Introduction
Acute myeloid leukemia (AML) is a malignant clonal
disease originating from hematopoietic stem progenitor cells
(1). Chromosomal heterogeneous t(8;
21) (q22; q22.1) translocations occur in certain patients with AML,
where the Runt-related transcription factor 1 (RUNX1) gene (also
known as AML1) at 21q22 is fused to the RUNX1 partner
transcriptional co-repressor 1 (RUNX1T1) gene (also known as ETO)
at 8q22, resulting in a RUNX1::RUNX1T1 fusion gene on chromosome 8.
This has a positive rate of 20–40% in AML with maturation (AML-M2)
and is found to a lesser degree in acute myelomonocytic leukemia
(AML-M4) and undifferentiated AML (AML-M1) (2). The 5th Edition of the World Health
Organization Classification of Hematolymphoid Tumors defines
RUNX1::RUNX1T1 fusion gene positivity as genetically abnormal AML;
that is, a diagnosis of AML should be considered when the
RUNX1::RUNX1T1 fusion gene is monitored, even if the blast cell
count is <20% (1).
The presence of RUNX1::RUNX1T1-positive AML is a
well-characterized karyotype associated with a favorable prognosis.
Remission induction through combination chemotherapy has shown a
complete response (CR) rate of 90%, with long-term disease-free
survival rates ranging from 50–70% after 5 years (2). It is crucial to regularly monitor the
quantitative levels of the RUNX1::RUNX1T1 fusion genes to assess
treatment efficacy and detect disease recurrence (1).
Monoclonal gammopathy of undetermined significance
(MGUS) refers to the presence of monoclonal immunoglobulins in
serum and urine; however, the proportion of plasma cells in the
bone marrow, clinical symptoms and biochemical indices is
insufficient to diagnose multiple myeloma and other plasma cell
diseases (3). MGUS is prevalent in
~3% of the population aged >50 years, and its incidence
increases with age. The progression rate from MGUS to multiple
myeloma (MM) is ~1% of patients per year, suggesting that the
majority of patients with MGUS remain undiagnosed and do not
develop symptomatic malignancies (3).
AML with a complex chromosomal translocation
t(8;17;21) (q22;q24;q22) is rarely reported, and at the same time,
its coexistence with MGUS is extremely uncommon. The present study
describes a case of AML with a RUNX1:RUNX1T1 fusion gene and the
complex chromosomal translocation (8; 17; 21), which was found to
be associated with MGUS during treatment. The therapeutic regimen
and prognosis of this patient are described and the relevant
literature on this rare type of AML is reviewed to improve
awareness of this disease.
Case report
A 57-year-old male with no history of hematological
diseases was admitted to the Second Hospital of Hebei Medical
University (Shijiazhuang, China) for treatment in October 2022 due
to a hemoglobin count decrease for >5 months and fatigue for 1
week. An assessment revealed the presence of anemia, devoid of any
apparent bleeding diathesis, superficial lymphadenopathy or
palpable enlargement of the liver and spleen. On admission, routine
peripheral blood analysis demonstrated the following results: White
blood cell count, 5.40×109/l (3.5–9.5×109/l);
hemoglobin level, 60 g/l (115–150 g/l); and platelet count,
29×109/l (125–300×109/l). The bone marrow
exhibited hypercellularity, with blast cells comprising 44.0% of
the total cell population. Bone marrow fluid was collected for flow
cytometry examination. The concentration of single-cell suspension
was 1×10^7/ml. EDTA anticoagulant were added to single-cell
suspension and antibodies were added: CD38 (FITC, Kuangbo Tongsheng
Biotechnology Co., Ltd., 103802), CD117 (P, Kuangbo Tongsheng
Biotechnology Co., Ltd., 111704), CD34 (PerCP/Cyanine5.5;
Biolegend, 343522), CD33 (PE-CY7, Biolegend, 366618), CD13 (APC,
Kuangbo Tongsheng Biotechnology Co., Ltd., A6009R12), CD123
(APC-A700, Biolegend, 306040), HLA-DR (APC/Cyanine7, Biolegend,
307618), CD11b (BV421, Biolegend, 301324), CD15 FITC, Biolegend,
301904), CD34 (PE, Kuangbo Tongsheng Biotechnology Co., Ltd.,
Z6410008), CD56 (PercpCy5.5, Kuangbo Tongsheng Biotechnology Co.,
Ltd., 105610), CD7 (APC, Kuangbo Tongsheng Biotechnology Co., Ltd.,
A6005R12), CD14 (APC/Fire 750, Biolegend, 301854), CD19 (BV421,
Biolegend, 302234), CD45 (QB500, Kuangbo Tongsheng Biotechnology
Co., Ltd., A6015V32) at 100 µl/tube, shaken, and kept in darkness
at room temperature for 30 min. A total of 3% paraformaldehyde was
added and incubated in the dark for 10 min at room temperature.1 ml
of cell membrane breaking buffer was added, and the solution was
incubated it at 4°C in the dark for 30 min subsequent to shaking.
The solution was centrifuged at 160 × g for 5 min at room
temperature, and the supernatant was discarded. A total of 5 ml of
PBS solution was added, shaken, and centrifuged at 160 × g for 5
min at room temperature. The above steps need to be repeated twice.
The supernatant was discarded, and 300 µl of PBS was added to form
a suspension. After adding FITC, phycoerythrin, peridinin chlorophy
II protion (PreCP) and allophycocyanin (APC), flow cytometer
(Beckman Coulter, Navios) to detect and analyze the cell
suspension. Data analysis was performed using Kaluza 2.1.1 software
(Beckman Coulter, Inc.). Flow cytometry analysis revealed abnormal
myeloid blasts accounting for 44.23% of all myeloid blast cells
(Fig. S1). We collect bone marrow
fluid for next-generation sequencing. The extraction steps for DNA
and RNA in next-generation sequencing were as aforementioned. The
DNA and RNA were extracted using commercial kits (Tiangen Biotech,
Co., Ltd.). The purities and concentrations of DNA and RNA were
confirmed using a Nanodrop 2000 (Thermo Scientific, Inc.) and a
Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Inc.). The
integrity of the DNA and RNA was evaluated with the Qsep400 nucleic
acid fragment analyzer (Hangzhou Houze Bio-Technology Co., Ltd.).
The DNA was transformed into libraries with the help of KAPA
EvoPlus kits (Kapa Biosystems; Roche Diagnostics;9420053001). The
libraries were analyzed on the Illumina sequencing platform
NextSeq550 using 150bp paired-end sequencing. Sequencing was
performed with the NextSeq 500/550 High Output Kit v2.5 (300
cycles; 20024908; Illumina Inc.). The final library concentration
was quantified using the Qubit 3.0 Fluorometer. The loading
concentration of the final library was ~14 pM. The mutation data
was screened through dbSNP (bioinfo.org.cn/relative/dbSNP%
20Home%20 Page.htm), gnomAD (ngdc.cncb.
ac.cn/databasecommons/database/id/6934), COSMIC
(cancer.sanger.ac.uk/cosmic/download), and Polyphen2 SIFT
(http://provean.jcvi.org/protein_batch_submit.php?species=human),
as well as the internal database of Tianjin Jiankang Huamei
Laboratory. Then the data was analyzed using BWA
(github.com/bwa-mem2/bwa-mem2), Sambamba (github.
com/biod/sambamba/releases), Bamdst (https://github.com/shiquan/bamdst), VarDict
(GitHub-AstraZeneca-NGS/VarDict: VarDict), CNV kit (cnvkit.
readthedocs.io/en/stable/), Pinde (gmt.genome.
wustl.edu/packages/pindel/), ANNOVAR (annovar. openbioinformatics.
org/en/latest/). Next-generation sequencing showed the presence of
Fms-related receptor tyrosine kinase 3 (Exon20. c2516>G.p.D839G)
and isocitrate dehydrogenase [NADP(+)] 2 (Exon4.c.419>
A.p.R140Q) mutations.200 µl of chloroform was added to the bone
marrow fluid and mixed it before placing it on ice for 5 min. The
mixture was centrifuged at 12,000 × g at 4°C for 15 min and were
used a pipette to aspirate 400–500 µl of the top supernatant. The
Specimens were transferred the supernatant to another new EP tube
and were added 400–500 µl of isopropanol (in a volume ratio of 1:1
to the supernatant). The mixture were placed on ice until the RNA
was completely precipitated. The Specimens were used by a
low-temperature high-speed centrifuge to centrifuge at 12,000 × g
for 10 min at 4°C, then discarded the supernatant and thoroughly
mixed it with 1 ml of 75% ethanol. The mixed liquid and discard the
supernatant. The supernatant was transferred into a DECP-treated EP
tube. The resuspended liquid was incubated at 65°C for 5 min and
mixed. RNA was obtained by centrifugation again at 2–8°C at 12,000
× g for 5 min.cDNA was synthesized from 2 µg of total RNA using a
1st Strand cDNA Synthesis Kit (Hifair III cDNA, Yeasen
Biotechnology (Shanghai) Co., Ltd., 11150ES10). The following
primer sequences were used: RUNX1::RUNX1T1, (forward)
5′-AML1-FCACCTACCACAGAGCCATCAAA-3′ and (reverse)
5′-ETO-RATCCACAGGTGAGTCTGGCATT-3′; housekeeping genes: ABL,
(forward) 5′-ENF1003TGGAGATAACACTCTAAGCATAACTAAAGGT-3′ and
(reverse) 5′-ENR1063GATGTAGTTGCTTGGGACCCA-3′ (SINO-US Diagnostics;
Tianjin Jiankang Huamei Medical Diagnostic Technology Co., Ltd.)
(4,5). The main sample for testing was fresh
anticoagulant bone marrow fluid, with a standard bone marrow
collection volume of 3–5 ml. The anticoagulant used was EDTA or
3.2% sodium citrate. The optimal concentration of RNA was 300 ng/µl
and the optimal purity was 1.8–2.0 for A260:A280 and 2.0–2.2 for
A260:A230. Amplification conditions were as follows (40 cycles: 2
min at 50°C, 10 min at 95°C, 15 sec at 95°C and 60 sec at 95°C. The
sample was then stored at 4°C. The results of reverse
transcription-PCR confirmed the presence of a RUNX1::RUNX1T1 fusion
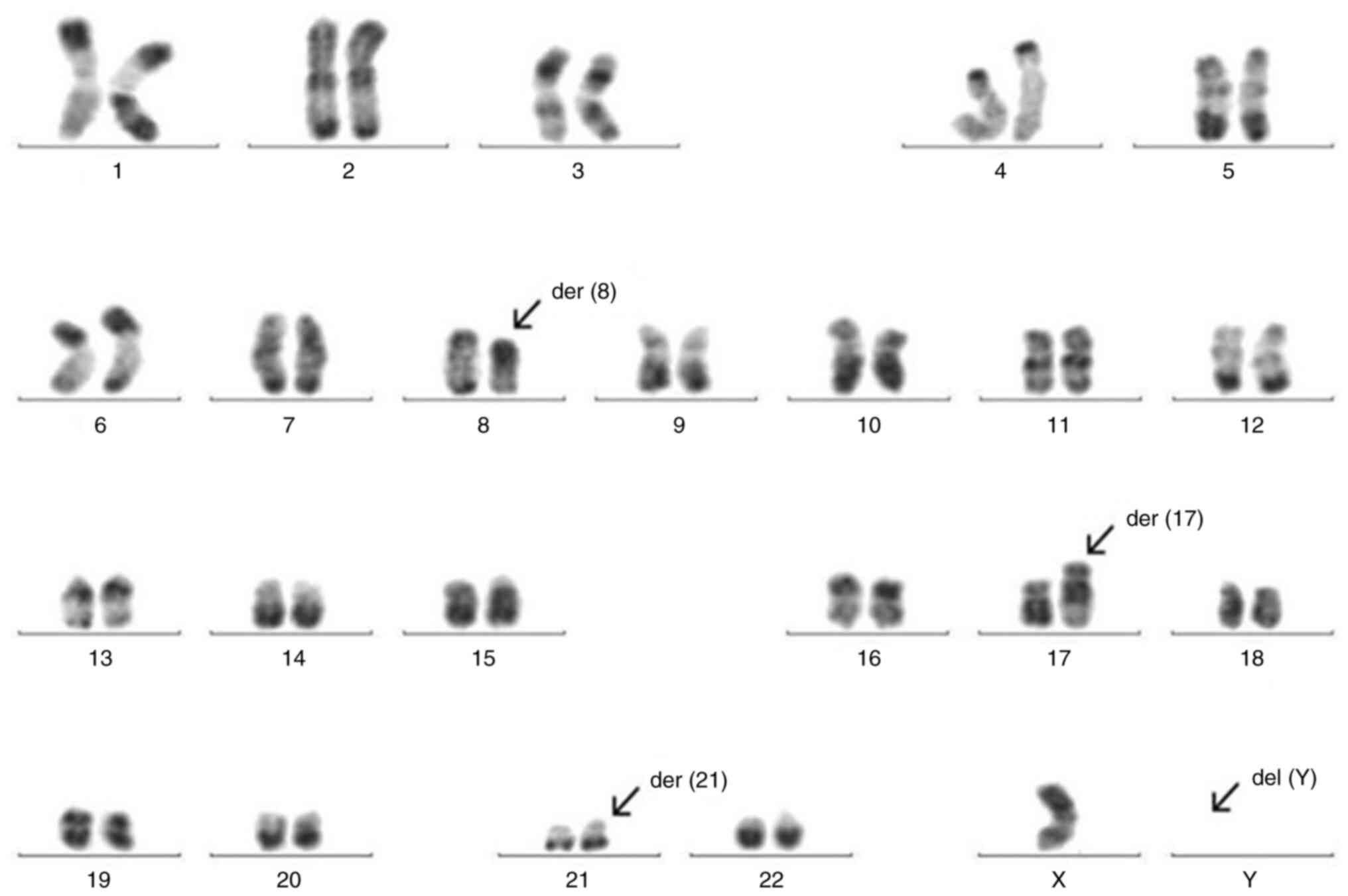
gene. The 20 metaphase chromosomes were subjected to R-banding
analysis, which revealed a complex karyotype with the following
abnormalities: 46,XY, t(8;17;21)(q22;q24;22.1)[9]/45,
idem,-Y[10]/46,XY[1] (Fig. 1).
Given that the chromosomal karyotype findings
deviated from the typical presentation associated with the
RUNX1::RUNX1T1 fusion gene, additional fluorescence in situ
hybridization (FISH) tests were performed to corroborate the
presence of the fusion gene and validate the chromosomal anomalies.
The samples were fixed three times in methanol and acetic acid
(3:1) following a prefixation step using a 10% solution. The fixed
cells were washed with a 2X sodium citrate saline buffer solution
at 37°C for 30 min, then dehydrated in 75, 85, and 100% ethanol for
1 min each, sequentially. The FISH probes for RUNX1 (Abnov, no.
FA0446) and RUNX1T1 (Abnov, no. H00000862-M01) were utilized with a
hybridization instrument (HANGZHOU RUICHENG INSTRUMENT CO., LTD.,
SH2000) for hybridization, denaturing at 78°C for 8 min, and
hybridizing at 42°C for 16 h. The next day, the samples were washed
with 0.3% NP40 detergent at 68°C for 2 min, and then were washed
with deionized water at 37°C for 1 mi. DAPI nuclear staining was
performed at room temperature for 20 min. FISH probe kits were
purchased from Wuhan Kanglu Biotechnology Co., Ltd. Results were
observed using a fluorescence microscope (Olympus; catalog number
BX63), and Metasystem ISIS V5.8.11 (Metasystem Co., Ltd.) FISH
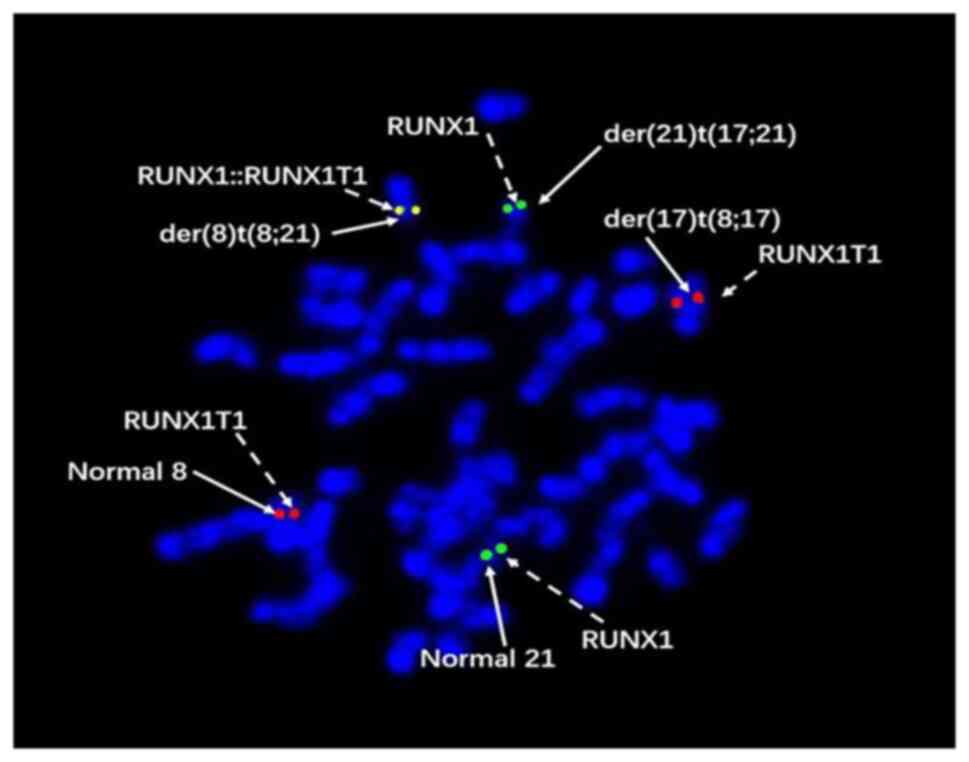
analysis software was used for photography and analysis. AML1
(21q22) gene labeled green and ETO (8q22) gene labeled in red. The
AML1-ETO fusion gene displayed a yellow or red-green superimposed
signal, with a normal signal characteristic of 2R2G and a positive
signal characteristic of 1R1G2F (G is a green signal, R is a red
signal, and F is a fusion signal). The FISH analysis revealed
distinct fluorescence patterns: The derivative chromosome 17
involving the t(8;17) translocation exhibited red fluorescence, the
derivative chromosome 21 involving the t(17;21) translocation
displayed green fluorescence and the derivative chromosome 8
involving the t(8;21) translocation showed a merged red-green
fluorescence, indicative of a complex chromosome translocation
event t(8;17;21) (Fig. 2). The
diagnosis was finally determined as AML with t(8;17;21)
(q22;q24;q22.1) chromosome karyotype.
After a course of idarubicin (12 mg/m2,
d1-3) + cytarabine (100 mg/m2, d1-7) regimen induction
treatment, the patient achieved a CR. However, due to financial
constraints, the family of the patient opted out of pursuing
intensive treatment involving high-dose cytarabine and declined
hematopoietic stem cell transplantation. The patient instead
requested a standard dose of chemotherapy. After 2 cycles of
consolidation chemotherapy with a daunorubicin (12
mg/m2, d1-3) + cytarabine (100 mg/m2, d1-7)
regimen, primitive and immature cells accounted for less than 5% of
bone marrow nucleated cells, indicating CR according to The 5th
edition of the World Health Organization Classification of
Haematolymphoid Tumours diagnostic criteria (1). But monitoring of the RUNX1::RUNX1T1
fusion gene of the patient showed positivity at low levels. After 4
months, bone marrow fluid was collected for bone marrow cytology
examination. The bone marrow smears were dried naturally before
addition of 2–3 drops of Wright Giemsa staining solution at room
temperature for 1–2 min. An equal amount of 0.01 mol/l sodium
dihydrogen phosphate was added at room temperature for 3–5 min. The
smear was washed with double distilled water, dried and examined
under a light microscope (Olympus Corporation). Bone marrow
cytology during the fourth hospitalization of the patient
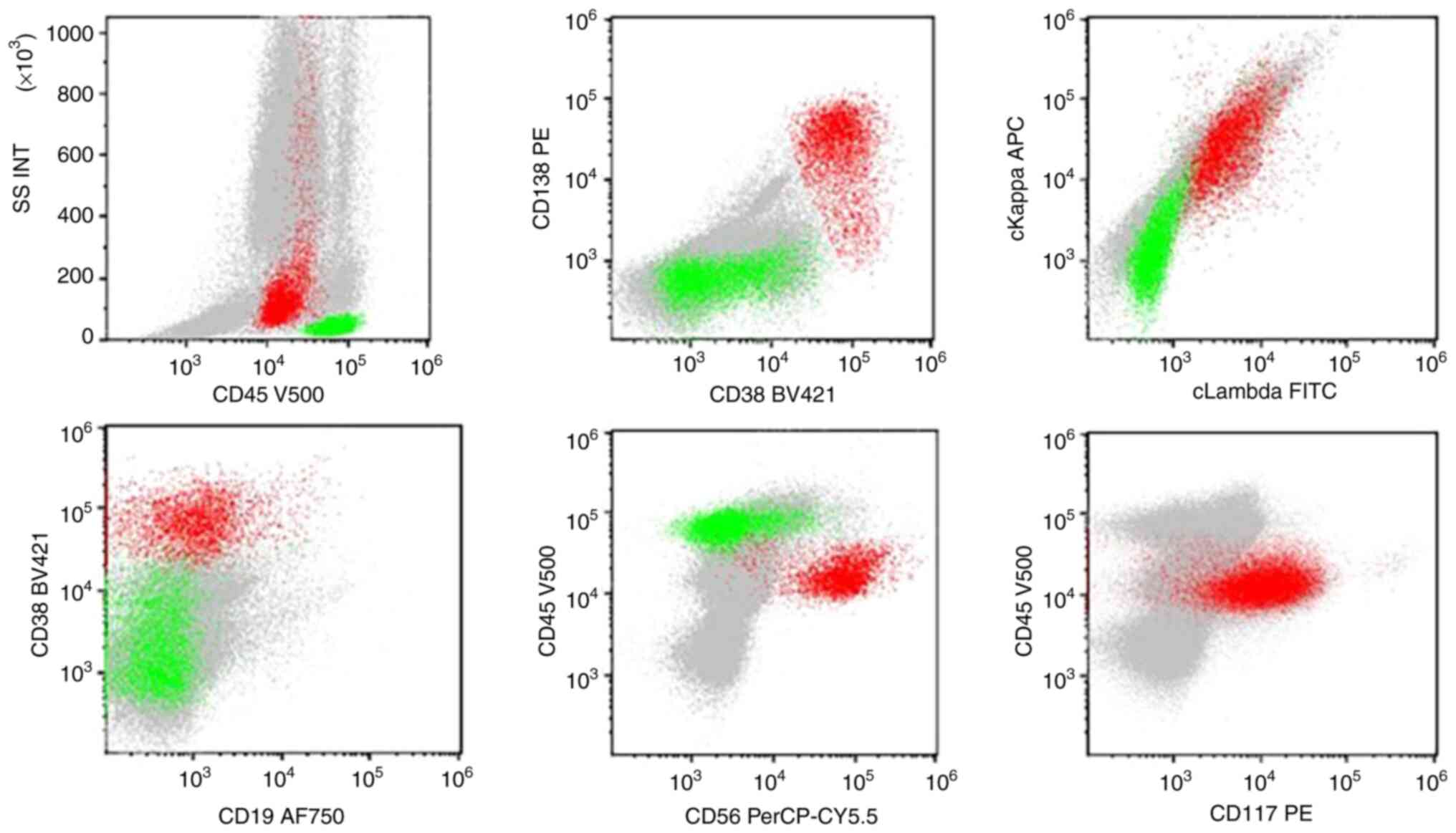
demonstrated 0.5% blasts and 4.5% mature plasma cells. Flow
cytology revealed 0.11% abnormal myeloid blasts and 2.07%
monoclonal plasma cells (expressing CD38, CD138, CD117 and CD56;
Fig. 3). Peripheral venous blood
and urine from patients were collected for quantitative detection
of immunoglobulin. A total of 100 µl of diluent was added to a 20
µl patient serum sample. Specimens were examined using
electrophoresis apparatus (Instrument and model: Sebia Hydrasys
2/Helena SPIFE). Urine samples do not require dilution.10 µl of
diluted serum or original urine samples were added into the wells
of the electrophoresis apparatus sampler, respectively. After the
last sample is added, the samples were diffused in the comb for 5
min. The buffer strips were removed from the electrophoresis
apparatus. The perforated plastic end of the sponge was inserted
into the pin on the electrode transporter, with the northern part
of the plastic end facing towards the transporter. The gels were
taken out and the excess liquid on their surface was gently
absorbed with a thin filter paper. A total of 120 µl of distilled
water was added to the bottom third box on the temperature control
board of the electrophoresis module. The adhesive side of the film
was placed upwards, and the edge of the film was pressed tightly
against the bottom edge of the frame. The film should be free of
bubbles, with distilled water evenly distributed across it, and
aligned with the frame. Mode selection electrophoresis program:
[HYDRAGEL 1 IF] select ‘1 IF SM/DM’; [HYDRAGEL IF 2/4] Select ‘2/4
IF SM/DM’; [HYDRAGEL 9 IF] Select ‘9 IF SM’. After placing the
dynamic mask mounting bracket on a flat surface, the immune
fixation was opened and set on the electrophoresis instrument. An
anti serum cup was placed on the dynamic mask mounting bracket.
Then, the reagent sample reference color card was placed on the cup
holder in front of the liquid cup well.12 µl of antibody was added
to each lane. The sample comb was removed and discarded. After
installing the two brackets, the buffer strip was removed and
discarded. Two transporters were removed. After wiping the
electrode wire, the gel sheet was placed on the electrophoresis
module. The cup holder of the anti-serum cup was in contact with
the film. The reagent diffused in the lane below. The program of
the electrophoresis instrument was initiated incubation. A thick
filter paper was placed on the film to dry the gel. The dried gel
holder was inserted into the dye vat for dyeing.
The specific anti immunoglobulin antiserum is
deposited on the gel to fix the immunoglobulin. The immunoglobulin
bands on the gel were stained. The detected data was analyzed by
Phoresis 9.3.0 (SEBIA). The results of the subsequent quantitative
immunoglobulin assessment of the blood and urine revealed the
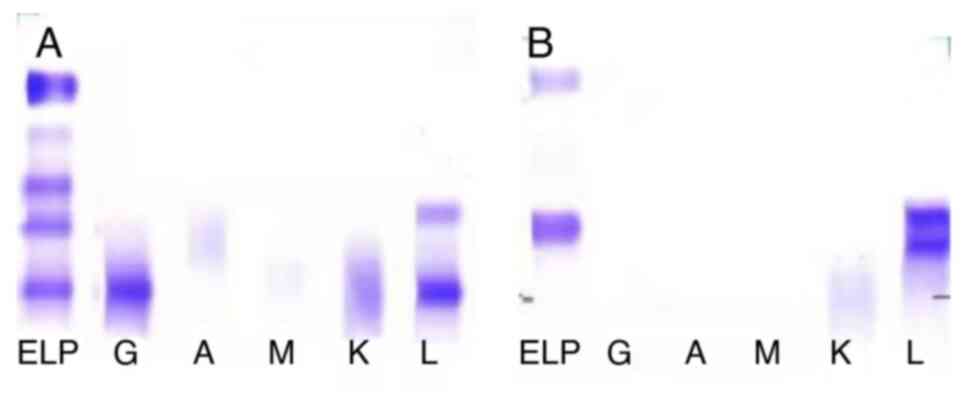
following: A serum-fixed quantitative immunoglobulin, monoclonal
IgG/λ component, M protein content of 3.2 g/l (reference, 0 g/l) in
the γ region (Fig. 4A) and
urine-fixed quantitative immunoglobulin, monoclonal light chain λ
component, M protein content of 0.26 g/24 h (The normal value is 0
g/24h) in the β region (Fig. 4B).
The patient was diagnosed with AML concomitant with MGUS.
Throughout this period, the patient was monitored for the
persistent presence of positive RUNX1::RUNX1T1 fusion genes.
Despite recommendations for intensive therapy involving
moderate-to-high doses of cytarabine or potential consideration of
allogeneic hematopoietic stem cell transplantation, the patient and
their family opted against such measures. Subsequently, the patient
was initiated on a regimen of maintenance therapy with standard
doses of chemotherapy for a total of four courses (daunorubicin 60
mg/m2, d1-3 + cytarabine 100 mg/m2, d1-5;
pirarubicin 25 mg/m2, d1-3 + cytarabine 100
mg/m2, d1-5; pirarubicin 25 mg/m2, d1-3 +
cytarabine 100 mg/m2; homoharringtonine 4
mg/m2, d1-3 + cytarabine 100 mg/m2, d1-5). In
August 2023, the patient was readmitted to Second Hospital of Hebei
Medical University with a hematological relapse. The family opted
to discontinue treatment and the patient died shortly
thereafter.
Discussion
AML characterized by the presence of the
RUNX1::RUNX1T1 fusion gene is classified under the genetic
aberration category of AML. This fusion gene is frequently
associated with the t(8;21)(q22;q22.1) translocation. Patients who
test negative for C-kit mutations in conjunction with this genetic
alteration are categorized into the low-risk group. Compared with
standard dosing regimen chemotherapy, treatment strategies such as
anthracycline drugs combined with cytarabine, high-dose cytarabine
and other regimens have demonstrated superior therapeutic efficacy
(1,6). Previous research has suggested that
patients with AML harboring a RUNX1::RUNX1T1 fusion gene may
exhibit complex chromosomal translocations involving abnormalities
affecting ≥3 chromosomes. Presence of a positive RUNX1::RUNX1T1
fusion gene can lead to chromosomal abnormalities not only on
chromosomes 8 and 21, but also on chromosomes 1–7, 11, 14, 15, 17,
and 20 (7–17).
Kim et al (8)
reported a patient with AML with a RUNX1::RUNX1T1 fusion gene with
a complex translocation involving chromosomes 1, 8, and 21. The
patient was treated using chemotherapy with norepinephrine and
cytarabine, and a CR was achieved. Additionally, they reviewed 24
patients with AML with the t(8;21) variant in which 7 patients
failed to achieve a CR and the remaining 17 patients achieved CR,
with only 3 patients experiencing a relapse. Of the 12 cases
included in the review of relevant literature in the present study
(Table I), 8 patients (67%)
achieved a CR following treatment. Only in 2/12 cases (17%) was the
presence of complex chromosomal abnormalities in the context of the
RUNX1::RUNX1T1 fusion gene suggested and was indicative of a poor
prognosis. In summary, trisomy abnormalities can be considered as
complex karyotypes to a certain extent. Furthermore, in terms of
treatment, previous research indicates that high-intensity
chemotherapy based on high-dose cytarabine and sequential
allogeneic hematopoietic stem cell transplantation may
fundamentally improve the prognosis of such patients.
 | Table I.Review of the literature on cases of
acute myeloid leukemia with a RUNX1::RUNX1T1 complex chromosomal
translocation. |
Table I.
Review of the literature on cases of
acute myeloid leukemia with a RUNX1::RUNX1T1 complex chromosomal
translocation.
| First author/s,
year | Sex | Age, years | Karyotype of AML | AML therapy | Response to AML
therapy | Outcome | (Refs.) |
|---|
| Kawakami et
al, 2008 | M | 37 |
45,X,-Y,der(8)t(8;21)(q22;q22),
der(9)(8;9)(q22;q34), and der(21)t (9;21)(q34;q22) | IA was not achieved
CR; HD-CA induced CR | CR | Relapsed after 3
months | (7) |
| Kim et al,
2011 | F | 63 | 46,XX,t(1; 21;
8)(q21;q22;q22) | IA was achieved
CR | CR | Clinically stable for
6 months | (8) |
| Udayakumar et
al, 2008 | F | 33 |
46,XX,t(8;13;21)(q22;q14;q22) | DA | CR | Alive after
allogeneic hematopoietic stem cell transplantation | (9) |
| Ihida et al,
2002 | M | 38 |
45,X,Y,del(3)(p25),t(8;21;14) (q22;
q22;q24),der(19)t(3;19)(?p25;p13) | IA was not achieved
CR; HD-CA induced CR | CR | NA | (11) |
| Gmidène et al,
2011 | M | 30 | 46,XY,t(1;21;8)(p34 ~
p35;q22;q22) | NA | NA | NA | (12) |
| Al Bahar et
al, 2009 | F | 25 |
46,XX,t(6;8;21)(p22;q22;q22) | Treated with
conventional chemotherapy regimen | CR | NA | (13) |
| Tay Za et al,
2019 | F | 23 | 46,
XX,t(8;22;21)(q22;q12;q22) | Treated with
conventional chemotherapy regimen | CR | NA | (14) |
| Mishra et al,
2021 | F | 19 |
46,XX,t(8;21;17)(q22;q22;p13) | DA | CR | CR >2 years of
diagnosis | (15) |
| Akhila Raj et
al, 2022 | F | 22 | 46, XX, der(8)
del(8q), der(13), t(8;21;13), der(21)t(13;21).ishder(8)
del(8q)(RUNX1T1+), der(13)t (8;21;13)(RUNX1T1+, RUNX1+),
der(21)(RUNX1+) | Cytarabine for
AML | CR | CR for 59 months | (16) |
|
| F | 68 | 45,
XX,+idicder(8)(q11.1) t(8;21) (q22;q11.1),-17, 21.ishidicder (8)
(q11.1) t(8;21)(RUNX1T1X2, RUNX1+) | Palliative
chemotherapy 7 days | NA | Died 2 months after
discharge |
|
|
| F | 25 | 45,X,-X, der(8)
t(8;21)(q22;q22), der(12) del(12q), der(21) t(8;12;21)
(q22;q?;q22)[7]/45,X,-X,t(8;21) (q22;q22)[3]/46,XX[10].ish der(8)
t(8;21)(q22;q22),(RUNX1T1+, RUNX1+),der(21) t(8;12;21)
(q22;q?;q22)(RUNX1+)[40/100] | No treatment | NA | NA |
|
| Han et al,
2024 | F | 57 |
46,XX,t(2;2;21;8)(p21;q37;
q22;q22)[18]/46,XX[2] | DA | NA | NA | (17) |
| Present case | M | 57 | 46,XY,
t(8;17;21)(q22;q24;22.1) [9]/45, idem,-Y[10]/46,XY[10] | DA | CR | Relapsed after 10
months | - |
In the present report, the case of a patient with
AML with a RUNX1::RUNX1T1 fusion gene is described. Further
investigation revealed a complex chromosomal translocation
t(8;17;21)(q22;q24;q22.1) and a CR was attained after a single
cycle of induction-remission chemotherapy. Nevertheless, during
subsequent consolidation-intensification therapy, the ongoing
presence of the RUNX1::RUNX1T1 fusion gene was detected, indicating
an unfavorable prognosis. The patient then experienced a disease
relapse 10 months later.
Due to the limited number of cases and the lack of
clinical information, the current evidence is insufficient to
distinguish the prognostic differences between variant complex
karyotype t(8;21) and classical t(8;21) chromosome translocations.
Traditionally, the prognosis for patients with AML with
RUNX1::RUNX1T1 positivity is favorable. However, complex
chromosomal karyotypes are a predictor of a poor prognosis
(1). In cases where intricate
chromosomal changes are present, we still recommended that these
patients should undergo allogeneic hematopoietic stem cell
transplantation as soon as possible after achieving a CR with
chemotherapy.
There is a paucity of literature documenting cases
of AML co-occurring with either MGUS or MM. Wu et al
(18) performed a retrospective
analysis on 14 elderly patients presenting with both MGUS and AML.
Luca and Almanaseer (19) described
a case involving an elderly patient with AML and MM, in which the
condition of the patient rapidly deteriorated post-diagnosis,
leading to their death from acute myocardial infarction and
congestive heart failure within a month. Wang et al
(20) reported the case of a
77-year-old male patient with AML complicated by MM, who underwent
treatment with a regimen comprising low-dose cytarabine, a
combination of aroubicin and granulocyte-stimulating factor (CAG
regimen), as well as bortezomib. Notably, these interventions
resulted in the patient achieving and maintaining a CR for >6
months until the end of the follow-up period. Additionally, the
case of a 51-year-old male patient with AML and MM was reported by
Kim et al (21). Despite
initial treatment failure with bortezomib and unsuccessful attempts
to achieve a CR with two cycles of induction-remission
chemotherapy, the patient achieved CR following allogeneic
hematopoietic stem cell transplantation.
AML and MGUS/MM originate in different cell lines.
The precise mechanisms underlying the co-occurrence of AML with
MGUS/MM are incompletely understood, suggesting a potential shared
etiological agent. Platanias (22)
reported that activation of the MAP kinase pathway was critically
implicated in the pathogenesis of AML, MDS and MM. Wu et al
(18) postulated a potential
association with underlying immune triggers, positing that these
stimuli may contribute to the development of AML and aberrant
plasma cell function. Lloyd et al (23) constructed a conceptual model of
tumors through Darwinian dynamics. They state that prior to the
onset of disease, the human body exists in a relatively stable
state. In a tumor, cancer cells may evolve to an evolutionary
stable state. Therefore, it was hypothesized that homeostasis of
the microenvironment is an important external factor for the
existence of AML.
In the present case report, MGUS presented during
the consolidation and intensification chemotherapy for AML, which
has not previously been documented in the literature, to the best
of our knowledge. Based on the hypothesis of Lloyd et al, we
hypothesize that the stable microenvironment of AML was disrupted,
driving the malignant transformation and proliferation of leukemia
cells, resulting in clonal the evolution of the leukemia cells in
that specific environment. This promoted the development of
monoclonal plasma cells and the coexistence of MGUS in the treated
patient. The management approach for such individuals poses a
dilemma of whether they be meticulously monitored for plasma cell
activity and monoclonal immunoglobulin levels without immediate
MGUS-specific intervention, or whether proactive inclusion of
anti-plasma cell agents, such as proteasome inhibitors and
immunomodulators should be considered at an early juncture. This
remains an unresolved query in clinical practice.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding was received from the Beijing Tianjin Hebei Basic
Research Cooperation Special Project (grant no. 2023206910) and
Hebei Province Central Guide Local Science and Technology
Development Fund Project (grant no. 246Z7729G).
Availability of data and materials
The high-throughput sequencing data generated in the
present study may be found in the NCBI SRA database under accession
number SRP542525 or at the following URL: https://www.ncbi.nlm.nih.gov/sra/?term=SRP542525.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
XQ interpreted the results and drafted the
manuscript. SQ designed the study and analyzed patient data. JZ and
TT confirm the authenticity of all the raw data. LG, JZ and TT
performed the experiments. XG advised on treatment of the patient.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent for publication of the case
report, including the clinical details and images was provided by a
relative of the patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Khoury JD, Solary E, Abla O, Akkari Y,
Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, et
al: The 5th edition of the World Health Organization classification
of haematolymphoid tumours: Myeloid and histiocytic/dendritic
neoplasms. Leukemia. 36:1703–1719. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanderson RN, Johnson PR, Moorman AV,
Roman E, Willett E, Taylor PR, Proctor SJ, Bown N, Ogston S and
Bowen DT: Population-based demographic study of karyotypes in 1709
patients with adult acute myeloid leukemia. Leukemia. 20:444–450.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kyle RA, Durie BGM, Rajkumar SV, Landgren
O, Blade J, Merlini G, Kröger N, Einsele H, Vesole DH, Dimopoulos
M, et al: Monoclonal gammopathy of undetermined significance (MGUS)
and smoldering (asymptomatic) multiple myeloma: IMWG consensus
perspectives risk factors for progression and guidelines for
monitoring and management. Leukemia. 24:1121–1127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gabert J, Beillard E, van der Velden VH,
Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela
JM, Cavé H, et al: Standardization and quality control studies of
‘real-time’ quantitative reverse transcriptase polymerase chain
reaction of fusion gene transcripts for residual disease detection
in leukemia-a Europe against cancer program. Leukemia.
17:2318–2357. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beillard E, Pallisgaard N, van der Velden
VHJ, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E,
Gottardi E, Saglio G, et al: Evaluation of candidate control genes
for diagnosis and residual disease detection in leukemic patients
using ‘real-time’ quantitative reverse-transcriptase polymerase
chain reaction (RQ-PCR)-a Europe against cancer program. Leukemia.
17:2474–2486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Wang J, Wei S, Zhao J, Xin B, Li
G, Zhao J, Wu D, Luo M, Zhao S, et al: The latest edition of WHO
and ELN guidance and a new risk model for Chinese acute myeloid
leukemia patients. Front Med (Lausanne). 10:11654452023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kawakami K, Nishii K, Hyou R, Watanabe Y,
Nakao M, Mitani H, Murata T, Monma F, Yamamori S, Hosokai N and
Miura I: A case of acute myeloblastic leukemia with a novel variant
of t(8;21)(q22;q22). Int J Hematol. 87:78–82. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim H, Moon HW, Hur M, Yun YM and Lee MH:
Acute myeloid leukemia with a RUNX1-RUNX1T1 t(1;21;8)(q21;q22;q22)
novel variant: A case report and review of the literature. Acta
Haematol. 125:237–241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Udayakumar AM, Alkindi S, Pathare AV and
Raeburn JA: Complex t(8;13;21)(q22;q14;q22)-a novel variant of
t(8;21) in a patient with acute myeloid leukemia (AML-M2). Arch Med
Res. 39:252–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue Y, Yu F, Xin Y, Lu D, Zou Z, Guo Y and
Xie X: t(8;20)(q22;p13): A novel variant translocation of t(8;21)
in acute myeloblastic leukaemia. Br J Haematol. 98:733–735. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ihida F, Ueno M, Tanaka H, Makishima H,
Suzawa K, Hosaka S, Hidaka E, Ishikawa M, Yamauchi K, Kitano K and
Kiyosawa K: t(8;21;14)(q22;q22;q24) is a novel variant of t(8;21)
with chimeric transcripts of AML1-ETO in acute myelogenous
leukemia. Cancer Genet Cytogenet. 132:133–135. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gmidène A, Frikha R, Sennana H, Elghezal
H, Elloumi M and Saad A: T(1;21;8)(p34;q22;q22): A novel variant of
t(8;21) in acute myeloblastic leukemia with maturation. Med Oncol.
28 (Suppl 1):S509–S512. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al Bahar S, Adriana Z and Pandita R: A
novel variant translocation t(6;8;21)(p22;q22;q22) leading to
AML/ETO fusion in acute myeloid leukemia. Gulf J Oncolog. 56–59.
2009.PubMed/NCBI
|
|
14
|
Tay Za K, Shanmugam H and Chin EFM: A new
complex translocation (8;22;21)(q22;q12;q22) in RUNX1/RUNX1T1 acute
myeloid leukaemia. Malays J Pathol. 41:333–338. 2019.PubMed/NCBI
|
|
15
|
Mishra SR, Rawal L, Othman MAK, Thatai A,
Sarkar A, Lal V and Bhattacharya SK: Complex rearrangement in acute
myeloid leukemia M2 with RUNX1/RUNX1T1 fusion involving chromosomes
8, 17 and 21. Mol Cytogenet. 14:282021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akhila Raj TV, Gopinath P, Geetha Raj JA,
Narayanan G, Nair SG, Joy Philip DS, Raveendran S, Geetha P and
Sreedharan H: Acute myeloid leukemia patients with variant or
unusual translocations involving chromosomes 8 and 21-A
comprehensive cytogenetic profiling of three cases with review of
literature. J Cancer Res Ther. 18:697–703. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han B, Jing Y, Bi X, Lin Y, Li H, Li H, Ru
K and Yang S: t(2;2;21;8)(p21;q37;q22;q22), a novel four-way
complex translocation involving variant t(8;21) in case of acute
myeloid leukemia: A case report and literature review. Cancer
Genet. 284-285:1–4. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu SP, Costello R, Hofmann JN, Korde N,
Mailankody S, Purdue M and Landgren O: MGUS prevalence in a cohort
of AML patients. Blood. 122:294–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luca DC and Almanaseer IY: Simultaneous
presentation of multiple myeloma and acute monocytic leukemia. Arch
Pathol Lab Med. 127:1506–1508. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang LQ, Li H, Li XX, Li FL, Wang LL, Chen
XL and Hou M: A case of simultaneous occurrence of acute myeloid
leukemia and multiple myeloma. BMC Cancer. 15:7242015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim D, Kwok B and Steinberg A:
Simultaneous acute myeloid leukemia and multiple myeloma
successfully treated with allogeneic stem cell transplantation.
South Med J. 103:1246–1249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Platanias LC: Map kinase signaling
pathways and hematologic malignancies. Blood. 101:4667–4679. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lloyd MC, Cunningham JJ, Bui MM, Gillies
RJ, Brown JS and Gatenby RA: Darwinian dynamics of intratumoral
heterogeneity: Not solely random mutations but also variable
environmental selection forces. Cancer Res. 76:3136–3144. 2016.
View Article : Google Scholar : PubMed/NCBI
|