cancer. Clin Transl Oncol.
23:418–423. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leon-Letelier RA, Sater AH, Chen Y,
Srejbhjv S, et al: Kynureninase upregulation is a prominent feature
of NFR2-activated cancers and is associated with tumor
immunosuppression and poor prognosis. Cancers (Basel). 15:8342023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu BX, Xie Y, Zhang J, Zeng S, Li J, Tao
Q, Yang J, Chen Y and Zeng C: SERPINB5 promotes colorectal cancer
invasion and migration by promoting EMT and angiogenesis via the
TNF-α/NF-κB pathway. Int Immunopharmacol. 131:1117592024.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rathod M, Franz H, Beyersdorfer V, Wanuske
MT, Leal-Fischer K, Hanns P, Stüdle C, Zimmermann A, Buczak K,
Schinner C and Spindler V: DPM1 modulates desmosomal adhesion and
epidermal differentiation through SERPINB5. J Cell Biol.
223:e2023050062024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He X, Ma Y, Huang Z, Wang G, Wang W, Zhang
R, Guo G, Zhang X, Wen Y and Zhang L: SERPINB5 is a prognostic
biomarker and promotes proliferation, metastasis and
epithelial-mesenchymal transition (EMT) in lung adenocarcinoma.
Thorac Cancer. 14:2275–2287. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Scopetti D, Piobbico D, Brunacci C,
Pieroni S, Bellezza G, Castelli M, Ludovini V, Tofanetti FR, Cagini
L, Sidoni A, et al: INSL4 as prognostic marker for proliferation
and invasiveness in non-small-cell lung cancer. J Cancer.
12:3781–3795. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang R, Li SW, Chen Z, Zhou X, Ni W, Fu
DA, Lu J, Kaye FJ and Wu L: Role of INSL4 signaling in sustaining
the growth and viability of LKB1-inactivated lung cancer. J Natl
Cancer Inst. 111:664–674. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bhattacharya D, Gawali VS, Kallay L,
Toukam DK, Koehler A, Stambrook P, Krummel DP and Sengupta S:
Therapeutically leveraging GABA(A) receptors in cancer. Exp Biol
Med (Maywood). 246:2128–2135. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu L, Yang C, Shen J, Huang L, Lin W,
Tang H, Liang W, Shao W, Zhang H and He J: GABRA3 promotes
lymphatic metastasis in lung adenocarcinoma by mediating
upregulation of matrix metalloproteinases. Oncotarget.
7:32341–32350. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Genova C, Dellepiane C, Carrega P,
Sommariva S, Ferlazzo G, Pronzato P, Gangemi R, Filaci G, Coco S
and Croce M: Therapeutic implications of tumor microenvironment in
lung cancer: Focus on immune checkpoint blockade. Front Immunol.
12:7994552021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song P, Li W, Guo L, Ying J, Gao S and He
J: Identification and validation of a novel signature based on NK
cell marker genes to predict prognosis and immunotherapy response
in lung adenocarcinoma by integrated analysis of single-cell and
bulk RNA-sequencing. Front Immunol. 13:8507452022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Terabe M and Berzofsky JA: Tissue-specific
roles of NKT cells in tumor immunity. Front Immunol. 9:18382018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rad SH, Monkman J, Warkiani ME, Ladwa R,
O'Byrne K, Rezaei N and Kulasinghe A: Understanding the tumor
microenvironment for effective immunotherapy. Med Res Rev.
41:1474–1498. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ge Z, Peppelenbosch MP, Sprengers D and
Kwekkeboom J: TIGIT, the next step towards successful combination
immune checkpoint therapy in cancer. Front Immunol. 12:6998952021.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Acharya N, Sabatos-Peyton C and Anderson
AC: Tim-3 finds its place in the cancer immunotherapy landscape. J
Immunother Cancer. 8:e0009112020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wojciechowicz K, Spodzieja M and Wardowska
A: The BTLA-HVEM complex-The future of cancer immunotherapy. Eur J
Med Chem. 268:1162312024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gebbia V, Lorusso V, Galetta D, Caruso MM,
Palomba G, Riccardi F, Borsellino N, Carrozza F, Leo S, Ferraù F,
et al: First-line cisplatin with docetaxel or vinorelbine in
patients with advanced non-small-cell lung cancer: A quality of
life directed phase II randomized trial of Gruppo Oncologico Italia
Meridionale. Lung Cancer. 69:218–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fuentes-Baile M, Ventero MP, Encinar JA,
García-Morales P, Poveda-Deltell M, Pérez-Valenciano E, Barberá VM,
Gallego-Plazas J, Rodríguez-Lescure Á, Martín-Nieto J and Saceda M:
Differential effects of IGF-1r small molecule tyrosine kinase
inhibitors BMS-754807 and OSI-906 on human cancer cell lines.
Cancers (Basel). 12:37172020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang C, Zhao X, Wang Z, Gong T, Zhao H,
Zhang D, Niu Y, Li X, Zhao X, Li G, et al: Dasatinib in combination
with BMS-754807 induce synergistic cytotoxicity in lung cancer
cells through inhibiting lung cancer cell growth, and inducing
autophagy as well as cell cycle arrest at the G1 phase. Invest New
Drugs. 41:438–452. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
DaCosta Byfield S, Major C, Laping NJ and
Roberts AB: SB-505124 is a selective inhibitor of transforming
growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol
Pharmacol. 65:744–752. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu TH, Chou YW, Chiu PH, Tang MJ, Hu CW
and Yeh ML: Validation of the effects of TGF-β1 on tumor recurrence
and prognosis through tumor retrieval and cell mechanical
properties. Cancer Cell Int. 14:202014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Stathis A and Bertoni F: BET proteins as
targets for anticancer treatment. Cancer Discov. 8:24–36. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang J, Xu Y, Rao X, Zhang R, Tang J,
Zhang D, Jie X, Zhu K, Wang X, Xu Y, et al: BRD4-IRF1 axis
regulates chemoradiotherapy-induced PD-L1 expression and immune
evasion in non-small cell lung cancer. Clin Transl Med.
12:e7182022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mrdjanovic J, Solajic S, Srdenovic-Conic
B, Bogdanović V, Dea KJ, Kladar N and Jurišić V: The oxidative
stress parameters as useful tools in evaluating the DNA damage and
changes in the complete blood count in hospital workers exposed to
low doses of antineoplastic drugs and ionizing radiation. Int J
Environ Res Public Health. 18:84452021. View Article : Google Scholar : PubMed/NCBI
|