Introduction
Esophageal cancer is the eighth most common cancer
and the sixth most common cause of cancer deaths worldwide
(1). Although Barrett’s
adenocarcinoma is the most rapidly increasing cancer in Western
countries (2), esophageal squamous
cell carcinoma (ESCC) is still dominant in East Asia, including
Japan (3). ESCC is often diagnosed
at a late stage; thus, the prognosis of affected patients is
unsatisfactory, despite the development of therapeutic options such
as surgery, chemotherapy and radiotherapy (4). Consequently, there is a need for
biomarkers to allow for a tailored, multimodality approach with
increased efficacy. However, efforts to identify molecular markers
in association with the pathogenesis of ESCC have been unsuccessful
thus far (5).
MicroRNAs (miRs) are small, non-coding RNAs that
negatively regulate gene expression via translational repression or
messenger RNA degradation. More than 700 miRs have been identified
and registered in humans, with each individual miR predicted to
target multiple genes based on the seed sequence matches in their
3′-untranslated regions (UTRs) (6).
MiRs are involved in biological and pathological processes,
including cell differentiation, proliferation, apoptosis and
metabolism (7), and they are
emerging as highly tissue-specific biomarkers with potential
clinical applicability for defining cancer type and origin
(8,9). Accumulating evidence indicates that
the deregulation of miRs is associated with human malignancies,
suggesting a causal role of miRs in tumor initiation and
progression (10) since they are
able to function as oncogenes or tumor suppressors (11).
Pioneering studies on chronic lymphocytic leukemia
(CLL) showed that miRs play a role in cancer pathogenesis and that
the expression of miR-15a and miR-16-1 was deleted in the majority
of CLL case analyses (12). Further
functional analysis identified an anti-apoptotic Bcl-2 as one of
the actual targets regulated by these miRs, implying that miR-15a
and miR-16-1 were tumor suppressor genes that deregulate cellular
survival (12). Human let-7
genes that map to regions are deleted in many cancer types, and the
let-7 family may also function as tumor suppressors (13). A possible mechanistic explanation
for this was provided by the discovery that RAS oncogenes are the
targets of let-7 members (14).
Similarly, the genomic locus encoding miR-34a is frequently lost in
certain malignancies (13), and
non-small cell lung tumors exhibit a reduced expression of miR-34b
and miR-34c (15). miR-34 members
were shown to be direct transcriptional targets of p53, a
representative tumor suppressor protein (13,16) in
that their ectopic expression induces p53 itself and its downstream
targets and reduces p53-dependent apoptosis. These data establish
the integration of certain down-regulated miRs into the tumor
suppressive pathways (11,13).
Currently, there is limited information on the
relationship between the pathogenesis of ESCC and miRs. Therefore,
the present study was designed to identify the miRs that are
specifically down-regulated in ESCC cells, possibly exerting
regulatory activities.
Materials and methods
Cell lines and cultures
Human ESCC cell lines OE21, TE5, TE8, TE10 and TE11;
1 non-malignant human esophageal squamous cell line immortalized by
SV40 infection (Het1A); 2 human Barrett’s adenocarcinoma cell lines
(Bic-1 and Seg-1); 3 human gastric adenocarcinoma cell lines (AGS,
AZ521 and KATOIII); 2 colorectal adenocarcinoma cell lines (Caco-2
and DLD1); 1 human cervix epithelioid carcinoma cell line (HeLa); 1
human lung adenocarcinoma cell line (A549) and human hematological
malignant cell lines (acute promyelotic leukemia, HL60; human T
cell lymphoblast-like cell line, Jurkat and histiocytic lymphoma,
U937) were cultured. The AZ521, KATOIII, DLD-1, HeLa, A549, HL60,
and U937 cells were purchased from the Japanese Collection of
Research Bioresources Foundation (Sennan, Japan). The OE21, Het-1A,
AGS and Caco-2 cells were obtained from the American Type Culture
Collection (Manassas, VA, USA). The TE5, TE8, TE10 and TE11 cells
were purchased from Riken Bioresource Center Cell Bank (Tsukuba,
Japan). Bic-1 and Seg-1 were kindly provided by Dr D.G. Beer (Ann
Arbor, MI, USA). The OE-21, TE5, TE8, TE10, TE11, Het-1A, U937,
HL-60, DLD-1, Jurkat and KATOIII cells were grown in RPMI 1640
medium, while the HeLa, A549, and Caco-2 cells were maintained in
Eagle’s minimal essential medium. Both media were supplemented with
10% fetal bovine serum, 1% penicillin/streptomycin and 1%
glutamine, and all cell lines were cultured in a humidified
incubator under 5% CO2 at 37°C.
Patients and clinical samples
Consecutively, 20 patients with ESCC or high-grade
intraepithelial neoplasm (HGIN), or controls without the tumor who
underwent esophagoscopy between June 2007 and May 2009, were
recruited. After obtaining their informed consent, three biopsy
samples were removed from the ESCC tumors and from normal-appearing
esophageal mucosa under endoscopic observation. Of these samples,
two were placed immediately into 1 ml of RNA (Applied Biosystems,
Foster City, CA, USA) for RNA isolation at a later time point. The
other sample was fixed in 10% formalin and embedded in paraffin for
histopathology. The paraffin-embedded biopsy samples were cut into
5-μm sections and stained with hematoxylin and eosin.
RNA extraction
Total RNA, including miR from the tissue samples and
cultured cells, was extracted using a mirVana RNA Isolation kit
(Applied Biosystems) according to the supplier’s instructions. The
quality of the total RNA was determined on a Bioanalyzer
(Bioanalyzer RNA Nano kit, Agilent, Santa Clara, CA, USA), and the
RNA was quantified using a Nanodrop-1000 spectrophotometer
(Nanodrop Technologies, Wilmington, DE, USA). The extracted RNA
samples were stored at −80°C until use.
MicroRNA array hybridization and
analysis
To find specific miRs for ESCC cells, total RNA was
extracted from OE21 and TE10 cells, representative well- and
moderately-differentiated human ESCC cell lines, respectively, and
the non-malignant human esophageal squamous cell line, Het1A. The
isolated RNA samples were subjected to a comprehensive analysis of
miRNA expression patterns with microarray-based technology, an
Agilent Human miRNA array chip version 1 (Agilent), containing
15,000 probes corresponding to 470 unique human miRs and 64 human
viral miRs catalogued in the Sanger database version 9.1. An
aliquot of 100 ng each of total RNA was treated with calf intestine
phosphatase (GE Healthcare, Chalfont St. Giles, UK), denatured
using DMSO (Sigma, St. Louis, MO, USA), and directly labeled with
Cy3 using T4 RNA ligase (GE Healthcare). Labeled samples were
hybridized to the miR array 8X15k (G4470A) platforms in SureHyb
chambers (Agilent), washed with the buffer supplied (Agilent),
according to the manufacturer’s instructions, and scanned using an
Agilent Scanner (G2505B). Data were extracted using Feature
Extraction software 9.3 and GeneSpring software (Agilent). To
identify miRs that were differentially expressed between the ESCC
cell lines and Het1A cells, a supervised analysis was performed
using significance analysis of microarrays (SAM, Stanford
University, Stanford, CA, USA). The differences in miR expression
were considered significant if the fold-change of expression values
was >2.0 and the p value was <0.05 using the t-test.
Quantitative reverse
transcription-polymerase chain reaction analysis for microRNAs
Expression levels of miRs that showed significant
differences based on the microarray results were analyzed by
quantitative reverse transcription-polymerase chain reaction
(RT-PCR) using various human malignant cell lines, including ESCC,
and non-malignant Het1A. cDNA was prepared from total RNA using a
Taq Man MicroRNA Reverse Transcription kit (Applied Biosystems).
Predesigned Taq Man MicroRNA assays including the primer set and
Taq Man probe were purchased from Applied Biosystems. The reverse
transcription reactions were performed in aliquots containing 50 ng
total RNA, 1.5 μl 1X RT primer, 1 μl 10X RT buffer, 0.15 μl 100 mM
dNTP, 1 μl reverse transcriptase and nuclease-free water added up
to 15 μl at 16°C for 30 min, followed by 42°C for 30 min and 85°C
for 5 min. PCR reactions were performed in 20-μl aliquots
containing 1.33 μl of miR RT products with 18.67 μl of PCR master
mixture (10 μl 2X Universal PCR master mix; 1 μl each primer; 1 μl
Taq Man probe; and 6.67 μl nuclease-free water), and run in
triplicate on the 7500 Real-Time PCR system (Applied Biosystems).
Thermal cycling was initiated with a first denaturation step at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min. The cycle passing threshold (Ct) was recorded for each
candidate miR, and a small RNA, U6B, was used as the endogenous
control for data normalization. Relative expression was calculated
using the formula 2−DCt = 2− (Ct, U6B −
Ct,Specific) as described in the ABI PRISM 7700 SDS relative
quantification of gene expression protocol by PE Applied
Biosystems. Similarly, total RNAs extracted from the neoplastic and
non-neoplastic samples (esophagoscopic biopsies) were subjected to
real-time quantitative RT-PCR for quantitation of miR-10a
expression levels.
5-Aza-2′-deoxycytidine and trichostatin A
(TSA) treatment
OE21 cells were incubated with or without
5-aza-2′-deoxycytidine (DAC) (1 or 5 μmol/l) for 96 h, followed by
treatment with 0.5 μmol/l trichostatin A or vehicle for an
additional 24 h. Total RNA was then extracted and subjected to the
quantitative RT-PCR for measurement of the cellular miR-10a
expression levels.
Statistical analysis
The differences between groups were analyzed using
the unpaired, one-tailed, Student’s t-test. Data are expressed as
means ± standard error. Differences were considered statistically
significant at p<0.05. All examinations were conducted according
to Good Clinical Practice and the Declaration of Helsinki, and were
approved by the Nagasaki University ethics committees.
Results
Specific down-regulation of microRNA-10a
in esophageal squamous cell carcinoma
Based on the miR microarray analysis, the expression
of miR-153, -100, -125b, -10a, -99a, -376a, -379, -651 and -146b
was significantly (>2-fold) down-regulated in the ESCC cell
lines compared to the non-malignant Het1A cells (Fig. 1A). On the other hand, miR-203, -429,
-205, -200c and -141 were significantly (>2-fold) overexpressed
in the two ESCC cell lines compared to non-malignant Het1A cells.
We focused on the significantly down-regulated miRs, considering
their possible regulatory actions for carcinogenesis (11,13).
Real-time RT-PCR was used to quantify expression levels of miRs
that showed significant reductions in expression on the microarray
analysis. Among the corresponding miRs, only the miR-10a expression
levels substantially decreased, respectively, in all of the ESCC
cell lines (OE21, TE5, TE8, TE10 and TE11) compared to Het1A cells
on quantitative RT-PCR (Fig. 1B).
However, the miR-10a expression levels did not necessarily decrease
only in the ESCC cells, and levels varied among the diverse
malignant cell types examined (Fig.
1C). MiR-10a expression was substantially down-regulated in
ESCC cells compared to the Barrett’s esophageal adenocarcinoma ones
(Fig. 1D). These results indicate
that miR-10a expression may be differentially down-regulated in SCC
of the esophagus.
 | Figure 1Based on microRNA (miR) microarray
analysis, the expression of miR-153, -100, -125b, -10a, -99a,
-376a, -379, -651, and -146b is significantly reduced in the two
esophageal squamous cell carcinoma (ESCC) cell lines (OE21 and
TE10) compared to Het1A cells (A). Quantitative reverse
transcription (RT)-PCR shows a substantial decrease in the relative
miR-10a expression levels in all ESCC cell lines (OE21, TE5, TE8,
TE10 and TE11) compared to Het1A (B) and the Barrett’s
adenocarcinoma cells (C and D). The miR-10a expression levels did
not necessarily decrease in the ESCC cells compared to those in the
remaining malignant cell types examined (C). |
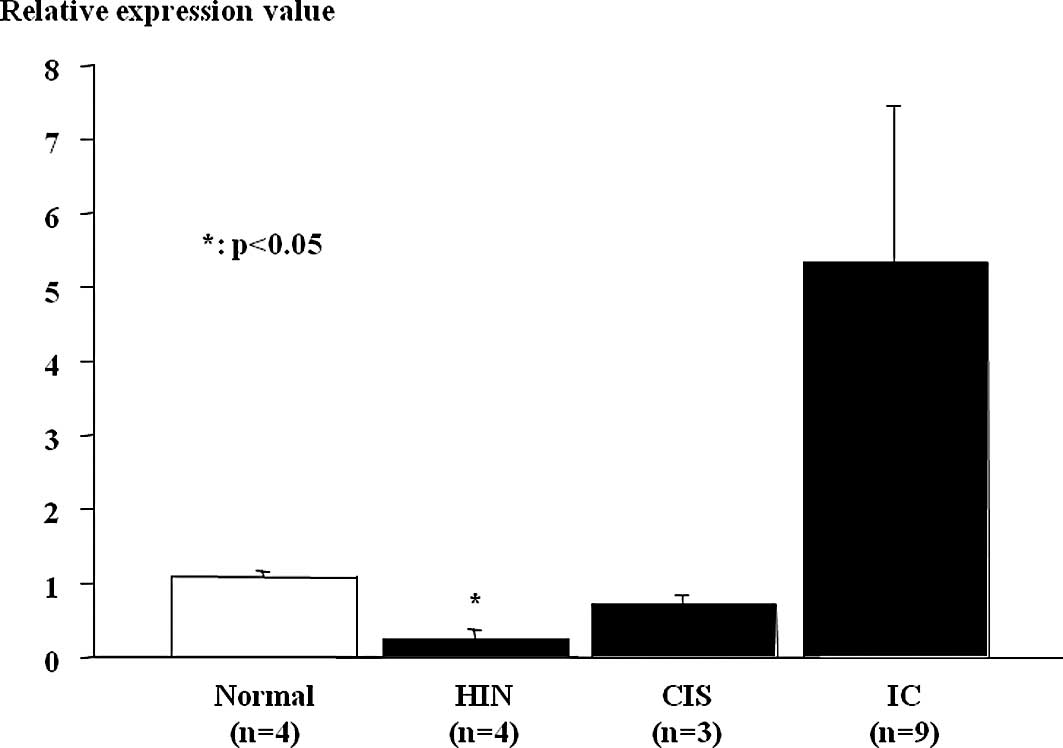
Down-regulation of microRNA-10a in
esophageal high-grade intraepithelial neoplasm and non-invasive
esophageal squamous cell carcinoma (carcinoma in situ)
MiR-10a expression in ESCC tumor samples and
non-cancerous tissues was assessed using real-time RT-PCR (Fig. 2). Relative miR-10a expression levels
were significantly lower in esophageal HGIN and tended to be low in
non-invasive ESCC (carcinoma in situ), which were
histopathologically classified according to the guidelines of the
Japanese Esophageal Society for the diagnosis and treatment of ESCC
(17), compared to the non-tumor
mucosa. MiR-10a expression was heightened in the invasive ESCCs
despite the results being insignificant.
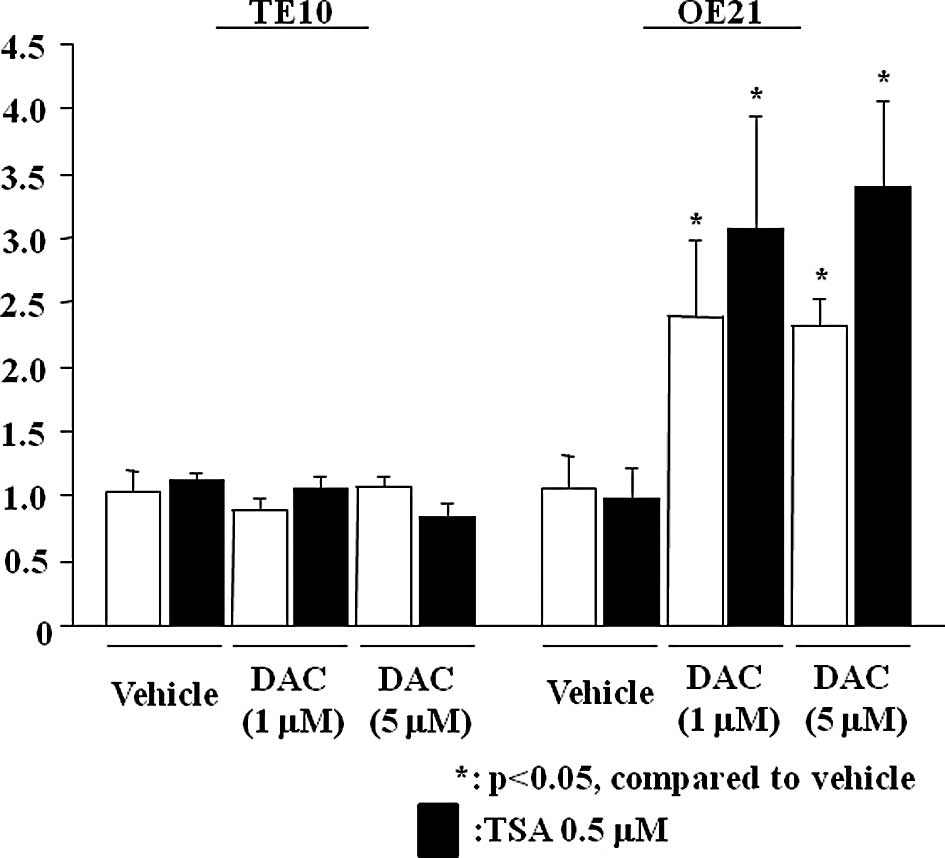
Restoration of microRNA-10a expression
with 5-aza-2′-deoxycytidine treatment
Relative expression levels of miR-10a significantly
increased in the presence of DAC, even at a concentration of 1
μmol/l in OE21 (Fig. 3). However,
no increase was noted in the TE10 cells (data not shown). No
significant effects on miR-10a expression were observed following
incubation with TSA at a sufficient concentration in the cell lines
(Fig. 3).
Discussion
In the present study, miR-10a was substantially
down-regulated in ESCC cells. The miR-10a expression level was
significantly lower in the ESCC cell lines examined compared to
non-malignant esophageal squamous cells. In addition, miR-10a was
substantially lower in the ESCC cells compared to those derived
from esophageal adenocarcinoma, another significant type of
esophageal cancer (2).
Nevertheless, miR-10a expression did not necessarily decrease when
compared with the remaining human malignant cell lines. These data
implicate miR-10a in ESCC pathogenesis, and further functional
analyses may shed light on the diagnostic and therapeutic potential
of miR-10a against this malignant disease.
Down-regulation of miR-10a has been reported in
chronic myeloid leukemia (CML) (18). Among 157 miRs tested using
quantitative RT-PCR, miR-10a, along with miR-150 and miR-151, was
listed in the significantly down-regulated miRs in CML cells
compared to CD34-positive cells taken from healthy controls
(18). The clinical relevance of
this finding was shown in a group of 85 newly-diagnosed patients
with CML in which the expression of miR-10a was down-regulated in
71% of cases (18). On the other
hand, previous studies showed an overexpression of miR-10a in other
cancers, including hepatocellular, pancreatic and urothelial
carcinomas and acute myeloid leukemia (19–21).
Based on the real-time PCR analysis and using RNA-extracted,
formalin-fixed, paraffin-embedded, archival liver tissue, miR-10a
expression levels were significantly increased in hepatitis C
virus-associated hepatocarcinoma compared to normal liver
parenchyma (19). Northern blot
analysis showed increased expression levels of miR-10a in
metastatic pancreatic adenocarcinoma (20). Weiss et al explored the
metastatic behavior of primary pancreatic tumors and cancer cell
lines in xenotransplantation experiments. These authors found that
miR-10a expression promoted metastasis of the tumor cells, and the
repression of this expression was sufficient to inhibit invasion
and metastasis formation (20). The
regulatory actions of miR-10a were mediated via its target
inhibition of HOXB1 and HOXB3 expression (20), implying that miR-10a is a
significant mediator of tumor metastasis. In the clinical settings,
the present study showed a comparable down-regulation of miR-10a in
HGIN and non-invasive ESCC, whereas it was highly expressed in the
invasive ESCCs. The exact reasons for this discrepancy remain
unknown, but the abundance of miR-10a expression may be involved in
ESCC development and progression. There may be differential miR-10a
expression in human cancers including ESCC, which may affect
cellular transformation, carcinogenesis and aggressive behavior and
act as an oncogene or tumor suppressor (11,13).
Transcriptional deregulation, epigenetic
alterations, mutations in miR sequences, DNA copy number
abnormalities and dysfunction in their biogenesis machinery may
contribute to the aberrant expression of miRs in human cancers
(22), though the underlying
mechanisms remain unknown. Recent evidence has shown that
epigenetic changes, including DNA methylation and histone
modification, play important roles in regulating expression of not
only protein-coding genes but also certain miRs (22,23).
In the present study, treatment with a demethylating agent restored
miR-10a expression in OE21 cells. Han et al compared miR
expression profiles between human colon cancer cell line HCT 116
and its derivative, DNA methyltransferase 1 and 3b double knockout
(DKO) cells, and found that the expression of approximately 10% of
miRs may be regulated by DNA methylation (23). Of note is that miR-10a was the most
strikingly up-regulated miR in the DKO HCT116 cells. Additionally,
well-defined CpG islands are located within 3 kb upstream of the
miR-10a gene locus. Bisulfite sequencing showed that the majority
of CpG sites proximal to miR-10a were hypermethylated in the parent
cell line, while DNA methylation was largely absent in the DKO
cells (23). Thus, epigenetic
regulatory mechanisms may be involved in miR-10a expression, at
least in certain human cancer cells.
In conclusion, based on the comprehensive microarray
analysis, following quantitative confirmation with the real-time
RT-PCR procedure, miR-10a expression was specifically
down-regulated in ESCC cells. miR-10a expression was comparably low
in HGIN and non-invasive ESCC cells, whereas the expression levels
increased in the invasive phenotypes, suggesting unique regulatory
mechanisms for the differential expression of miR-10a. In this
context, miR-10a expression is likely to be regulated via DNA
methylation in the CpG islands proximal to its gene locus in
certain ESCC cells.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Crew KD and Neugut AI: Epidemiology of
upper gastrointestinal malignancies. Semin Oncol. 31:450–464. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mathé EA, Nguyen GH, Bowman ED, et al:
MicroRNA expression in squamous cell carcinoma and adenocarcinoma
of the esophagus: associations with survival. Clin Cancer Res.
15:6192–6200. 2009.PubMed/NCBI
|
|
4
|
Orringer MB: Multimodality therapy for
esophageal carcinoma – an update. Chest. 103:S406–S409. 1993.
|
|
5
|
Fareed KR, Kaye P, Soomro IN, et al:
Biomarkers of response to therapy in oesophago-gastric cancer. Gut.
58:127–143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmittgen TD: Regulation of microRNA
processing in development, differentiation and cancer. J Cell Mol
Med. 12:1811–1819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosenfeld N, Aharonov R, Meiri E, et al:
MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol.
26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang Y, Ridzon D, Wong L and Chen C:
Characterization of microRNA expression profiles in normal human
tissues. BMC Genomics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lotterman CD, Kent OA and Mendell JT:
Functional integration of microRNAs into oncogenic and tumor
suppressor pathways. Cell Cycle. 7:2493–2499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of microRNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Medina PP and Slack FJ: microRNAs and
cancer: an overview. Cell Cycle. 7:2485–2492. 2007. View Article : Google Scholar
|
|
14
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bommer GT, Gerin I, Feng Y, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ono S, Fujishiro M, Niimi K, et al:
Predictors of postoperative stricture after esophageal endoscopic
submucosal dissection for superficial squamous cell neoplasms.
Endoscopy. 41:661–665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agirre X, Jiménez-Velasco A, San
José-Enériz E, et al: Down-regulation of hsa-miR-10a in chronic
myeloid leukemia CD34+ cells increases USF2-mediated
cell growth. Mol Cancer Res. 6:1830–1840. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Varnholt H, Drebber U, Schulze F, et al:
MicroRNA gene expression profile of hepatitis C virus-associated
hepatocellular carcinoma. Hepatology. 47:1223–1232. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weiss FU, Marques IJ, Woltering JM, et al:
Retinoic acid receptor antagonists inhibit miR-10a expression and
block metastatic behavior of pancreatic cancer. Gastroenterology.
137:2136–2145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garzon R, Garofalo M, Martelli MP, et al:
Distinctive microRNA signature of acute myeloid leukemia bearing
cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA.
105:3945–3950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng S, Calin GA, Croce CM, Coukos G and
Zhang L: Mechanisms of microRNA deregulation in human cancer. Cell
Cycle. 7:2643–2646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han L, Witmer PD, Casey E, Valle D and
Sukumar S: DNA methylation regulates microRNA expression. Cancer
Biol Ther. 6:1284–1288. 2007.PubMed/NCBI
|