Introduction
In the tumor microenvironment, cancer cells exhibit
a greater ability to survive than normal cells, even under harsh
conditions such as glucose deprivation or hypoxia. Whilst previous
studies have focused primarily on elucidating the genetic and
molecular basis of cancer, recent research has focused on the
metabolic reprogramming of cancer cells (1). In particular, the concept of the
‘Warburg effect’, which states that cancer cells take up glucose as
their primary energy sources to activate aerobic glycolysis, has
led to important advances in understanding cancer metabolism
(2). However, experimental evidence
from previous research has led to the hypothesis that cancer cells
utilize lipids instead of glucose as their primary energy source
(3). This has led to a growing need
for research to understand lipid metabolism in cancer cells,
particularly in hypoxic environments (4).
Globally, breast cancer is a major contributor to
cancer-associated deaths, with a notable increase of 20.9% in
mortality rate from 2010 to 2019 (5). Breast cancer is divided into different
subtypes based on hormone receptor status, of which triple-negative
breast cancer (TNBC) has a worse prognosis compared with other
subtypes due to the ineffectiveness of endocrine therapy (6). Currently, cytotoxic chemotherapy is
the mainstay of treatment for TNBC, but resistance to chemotherapy
frequently develops, leading to a growing need for the development
of new therapies. In addition, women with obesity-related risk
factors are more likely to develop TNBC (7,8),
emphasizing the importance of investigating lipid metabolism
reprogramming in this subtype (9,10).
Genes related to lipid metabolism are dysregulated
in breast tissue before cancer diagnosis (11). Fatty acid synthase (FASN), a key
enzyme involved in de novo fatty acid synthesis, is
upregulated in TNBC, and FASN inhibitors have demonstrated
anticancer effects against chemoresistant TNBC (12). Clinical studies are currently
exploring the potential of combining the inhibitors of
3-hydroxy-3-methylglutaryl-coenzyme A reductase, a key enzyme in
cholesterol synthesis, with conventional therapies for the
treatment of TNBC (13). Sterol
regulatory element-binding protein 1 (SREBP1), a key transcription
factor, is gaining increasing attention for its role in regulating
the genes involved in fatty acid and cholesterol production
(13–15). The expression of SREBP1 increases
upon exposure to a hostile environment in glioblastoma, prostate,
nasopharyngeal, endometrial and breast cancers (16–19),
and recent studies have highlighted the important role of SREBP1 in
TNBC cells, suggesting the need to explore the mechanisms of SREBP1
metabolism, especially under hypoxic conditions (20,21).
In hypoxic environments, increased hypoxia-inducible
factor (HIF)-1α promotes the expression of de novo lipid
synthesis genes, such as FASN and stearoyl-CoA desaturase 1, and
lipid uptake-related genes, such as fatty acid binding proteins
(FABPs). This leads to the accumulation of lipid droplets (LDs) in
cancer cells (22), which serves as
a protective mechanism to alleviate endoplasmic reticulum stress
(23,24). Recently, researchers have
demonstrated that fatty acids serve as a major fuel source of ATP
for cancer cell growth, raising an important question of why fatty
acids can function as a source of ATP without causing lipotoxicity,
especially in oxygen-poor tumor environments. Further research is
required to elucidate the mechanisms under hypoxic conditions
(25,26).
The present study aimed to assess the effects of
hypoxia on cell survival via lipid reprogramming in breast cancer
cells. The results demonstrated that SREBP1-mediated lipid
reprogramming, along with autophagy, promotes cell survival under
hypoxic conditions by facilitating ATP production via fatty acid
oxidation (FAO) in TNBC cells.
Materials and methods
Cell culture and reagents
The non-cancerous human breast epithelial MCF-10A
cell line was donated by Dr Je-Yoel Cho (Seoul National University;
Seoul, South Korea); the estrogen receptor (ER)-positive human
breast cancer MCF-7 cell line was donated by Dr So Yeong Lee (Seoul
National University); and the TNBC MDA-MB-231 cell line was
purchased from the Korean Cell Line Bank (KCLB). MCF-10A was
cultured in mammary epithelial cell growth medium
BulletKit™ (Lonza Group, Ltd.) supplemented with 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
MCF-7 and MDA-MB-231 cells were cultured in RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 1%
penicillin/streptomycin, 1 mM sodium pyruvate (Sigma-Aldrich; Merck
KGaA), 10 mM sodium bicarbonate (Sigma-Aldrich; Merck KGaA), 10 mM
HEPES (Sigma-Aldrich; Merck KGaA) and 10% fetal bovine serum. The
authenticity of the cell lines used in the present study was
confirmed using short tandem repeat markers at the KCLB. All cells
were tested using e-Myco™ Plus Mycoplasma PCR Detection
Kit (Intron Biotechnology, Inc.). For hypoxic conditions, cells
were placed in a sealed hypoxic incubator chamber (Stemcell
Technologies, Inc.) containing 1% O2, 5% CO2
and 95% N2 at 37°C for 48 h and monitored using an
oxygen meter (GOX-100; GHM Messtechnik GmbH). For the control
group, cells were maintained under normoxic conditions with 21%
O2 at 37°C. Fatostatin, an SREBP1 inhibitor, and
rapamycin, an autophagy inducer, were purchased from Sigma-Aldrich
(Merck KGaA) and Selleck Chemicals, respectively. Fatostatin at
concentrations of 5, 10, and 15 µM and rapamycin at concentrations
of 0.1, 1, and 10 µM were administered at 37°C for 48 h.
Cell viability and proliferation
assays
Cell viability was assessed using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT;
Sigma-Aldrich; Merck KGaA) assay. MCF-10a, MCF-7, and MDA-MB-231
cells were seeded in 96-well plates at a concentration of
0.5×104 cells/well. After 24 h incubation, the cells
were maintained under normoxic or hypoxic conditions at 37°C for 48
h. MTT solution dissolved in phosphate-buffered saline (PBS) at 5
mg/ml was added to each well and incubated for 1 h at 37°C.
Subsequently, 100 µl of a solution composed of dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) and 2-propanol (Merck KGaA) in a 9:1
ratio was added to each well. Absorbance was determined at 570 nm
using a BioTek Epoch microplate spectrophotometer (Agilent
Technologies, Inc.).
For the cell proliferation assay, 3×105
MCF-10A, MCF-7, and MDA-MB-231 cells/well were seeded into 60-mm
dishes. After 24 h incubation, the cells were further incubated
under normoxic or hypoxic conditions at 37°C for 48 h. The cells
were stained with 0.4% trypan blue dye (Gibco; Thermo Fisher
Scientific, Inc.) at room temperature for 5 min and surviving cells
were counted manually using an inverted microscope (cat. no.
KS-TCM4; Korea Scope).
Apoptosis assay
The apoptotic rate was measured using an EzWay
Annexin V-FITC apoptosis Kit (Koma Biotech Inc.) according to the
manufacturer's instructions. The harvested MCF-10A, MCF-7, and
MDA-MB-231 were washed with a binding buffer and incubated with
FITC-Annexin V reagent in binding buffer for 15 min at room
temperature in the dark, protected from light. After washing with
the binding buffer, the cells were resuspended in PI/RNase staining
solution (BD Biosciences) and immediately analyzed using FACSVerse
(BD Biosciences). The data were gated based on forward and side
scatter to exclude cell debris using BD FACSuite™
Application software version 1.6 (BD Biosciences). Total apoptosis
included late apoptotic cells (double positive for annexin V and
PI) and early apoptotic cells (positive for annexin V and negative
for PI).
Immunofluorescence
MCF-7 and MDA-MB-231 cells were cultured in 35-mm
confocal dishes (SPL Life Sciences), fixed with 4% formaldehyde and
permeabilized with 0.1% Triton X-100, each for 20 min at room
temperature. They were first blocked with 5% bovine serum albumin
(Bovostar) for 1 h at room temperature and then incubated with 5
µg/ml Nile Red (Sigma-Aldrich; Merck KGaA) or microtubule
associated protein 1 light chain 3-β (LC3B; cat no. NB100-2220;
Novus Biologicals, LLC; Bio-Techne) and lysosomal associated
membrane protein 2 (LAMP2; cat. no. CSB-PA012740EA01HU; Cusabio
Technology, LLC) antibodies, each diluted at 1:200, for 30 min at
room temperature in the dark. The secondary antibody, Alexa
Fluor™ 488 (cat. no. A-11008; Thermo Fisher Scientific,
Inc.), was then added at a 1:1,000 dilution and incubated for 1 h
at room temperature in the dark. Nuclei were stained with DAPI
Fluoromount-G (SouthernBiotech) for 5 min at room temperature.
Fluorescence images of the control and experimental groups were
captured under identical conditions including magnification,
brightness, laser intensity and channel settings, using the EVOS
M7000 Imaging System (cat. no. AMF7000; Thermo Fisher Scientific,
Inc.). For enhanced clarity, contrast for all group images was
adjusted to the same values using the Celleste™ 6 Image
Analysis Software (Thermo Fisher Scientific, Inc.) according to the
guidelines (27).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from MCF-7 and MDA-MB-231
cells using TRIzol™ reagent (Invitrogen™;
Thermo Fisher Scientific, Inc.). A total of 500 ng RNA was
reverse-transcribed into single-stranded complementary DNA for 30
min at 50°C using TOPscript™ RT DryMIX (dT 18plus;
Enzynomics Co., Ltd.). RT-qPCR was performed to evaluate the
expression levels of very low-density lipoprotein receptor (VLDLR),
CD36, FABP3 and FABP7, perilipin 2, hypoxia-inducible lipid droplet
associated, patatin-like phospholipase domain containing 2, lipin
1, diacylglycerol O-acyltransferase 1, sterol regulatory element
binding transcription factor 1 (SREBF1) and FASN using the SYBR
Green Real-Time PCR Kit (Enzynomics Co., Ltd.) according to the
manufacturer's instructions. Using β-actin as a control, the level
of the target product relative to that of the internal control was
calculated using the 2−ΔΔCq method (28). The primer sequences for these genes
are listed in Table SI.
Western blot analysis
Whole-cell lysates from MCF-7 and MDA-MB-231 cells
were prepared by treatment of the cells with EzRIPA buffer (ATTO
Corporation) and protein concentration was measured using the
Bradford assay (Bio-Rad Laboratories, Inc.). A total of 10 µg total
lysate was applied to a 10% sodium dodecyl sulfate-polyacrylamide
gel and subsequently transferred onto a nitrocellulose membrane
(Cytiva). For blocking, the membranes were incubated with 5%
skimmed milk in PBS containing 0.05% Tween 20 (PBST) at 37°C for 60
min. After washing with PBST three times, the membrane was
incubated overnight at 4°C using primary antibodies in PBST
containing 5% bovine serum albumin. The primary antibodies used
were against HIF-1α (cat. no. 3716S; Cell Signaling Technology,
Inc.), SREBP1 (cat. no. SC-365513; Santa Cruz Biotechnology, Inc.),
LC3B (cat. no. NB100-2220; Novus Biologicals, LLC; Bio-Techne) and
β-actin (cat. no. SC-47778; Santa Cruz Biotechnology, Inc.), all
1:1,000 dilution. Secondary horseradish peroxidase-conjugated
anti-rabbit (cat. no. SC-2004; Santa Cruz Biotechnology Inc.) or
anti-mouse (cat. no. SC-2357; Santa Cruz Biotechnology Inc.)
antibodies were added at a 1:4,000 dilution and incubated for 2 h
at room temperature. Immunocomplexes were detected using an
enhanced chemiluminescence detection system (Bio-Rad Laboratories,
Inc.), and image acquisition was performed with the
ImageQuant™ LAS-4000 mini (FUJIFILM Corporation).
Densitometry analysis was performed using the ImageJ software
version 1.50i (National Institutes of Health) based on a previous
reference (29).
Cyto-ID autophagy detection
Autophagy activation was assessed using the Cyto-ID
Autophagy Kit (Enzo Life Sciences, Inc.) following the
manufacturer's instructions. Briefly, MCF-7 and MDA-MB-231 cells
were washed with assay buffer and subsequently incubated with
Cyto-ID (cat. no. ENZ-51031; Enzo Life Sciences) at a 1:2,000
dilution in assay buffer for 30 min at 37°C in the dark. After
washing and resuspending the cells in assay buffer, they were
immediately analyzed by flow cytometry using a FACSVerse instrument
(BD Biosciences). As a positive control for autophagy, cells were
treated with 20 µM chloroquine (Sigma-Aldrich; Merck KGaA) for 16 h
at 37°C for confirming the accumulation of autophagosomes as
previously described (30). Data
were gated based on forward and side scatter to exclude cell
debris, using BD FACSuite™ Application version 1.6 (BD
Biosciences).
Fatty acid oxidation assay
The key FAO enzymes [acyl-CoA dehydrogenase very
long chain (ACADVL), acyl-CoA dehydrogenase medium chain (ACADM)
and hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex
subunit a (HADHA)] were assessed using an FAO kit (cat. no.
ab118183; Abcam) following the manufacturer's protocol. Briefly,
the MDA-MB-231 cells were harvested, fixed with 4% formaldehyde and
permeabilized with Triton X-100. They were incubated with primary
antibodies for 1 h at room temperature, followed by incubation with
the secondary antibody Alexa Fluor 488 (cat. no. A-11001; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature in the dark.
Finally, the samples were analyzed by flow cytometry using
FACSVerse (BD Biosciences). Data were gated based on forward and
side scatter to exclude cell debris, using BD FACSuite™
Application software version 1.6 (BD Biosciences).
Assessment of ATP concentration
ATP production was measured using a commercial ATP
assay kit (cat. no. BM-ATP-100; Biomax Co., Ltd.) according to the
manufacturer's recommendations. Briefly, a mixture containing ATP
assay buffer, probe, converter and developer was added to cell
lysates prepared from 1×106 cells of MDA-MB-231. The
samples were incubated in this solution in a 96-well plate for 30
min at room temperature. The absorbance was measured at 570 nm
using a BioTek Epoch microplate spectrophotometer (Agilent
Technologies, Inc.), and ATP concentration of each sample was
calculated based on a standard curve.
Small interfering (si)RNA
transfection
Gene knockdown experiments were performed by
transfection with AccuTarget negative control siRNA (cat. no.
SN-1011; Bioneer Corporation) and AccuTarget Genome-wide
Predesigned siRNA targeting Human SREBF1 (Bioneer Corporation). The
sense and antisense sequences of the SREBP1 siRNAs were
5′-CCACCGUUUCUUCGUGGAU-3′ and 5′-AUCCACGAAGAAACGGUGG-3′,
respectively. Control and SREBP1 siRNAs (100 nM were mixed with
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and transfected into MCF-7 and MDA-MB-231 cells
according to the manufacturer's recommendations. MCF-7 and
MDA-MB-231 cells were incubated for 24 h at 37°C in RPMI 1640
medium supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.). After 24 h, the adhered cells were
transfected with siRNAs using a Lipofectamineâ 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) in Opti-MEM
reduced-serum medium (Gibco; Thermo Fisher Scientific, Inc.). After
6 h of incubation, the Opti-MEM was replaced by a fresh culture
medium and the cells were further cultured for 48 h at 37°C under
normoxic and hypoxic conditions.
Kaplan-Meier analysis
Kaplan-Meier plotter tools were used to assess the
association between SREBF1 mRNA expression and recurrence-free
survival (RFS) and distant metastasis-free survival (DMFS) in
patients with ER+/progesterone receptor (PR)-/human epidermal
growth factor receptor 2 (HER2)- and ER-/PR-/HER2-subtypes of
breast cancer (http://kmplot.com/analysis/index.php?p=service&cancer=breast).
The analysis was conducted with ‘auto-select best cut-off’ option
to determine the cut-off value and patients were stratified based
on hormone receptor and HER2 status for the dichotomization of
SREBF1 mRNA expression level. Hazard ratios along with their
corresponding 95% confidence intervals and log-rank P-values were
reported for each result.
Statistical analysis
Data are presented as the mean ± standard error of
the mean based on a minimum of three independent experiments.
Statistical analyses were performed using GraphPad PRISM software
version 5.01 (Dotmatics). Datasets with two groups were analyzed
using one-way analysis of variance (ANOVA) and two-way ANOVA was
used for multiple group comparisons with Bonferroni post hoc to
identify significant differences. P<0.05 was considered to
indicate a statistically significant difference.
Results
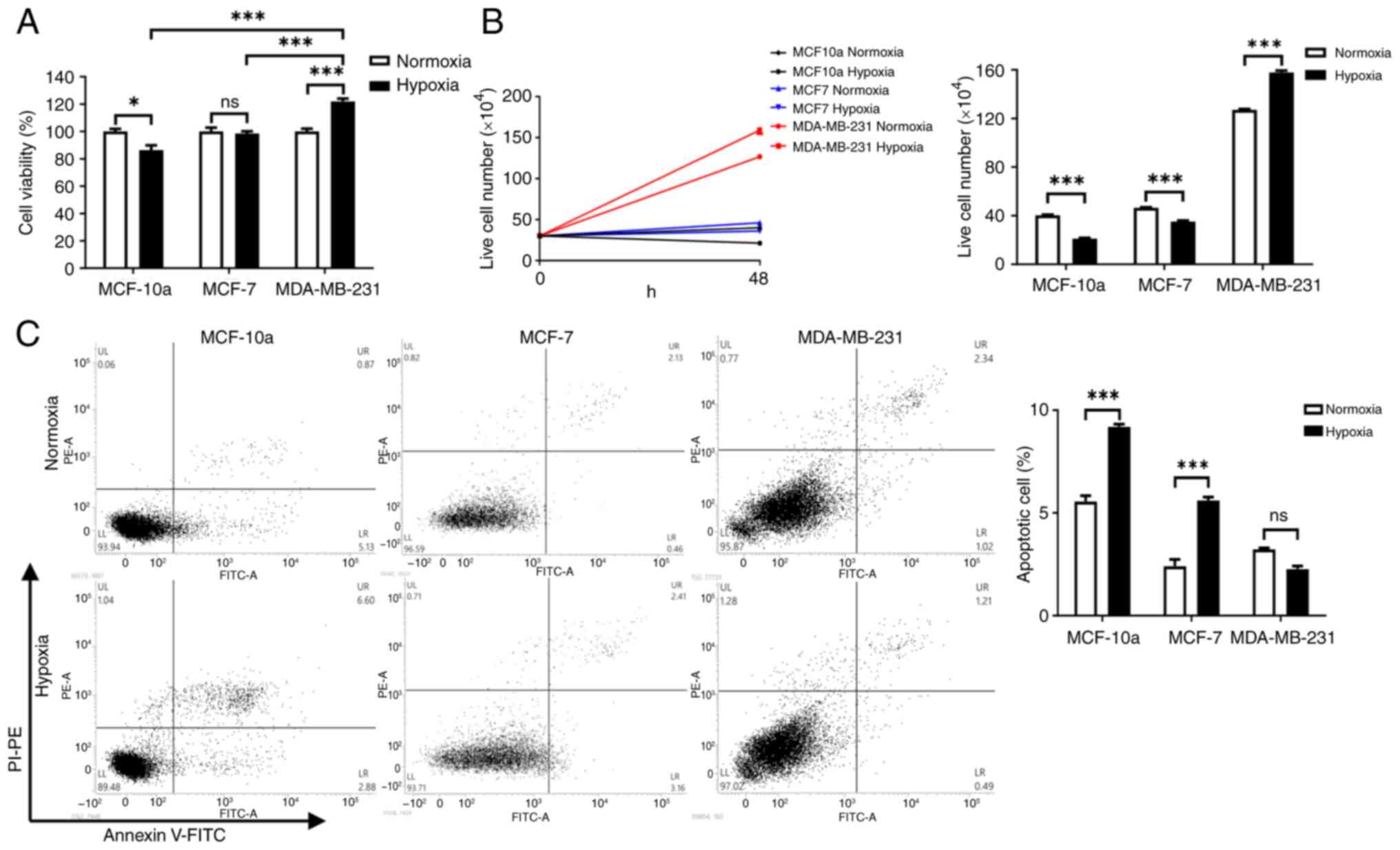
Hypoxia increases survival of
MDA-MB-231 cells and decreases the viability of MCF-10a and MCF-7
cells
The effect of hypoxia on cell viability was
evaluated using MTT, trypan blue and apoptosis assays. MTT assay
revealed that cell viability significantly increased in MDA-MB-231
cells under hypoxic conditions compared with under normoxic
conditions (Fig. 1A). By contrast,
compared with under normoxic conditions, cell viability
significantly decreased in MCF-10a cells under hypoxic conditions;
however, no significant change was observed in MCF-7 cells. In the
trypan blue assay, the live cell number significantly increased in
MDA-MB-231 cells under hypoxic conditions compared with under
normoxic conditions; however, the live cell number was
significantly reduced in both MCF-10a and MCF-7 cells (Fig. 1B). The apoptosis assay demonstrated
that the apoptotic rates of MCF-10a and MCF-7 cells were
significantly increased under hypoxic conditions compared with
under normoxic conditions; however, no significant change was
observed in MDA-MB-231 cells (Fig.
1C).
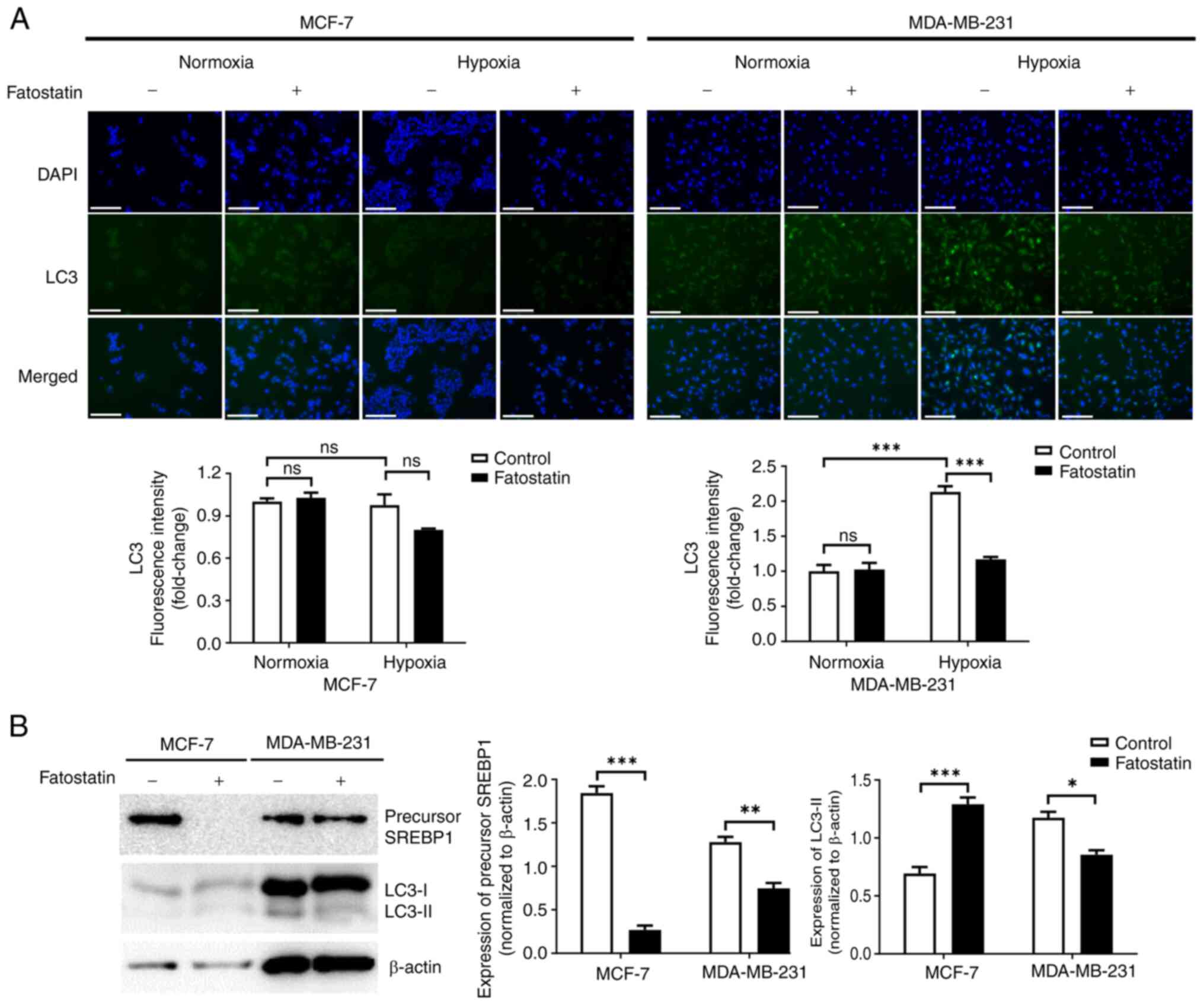
SREBP1 is required for lipogenesis in
MDA-MB-231 cells under hypoxic conditions
To determine whether lipid reprogramming is involved
in cell survival under hypoxic conditions, the number of lipid
droplets was evaluated using Nile Red staining. Under hypoxic
conditions, the fluorescence intensity of Nile Red significantly
increased in MDA-MB-231 cells, but not in MCF-7 cells (Fig. 2A). Using RT-qPCR, the expression of
lipid-related genes was assessed in MCF-7 and MDA-MB-231 cells
under normoxic and hypoxic conditions. Under hypoxic conditions,
the expression levels of lipid-related genes were significantly
downregulated in MCF-7 cells, whereas the expression levels of
SREBF1, VLDLR and FABP3 were significantly upregulated in
MDA-MB-231 cells, compared with those under normoxic conditions
(Fig. S1). Notably, expression of
the lipid synthesis-related gene FASN was significantly
downregulated in both MCF-7 and MDA-MB-231 cells under hypoxic
conditions, compared with under normoxic conditions. Given that
SREBP1 is a transcription factor that upregulates the synthesis or
uptake of enzymes involved in lipid metabolism (15), SREBP1 levels were evaluated using
western blotting. Expression of SREBP1 was significantly increased
in MDA-MB-231 cells under hypoxic conditions compared with under
normoxic conditions, whereas no change was observed in MCF-7 cells
(Fig. 2B). Treatment with
fatostatin, an SREBP1 inhibitor, markedly reduced SREBP1 levels in
both MCF-7 and MDA-MB-231 cells under hypoxic conditions compared
with normoxic conditions (Fig. 2C).
Moreover, the treatment significantly decreased the fluorescence
intensity of Nile Red in MDA-MB-231 cells under hypoxic conditions
compared with under normoxic conditions (Fig. 2A). The data collectively indicate
that SREBP1 is crucial for lipogenesis under hypoxic conditions in
TNBC cells.
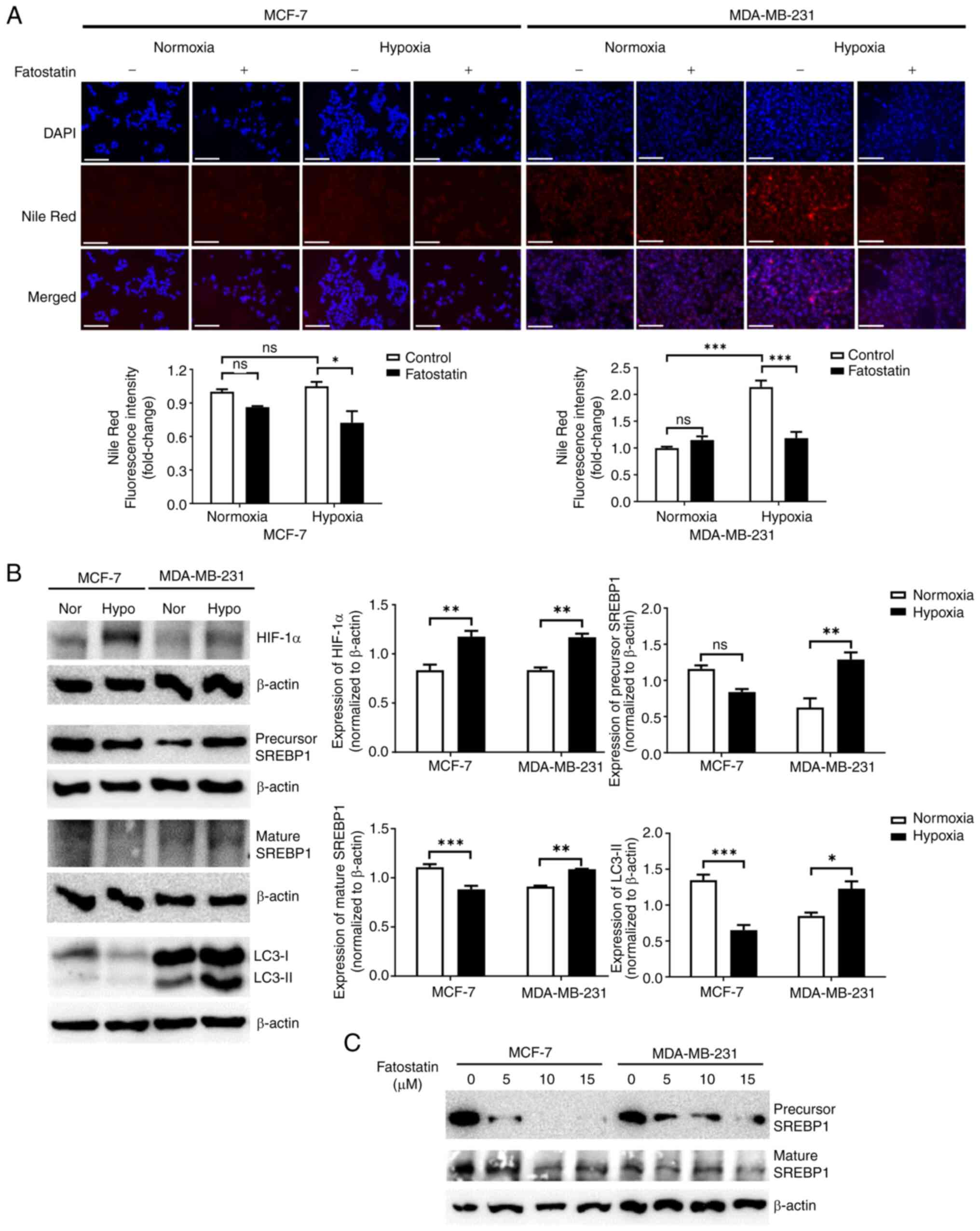 | Figure 2.SREBP1 enhances lipogenesis in
MDA-MB-231 cells under hypoxic conditions. (A) Assessment of the
amount of lipids using Nile Red staining by immunofluorescence with
or without fatostatin in MCF-7 and MDA-MB-231 cells under normoxic
and hypoxic conditions for 48 h. Scale bar, 150 µm. The graphs
indicate the quantitative analysis of fluorescence intensity. (B)
Expression levels of hypoxia indicators, HIF-1α, autophagy-related
markers, LC3-I/II and lipogenesis-related protein, SREBP1, in MCF-7
and MDA-MB-231 cells under normoxic and hypoxic conditions for 48
h. The graphs indicate the semi-quantitative analysis of HIF-1α,
LC3-II and SREBP1 protein levels normalized to β-actin expression.
(C) Expression levels of SREBP1 and β-actin proteins following
treatment with different concentrations of fatostatin in MCF-7 and
MDA-MB-231 cells under hypoxic conditions for 48 h. *P<0.05;
**P<0.01; ***P<0.001. HIF-1α, hypoxia inducible factor-1α;
LC3, microtubule associated protein 1 light chain 3; SREBP1, sterol
regulatory element-binding protein 1; Nor, normoxia; Hypo, hypoxia;
ns, not significant. |
SREBP1 enhances autophagy in
MDA-MB-231 cells under hypoxic conditions
Autophagy is a key process of cell survival in a
hostile microenvironment (31,32).
Under hypoxic conditions, the level of LC3, a marker of autophagy,
was significantly increased in MDA-MB-231 cells and decreased in
MCF-7 cells, in comparison with under normoxic conditions (Fig. 2B). To evaluate the effect of SREBP1
on autophagy under hypoxic conditions, the initial staining with
Nile Red (Fig. 2A) was re-stained
for LC3. Under hypoxic conditions, the fluorescence intensity of
LC3 significantly increased in MDA-MB-231 cells comparison with
normoxic conditions and the effect was reversed by treatment with
fatostatin; however, no significant change was observed in MCF-7
cells (Fig. 3A). The conversion of
LC3 to its lower migrating form, LC3-II, is commonly used as a
marker of autophagy (30). Western
blot analysis revealed significantly decreased LC3-II protein
levels following treatment with fatostatin in MDA-MB-231 cells
under hypoxic conditions (Fig. 3B).
This suggests that SREBP1 regulates autophagy in MDA-MB-231 cells
under hypoxic conditions.
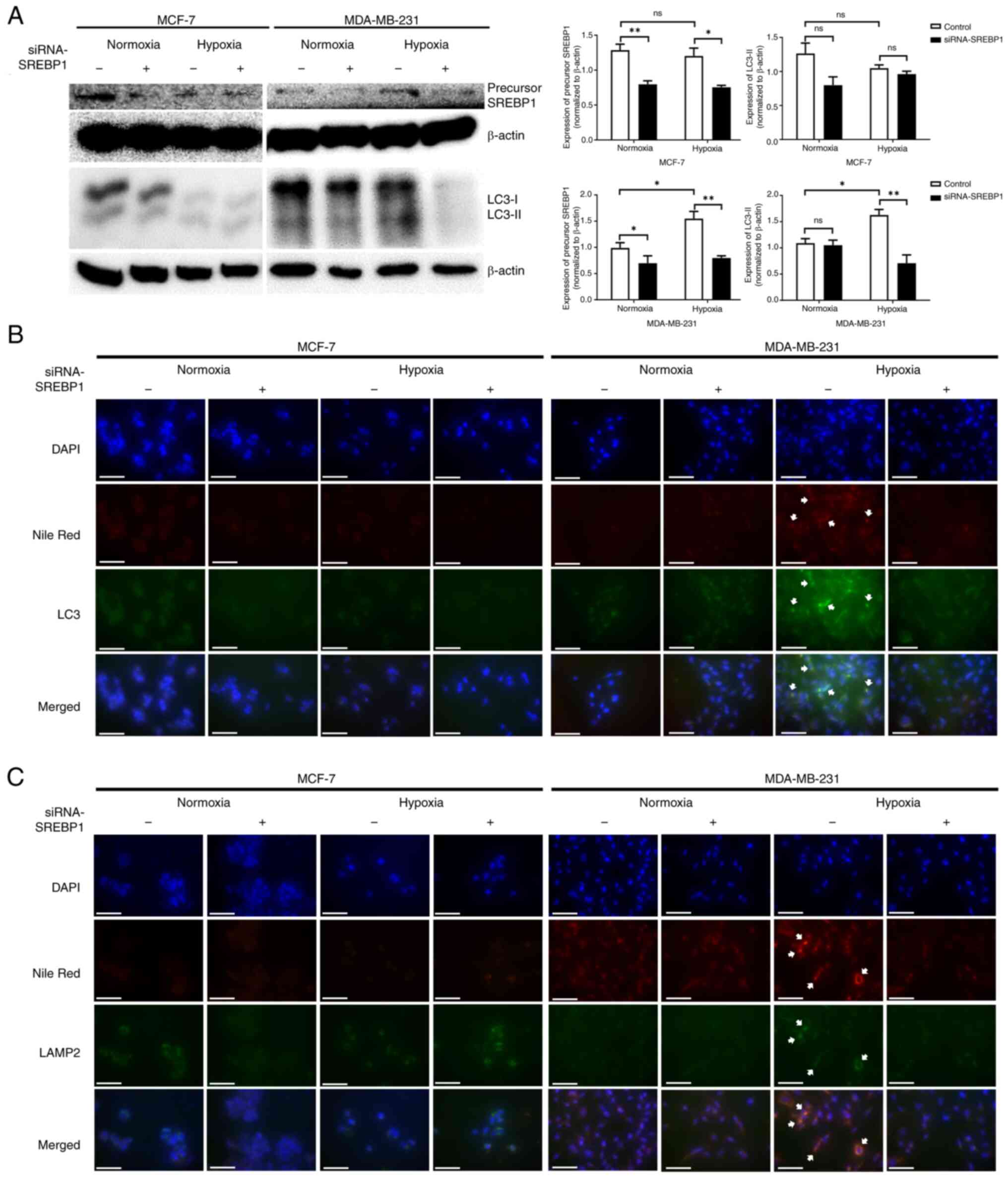
SREBP1 knockdown simultaneously alters
lipogenesis and autophagy in MDA-MB-231 cells under hypoxic
conditions
To evaluate whether SREBP1 directly mediates
autophagy and lipid reprogramming in cancer cells under hypoxic
conditions, siRNA was used to knockdown SREBP1 expression.
Transfection with SREBP1 siRNA significantly reduced SREBP1 protein
expression in both MCF-7 and MDA-MB-231 cells under normoxic and
hypoxic conditions compared with the negative control siRNA
(Fig. 4A). The expression of LC3-II
protein was significantly increased in MDA-MB-231 cells under
hypoxic conditions compared with under normoxic conditions,
although it was reversed by SREBP1 siRNA treatment. However, no
significant changes were observed in the MCF-7 cells after siRNA
treatment under normoxic or hypoxic conditions.
To confirm whether lipids are directly involved in
the autophagy process, immunofluorescence staining was performed
using the autophagosome marker LC3 and lysosome marker LAMP2, and
their co-localization with lipids was analyzed using Nile Red
staining following SREBP1 siRNA transfection. The co-localization
of LC3 and LAMP2 with Nile Red notably increased in MDA-MB-231
cells under hypoxic conditions compared with normoxic conditions
but decreased upon transfection with SREBP1 siRNAs (Fig. 4B and C). However, in MCF-7 cells, no
changes in the colocalization of LC3 and LAMP2 with Nile Red were
observed under varying oxygen concentrations or siRNAs treatments,
suggesting that SREBP1-mediated lipogenesis serves a direct role in
autophagy in MDA-MB-231 cells under hypoxic conditions.
SREBP1-mediates autophagy-enhanced
cell viability in MDA-MB-231 cells under hypoxic conditions
To determine the impact of SREBP1-mediated autophagy
on cancer cell survival under hypoxic conditions, cell viability
was evaluated using MTT and trypan blue assays in cells treated
with fatostatin and rapamycin. To determine the appropriate
concentration of rapamycin, cells were treated with several
concentrations of rapamycin, followed by the measurement of
autophagy using a Cyto-ID detection kit. In MCF-7 cells under
hypoxic conditions, autophagy was significantly induced by 1 µM
rapamycin but decreased upon treatment with 10 µM rapamycin.
However, autophagy was significantly induced upon treatment of
MDA-MB-231 cells with 0.1, 1 and 10 µM of rapamycin. Based on these
findings, a concentration of 1 µM rapamycin was selected, as it
induced autophagy maximally in both the cell lines (Fig. S2).
The MTT and trypan blue assays demonstrated that
fatostatin or rapamycin did not significantly reduce the viability
or live cell number of MCF-7 cells under hypoxic conditions
compared with the untreated control (Fig. 5). However, significant decreases in
viability and cell number were observed with both treatments
together under hypoxic conditions compared with the untreated
control group. Treatment of hypoxic MDA-MB-231 cells with
fatostatin and rapamycin significantly reduced cell viability and
cell number compared with the control (Fig. 5). Notably, combination treatment led
to a significant increase in viability and live cell number
compared with fatostatin treatment alone. Taken together, the
findings indicate that SREBP1-mediated autophagy serves a crucial
role in the survival of MDA-MB-231 cells under hypoxic conditions,
as evidenced by decreased cell viability upon SREBP1 inhibition,
followed by a subsequent increase in autophagy induction.
ATP production is induced via
SREBP1-mediated FAO in MDA-MB-231 cells under hypoxic
conditions
To assess whether fatty acids derived from
SREBP1-mediated autophagy serve as energy sources for cell
survival, the expression of FAO-related enzymes was assessed in
MDA-MB-231 cells treated with fatostatin and rapamycin under
hypoxic conditions. The expression levels of ACADVL, ACADM and
HADHA were significantly increased in MDA-MB-231 cells under
hypoxic conditions compared with under normoxic conditions, but the
expression was significantly decreased with fatostatin treatment
(Fig. 6A). Notably, the combined
treatment led to a significant increase in the expression levels of
all three enzymes compared with fatostatin treatment alone under
hypoxic conditions. The results suggest that FAO is regulated by
SREBP1-mediated autophagy.
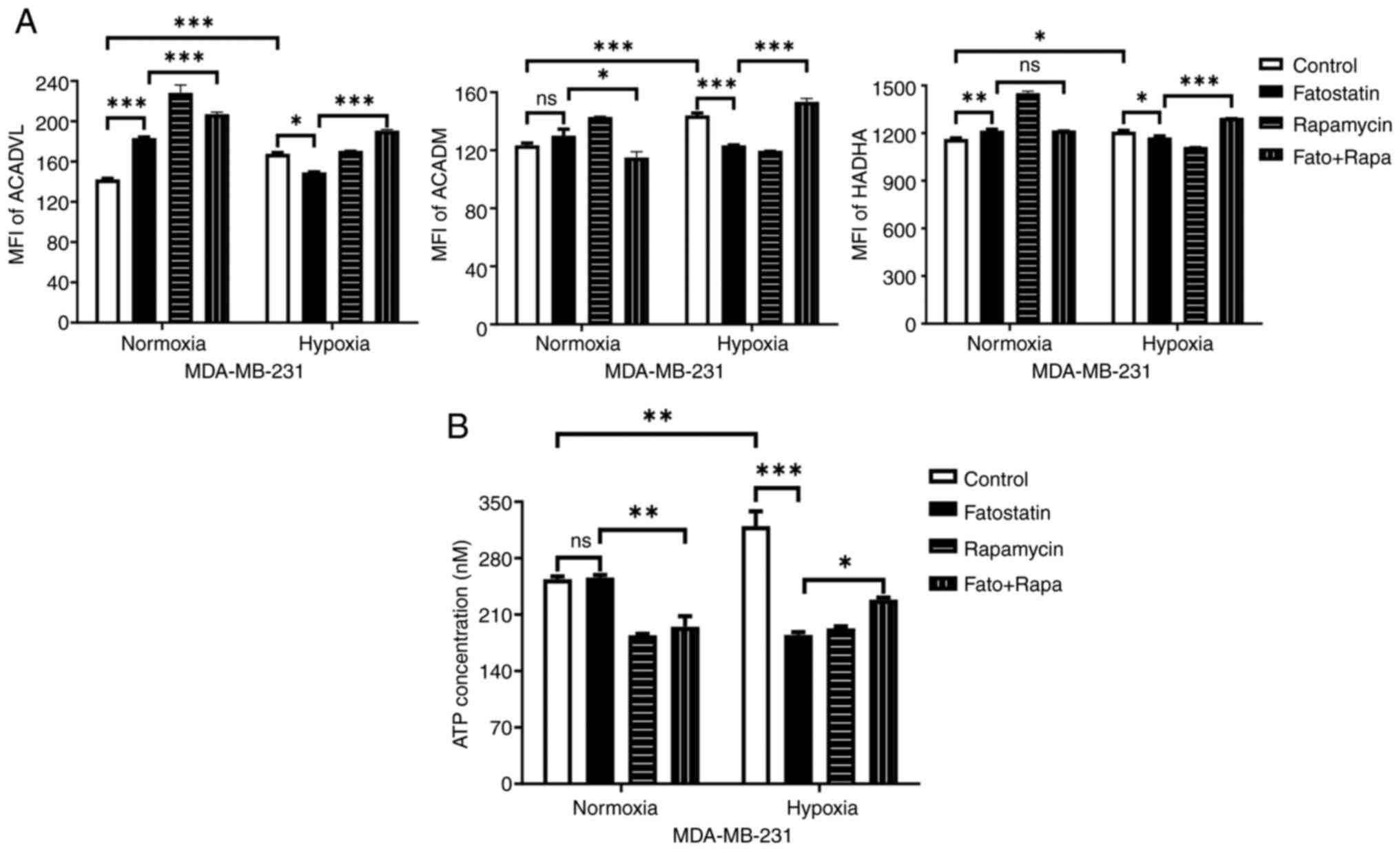 | Figure 6.ATP production via sterol regulatory
element-binding protein 1-mediated FAO observed in MDA-MB-231 cells
under hypoxic conditions. Assessment of (A) FAO-related enzymes,
ACADVL, ACADM and HADHA staining using flow cytometry and (B) ATP
concentration with fatostatin and rapamycin in MDA-MB-231 cells
under normoxic and hypoxic conditions for 48 h. *P<0.05;
**P<0.01; ***P<0.001. FAO, fatty acid oxidation; ACADVL,
acyl-CoA dehydrogenase very long chain; ACADM, acyl-CoA
dehydrogenase medium chain; HADHA, hydroxyacyl-CoA dehydrogenase
trifunctional multienzyme complex subunit a; MFI, mean fluorescence
intensity; Fato, fatostatin; Rapa, rapamycin; ns, not
significant. |
ATP concentration was measured to determine whether
ATP production occurs through FAO in MDA-MB-231 cells under hypoxic
conditions. ATP levels were significantly higher in MDA-MB-231
cells under hypoxic conditions compared with under normoxic
conditions, but treatment with fatostatin and rapamycin
significantly reduced the ATP levels. Notably, the combined
treatment significantly increased ATP levels compared with
fatostatin alone under hypoxic conditions (Fig. 6B).
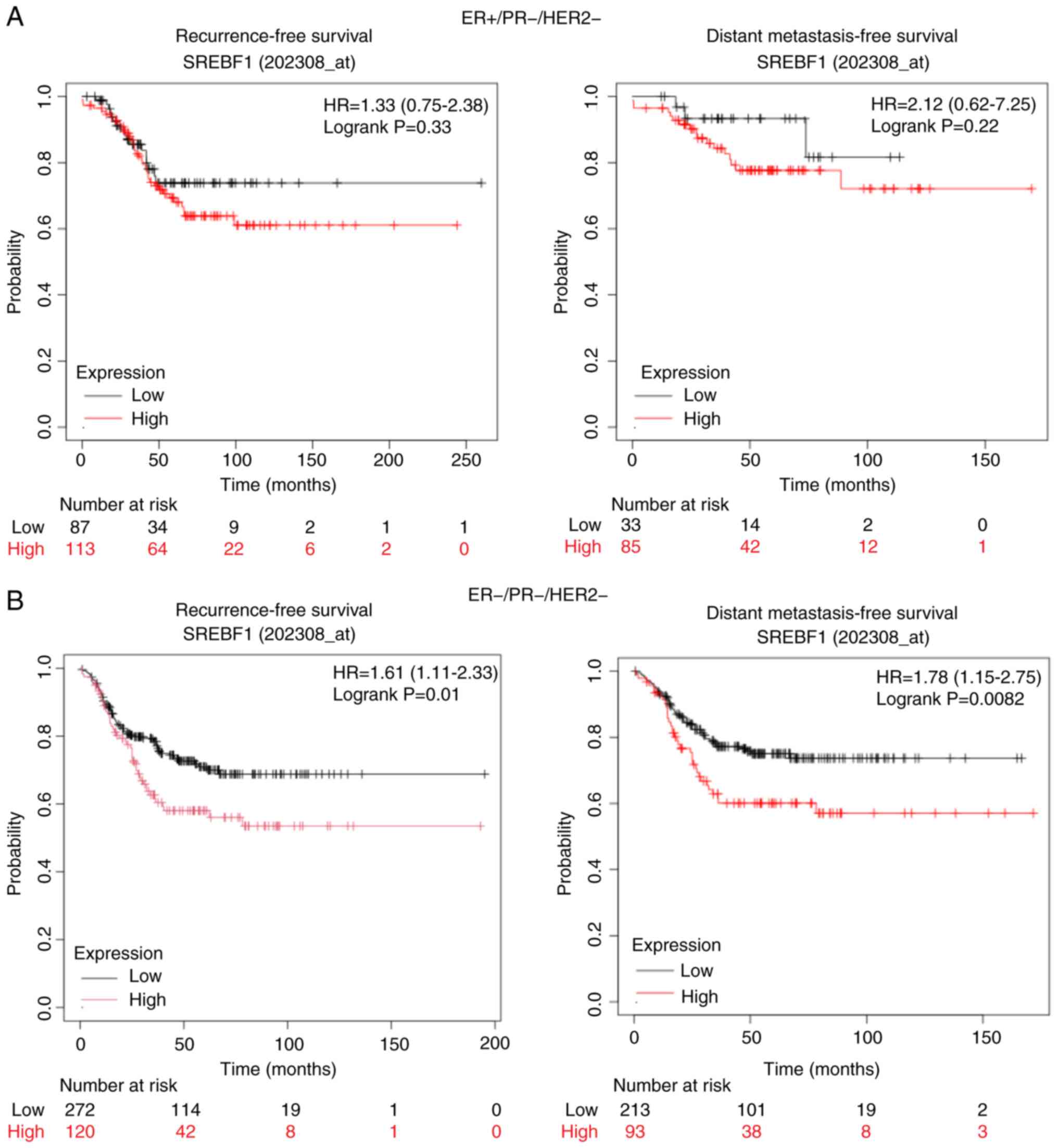
Expression of SREBF1 mRNA is
negatively associated with the survival of patients with TNBC
To evaluate the prognostic value of SREBP1 based on
SREBF1 mRNA expression for survival analysis, Kaplan-Meier plots
were generated for patients with breast cancer with ER-positive
(ER+/PR-/HER2-) and TNBC (ER-/PR-/HER2-) subtypes with a follow-up
period of 250 months. In patients with the ER-positive subtype, RFS
and DMFS were independent of SREBF1 mRNA expression (Fig. 7A). However, in patients with the
TNBC subtype, the patients with low SREBF1 mRNA expression had a
significantly longer RFS and DMFS compared with those with high
SREBF1 mRNA expression (Fig. 7B).
Specifically, the upper quartile survival of patients with TNBC
with low SREBF1 mRNA expression was 38.7 and 67.09 months for RFS
and DMFS, respectively; whereas for those with high SREBF1 mRNA
expression, both RFS and DMFS were 25 months.
Discussion
Oxygen scarcity in the tumor microenvironment poses
a notable challenge for cancer cells in achieving their metabolic
requirements. In response to hypoxia, cancer cells mobilize an
adaptive mechanism that activates less oxygen-dependent metabolic
pathways via HIF-1α activation (33). HIF-1α regulates the transcription of
the glucose transporter and corresponding enzyme genes, thereby
promoting ATP production through these processes. Previous studies
have reported that cancer cells surpass this process by activating
additional pathways, including nucleotide, amino acid and lipid
metabolism, to enhance tumor malignancy (3). The results of the present study
revealed that different breast cancer cell types survive under
hypoxic conditions in different ways and that SREBP1-mediated
lipogenesis and autophagy may serve a key role in MDA-MB-231 cells.
The findings also highlighted that the survival of MDA-MB-231 cells
is maintained by ATP production through FAO.
In cancer research, higher survival rates under
hypoxic conditions are positively associated with cancer severity
(34). In particular, TNBCs exhibit
higher HIF-1α-mediated activity than other breast cancer subtypes,
resulting in higher malignancy in molecular and prognostic terms
(35,36). The results of the present study are
consistent with these findings and demonstrate that MDA-MB-231
cells are more proliferative under hypoxic conditions compared with
MCF-10a and MCF-7 cells.
In the present study, SREBP1 expression in
MDA-MB-231 cells was demonstrated to be significantly upregulated
in the regulation of lipogenesis under hypoxic conditions. First
discovered in yeast, the role of SREBP1 in the hypoxic environment
has gained increasing importance in cancer research (37–39).
Given the ability of SREBP1 to promote the expression of lipid
metabolism-related genes, studying SREBP1-related lipid metabolism
in cancer has become increasingly important. Bensaad et al
(38) reported that LD accumulation
was higher in TNBC cell lines than in ER-positive cell lines under
hypoxic conditions, which aligns with the results of the present
study. Furthermore, the authors suggested that LD accumulation in
TNBC under hypoxic conditions occurs mainly through fatty acid
uptake by FABP rather than de novo lipogenesis via FASN. The
present study showed similar results in MDA-MB-231 cells under
hypoxic conditions. By contrast, Bensaad et al (38) identified an oxygen-independent role
of SREBP1 in MCF-7 cells but did not investigate it in MDA-MB-231
cells.
The importance of SREBP1-mediated lipid metabolism
in TNBC has been increasingly recognized (20,21).
The Kaplan-Meier analysis in the present study demonstrated that
high SREBF1 mRNA expression was associated with poor survival in
patients with TNBC, suggesting that SREBP1 may serve as a potential
prognostic biomarker. Therefore, elucidating the specific role of
SREBP1 for the survival of MDA-MB-231 cells under hypoxic
conditions is an important research challenge.
The present study also demonstrated that SREBP1
promotes autophagy in MDA-MB-231 cells under hypoxic conditions.
Autophagy is a highly conserved process that supplies energy and
macromolecular precursors essential for cell survival and can be
induced by several stressors, including radiation, drugs and
glucose and oxygen deprivation (31). Previous studies have highlighted
that hypoxia-induced autophagy is particularly important for cell
survival, including selective forms of autophagy such as mitophagy,
pexophagy, endoplasmic reticulum-phagy and lipophagy (32,40).
Although the relationship between lipid metabolism and autophagy is
controversial (41–43), lipophagy, which involves the
degradation of LDs, is considered to serve an important role in
cancer research (44). The results
of the present study revealed that SREBP1-mediated lipogenesis
increased the expression of both autophagy and lysosome markers,
suggesting the possibility of lipophagy in MDA-MB-231 cells under
hypoxic conditions.
Moreover, the present study demonstrated that
inhibiting SREBP1 decreased the survival of MDA-MB-231 cells;
however, combining SREBP1 inhibition with autophagy induction under
hypoxic conditions increased survival and regulated FAO and ATP
production. This indicated that autophagy via SRBP1 may serve an
important role in cell survival and as a source of fatty acids for
ATP production. The results also suggest that free fatty acids can
provide the necessary fuel for FAO through autophagy induction,
even when SREBP1-mediated lipogenesis is inhibited. However,
treatment with the autophagy inducer rapamycin alone resulted in
decreased FAO-related enzymes and ATP production along with
decreased survival, likely because excessive autophagy can cause
cell death (31). With
SREBP1-mediated autophagy already activated under hypoxic
conditions, further autophagy induction likely promoted cell death.
Although research on this is still limited, other studies have
reported that leptin-induced autophagy simultaneously activates FAO
and ATP production and increases SREBP1 expression, promoting
lipogenesis (45). However, this
effect was observed only in ER-positive cells under normoxic
conditions and was not assessed in TNBC cells under hypoxic
conditions. Thus, the present findings contribute to understanding
the mechanisms linking SREBP1-mediated lipogenesis, autophagy and
FAO in TNBC cells under hypoxia, suggesting new avenues for
investigation.
The present study also demonstrated that SREBP1
increases mitochondrial FAO-related enzymes and ATP production in
MDA-MB-231 cells under hypoxic conditions. Although oxidative
phosphorylation is generally considered to be inhibited under
hypoxic conditions, several studies have reported that ATP
production by oxidative phosphorylation continues to occur even at
low oxygen levels (46,47). Previous studies have provided a new
perspective by revealing that the primary energy source for cancer
cell survival in hypoxia is fatty acids rather than glucose
(25). This suggests that FAO is an
important metabolic pathway and a potential target for cancer
therapy. The results of the present study also support this,
demonstrating that FAO remains active and generates ATP even under
hypoxic conditions in MDA-MB-231 cells.
The present study has some limitations. First, the
study focused on three representative cell lines for each breast
cancer subtype. While this approach has significantly enhanced our
understanding of the unique role of SREBP1 in MDA-MB-231 cells
under hypoxic conditions, further studies using additional TNBC
cell lines and tissues are needed to validate these findings.
Second, the study lacks detailed mechanistic data for MCF-7 cells
under hypoxia. In this cell line, precursor SREBP1 levels remained
unchanged, while mature SREBP1 levels significantly decreased,
suggesting differential regulation of SREBP1 processing or
stability compared to MDA-MB-231 cells, where both forms were
increased. Moreover, discrepancies between LC3 levels of IF and
western blot in MCF-7 cells may reflect differences in LC3 spatial
distribution or dynamics under hypoxia and fatostatin treatment. In
contrast, MDA-MB-231 cells showed consistent results across both
methods, suggesting more straightforward regulatory mechanisms.
These findings underscore the complexity of SREBP1 and LC3
regulation in MCF-7 cells, warranting further investigation to
fully understand these processes.
In conclusion, the present study revealed that
SREBP1-mediated lipogenesis and autophagy significantly increase
under hypoxic conditions, which is essential for the survival of
MDA-MB-231 cells. The interaction of SREBP1-mediated lipogenesis
and autophagy is likely to promote ATP production via FAO to
support cell survival. Furthermore, the results suggest that SREBP1
may serve as a promising prognostic marker for TNBC. These findings
demonstrate that a therapeutic strategy that targets
SREBP1-mediated lipid reprogramming and autophagy together may
offer a promising approach to address the limitations of existing
therapies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Je-Yoel Cho
(Laboratory of Biochemistry, College of Veterinary Medicine, Seoul
National University, Seoul, Republic of Korea) and Dr. So Yeong Lee
(Laboratory of Pharmacology, College of Veterinary Medicine, Seoul
National University, Republic of Korea) for providing cell
lines.
Funding
The present study was supported by BK21 FOUR Future Veterinary
Medicine Leading Education and Research Center and the Basic
Science Research Program through the National Research Foundation
of Korea funded by the Ministry of Education, Science and
Technology (grant nos. 2020R1A2C1010215 and RS-2024-00351740).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JJ performed the majority of the experiments,
analyzed the data and was a major contributor to writing the
manuscript. YY analyzed and interpreted the data. YK supervised the
project, assisted with the research design and manuscript
preparation. YY and YK confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ANOVA
|
analysis of variance
|
|
DMFS
|
distant metastasis-free survival
|
|
ER
|
estrogen receptor
|
|
FABP
|
fatty acid binding protein
|
|
FAO
|
fatty acid oxidation
|
|
FASN
|
fatty acid synthase
|
|
HIF-1α
|
hypoxia inducible factor-1α
|
|
LD
|
lipid droplet
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
|
|
PBS
|
phosphate-buffered saline
|
|
PBST
|
PBS containing 0.05% Tween 20
|
|
RFS
|
recurrence-free survival
|
|
SREBF1
|
sterol regulatory element-binding
factor 1
|
|
SREBP1
|
sterol regulatory element-binding
protein 1
|
|
TNBC
|
triple-negative breast cancer
|
References
|
1
|
Liu S, Zhang X, Wang W, Li X, Sun X, Zhao
Y, Wang Q, Li Y, Hu F and Ren H: Metabolic reprogramming and
therapeutic resistance in primary and metastatic breast cancer. Mol
Cancer. 23:2612024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Munir R, Lisec J, Swinnen JV and Zaidi N:
Lipid metabolism in cancer cells under metabolic stress. Br J
Cancer. 120:1090–1098. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadfield LA, Pane AA, Talebi A, Swinnen
JV and Fendt SM: Lipid metabolism in cancer: New perspectives and
emerging mechanisms. Dev Cell. 56:1363–1393. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Global Burden of Disease 2019 Cancer
Collaboration, . Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL,
Harvey JD, Henrikson HJ, Lu D, Pennini A, et al: Cancer incidence,
mortality, years of life lost, years lived with disability, and
disability-adjusted life years for 29 cancer groups from 2010 to
2019: A systematic analysis for the global burden of disease study
2019. JAMA Oncol. 8:420–444. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zipinotti dos Santos D, de Souza JC,
Pimenta TM, da Silva Martins B, Junior RSR, Butzene SMS, Tessarolo
NG, Cilas PML Jr, Silva IV and Rangel LBA: The impact of lipid
metabolism on breast cancer: A review about its role in
tumorigenesis and immune escape. Cell Commun Signal. 21:1612023.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harborg S, Zachariae R, Olsen J, Johannsen
M, Cronin-Fenton D, Bøggild H and Borgquist S: Overweight and
prognosis in triple-negative breast cancer patients: A systematic
review and meta-analysis. NPJ Breast Cancer. 7:1192021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kaul K, Misri S, Ramaswamy B and Ganju RK:
Contribution of the tumor and obese microenvironment to triple
negative breast cancer. Cancer Lett. 509:115–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun X, Wang M, Wang M, Yu X, Guo J, Sun T,
Li X, Yao L, Dong H and Xu Y: Metabolic reprogramming in
triple-negative breast cancer. Front Oncol. 10:4282020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Jiang Q and Dong C: Metabolic
reprogramming in triple-negative breast cancer. Cancer Biol Med.
17:44–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marino N, German R, Rao X, Simpson E, Liu
S, Wan J, Liu Y, Sandusky G, Jacobsen M, Stoval M, et al:
Upregulation of lipid metabolism genes in the breast prior to
cancer diagnosis. NPJ Breast Cancer. 6:502020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giró-Perafita A, Palomeras S, Lum DH,
Blancafort A, Viñas G, Oliveras G, Pérez-Bueno F, Sarrats A, Welm
AL and Puig T: Preclinical evaluation of fatty acid synthase and
EGFR inhibition in triple-negative breast cancer. Clin Cancer Res.
22:4687–4697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Zhang P, Xu J, Lv G and Li Y: Lipid
metabolism in tumor microenvironment: Novel therapeutic targets.
Cancer Cell Int. 22:2242022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo D, Bell EH, Mischel P and Chakravarti
A: Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr
Pharm Des. 20:2619–2626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38:272018.PubMed/NCBI
|
|
16
|
Griffiths B, Lewis CA, Bensaad K, Ros S,
Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, et al:
Sterol regulatory element binding protein-dependent regulation of
lipid synthesis supports cell survival and tumor growth. Cancer
Metab. 1:32013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lo AKF, Lung RWM, Dawson CW, Young LS, Ko
CW, Yeung WW, Kang W, To KF and Lo KW: Activation of sterol
regulatory element-binding protein 1 (SREBP1)-mediated lipogenesis
by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1)
promotes cell proliferation and progression of nasopharyngeal
carcinoma. J Pathol. 246:180–190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi Z, Zhou Q, Gao S, Li W, Li X, Liu Z,
Jin P and Jiang J: Silibinin inhibits endometrial carcinoma via
blocking pathways of STAT3 activation and SREBP1-mediated lipid
accumulation. Life Sci. 217:70–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang N, Zhang H, Liu Y, Su P, Zhang J,
Wang X, Sun M, Chen B, Zhao W, Wang L, et al: SREBP1, targeted by
miR-18a-5p, modulates epithelial-mesenchymal transition in breast
cancer via forming a co-repressor complex with Snail and HDAC1/2.
Cell Death Differ. 26:843–859. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mahmud I, Tian G, Wang J, Hutchinson TE,
Kim BJ, Awasthee N, Hale S, Meng C, Moore A, Zhao L, et al: DAXX
drives de novo lipogenesis and contributes to tumorigenesis. Nat
Commun. 14:19272023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu P, Zhou P, Sun T, Liu D, Yin J and Liu
L: Therapeutic protein PAK restrains the progression of triple
negative breast cancer through degrading SREBP-1 mRNA. Breast
Cancer Res. 25:1512023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Azam A and Sounni NE: Lipid metabolism
heterogeneity and crosstalk with mitochondria functions drive
breast cancer progression and drug resistance. Cancers (Basel).
14:62672022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Listenberger LL, Han X, Lewis SE, Cases S,
Farese RV Jr, Ory DS and Schaffer JE: Triglyceride accumulation
protects against fatty acid-induced lipotoxicity. Proc Natl Acad
Sci USA. 100:3077–3082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoon H, Shaw JL, Haigis MC and Greka A:
Lipid metabolism in sickness and in health: Emerging regulators of
lipotoxicity. Mol Cell. 81:3708–3730. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee H, Woo SM, Jang H, Kang M and Kim SY:
Cancer depends on fatty acids for ATP production: A possible link
between cancer and obesity. Semin Cancer Biol. 86:347–357. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ping P, Li J, Lei H and Xu X: Fatty acid
metabolism: A new therapeutic target for cervical cancer. Front
Oncol. 13:11117782023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miller DM and Shakes DC:
Immunofluorescence microscopy. Methods Cell Biol. 48:365–394. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghosh R, Gilda JE and Gomes AV: The
necessity of and strategies for improving confidence in the
accuracy of western blots. Expert Rev Proteomics. 11:549–560. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy research. Cell. 140:313–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mulcahy Levy JM and Thorburn A: Autophagy
in cancer: Moving from understanding mechanism to improving therapy
responses in patients. Cell Death Differ. 27:843–857. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Daskalaki I, Gkikas I and Tavernarakis N:
Hypoxia and selective autophagy in cancer development and therapy.
Front Cell Dev Biol. 6:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Finger EC and Giaccia AJ: Hypoxia,
inflammation, and the tumor microenvironment in metastatic disease.
Cancer Metastasis Rev. 29:285–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Zhang H, Wang M, Schmid T, Xin L,
Kozhuharova L, Yu WK, Huang Y, Cai F and Biskup E: Hypoxia in
breast cancer-scientific translation to therapeutic and diagnostic
clinical applications. Front Oncol. 11:6522662021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Todd BL, Stewart EV, Burg JS, Hughes AL
and Espenshade PJ: Sterol regulatory element binding protein is a
principal regulator of anaerobic gene expression in fission yeast.
Mol Cell Biol. 26:2817–2831. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bensaad K, Favaro E, Lewis CA, Peck B,
Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, et al:
Fatty acid uptake and lipid storage induced by HIF-1α contribute to
cell growth and survival after hypoxia-reoxygenation. Cell Rep.
9:349–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lewis C, Brault C, Peck B, Bensaad K,
Griffiths B, Mitter R, Chakravarty P, East P, Dankworth B, Alibhai
D, et al: SREBP maintains lipid biosynthesis and viability of
cancer cells under lipid- and oxygen-deprived conditions and
defines a gene signature associated with poor survival in
glioblastoma multiforme. Oncogene. 34:5128–5140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li W, He P, Huang YF, Li Y, Lu J, Li M,
Kurihara H, Luo Z, Meng T, Onishi M, et al: Selective autophagy of
intracellular organelles: Recent research advances. Theranostics.
11:222–256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Soto-Avellaneda A and Morrison BE:
Signaling and other functions of lipids in autophagy: A review.
Lipids Health Dis. 19:2142020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yan Q, Song Y, Zhang L, Chen Z, Yang C,
Liu S, Yuan X, Gao H, Ding G and Wang H: Autophagy activation
contributes to lipid accumulation in tubular epithelial cells
during kidney fibrosis. Cell Death Discov. 4:22018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou C, Qian W, Li J, Ma J, Chen X, Jiang
Z, Cheng L, Duan W, Wang Z, Wu Z, et al: High glucose
microenvironment accelerates tumor growth via SREBP1-autophagy axis
in pancreatic cancer. J Exp Clin Cancer Res. 38:3022019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Singh R, Kaushik S, Wang Y, Xiang Y, Novak
I, Komatsu M, Tanaka K, Cuervo AM and Czaja MJ: Autophagy regulates
lipid metabolism. Nature. 458:1131–1135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pham DV, Tilija Pun N and Park PH:
Autophagy activation and SREBP-1 induction contribute to fatty acid
metabolic reprogramming by leptin in breast cancer cells. Mol
Oncol. 15:657–678. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chandel NS, Budinger GR, Choe SH and
Schumacker PT: Cellular respiration during hypoxia. Role of
cytochrome oxidase as the oxygen sensor in hepatocytes. J Biol
Chem. 272:18808–18816. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ashton TM, McKenna WG, Kunz-Schughart LA
and Higgins GS: Oxidative phosphorylation as an emerging target in
cancer therapy. Clin Cancer Res. 24:2482–2490. 2018. View Article : Google Scholar : PubMed/NCBI
|