Introduction
Helicobacter pylori (H. pylori) is a
microaerophilic gram-negative bacterium known to be associated with
chronic gastritis, peptic ulcer and gastric adenocarcinoma
(1). It is of great clinical
importance to identify the organism in gastric specimens.
Subsequently, several diagnostic assays exist. H. pylori
infection in gastric specimens can be demonstrated through the use
of culture, histological examination of biopsy specimens using
different stains, assaying for urease activity and PCR assay with
the aim of specifically detecting H. pylori DNA (2). Assays based on the use of PCR to
detect the presence of H. pylori DNA using several different
gene targets have been described (2–10).
Moreover, it is well known that the PCR assay is highly reliable in
the detection of H. pylori. The pyrosequencing analysis was
employed to identify H. pylori by sequencing a part of the
16S rRNA gene covering the H. pylori signature sequence
(6). The H. pylori signature
sequence allows for the distinction of the organism from a set of
other bacterial species (11). This
study investigated the possibility of using pyrosequencing
technology to verify the species identity of H. pylori from
paraffin-embedded tissues of resected gastric adenocarcinomas by
amplifying a part of the 16S rRNA gene using broadly reactive
primers followed by the sequencing of a 20-bp sequence unique to
the 16S rRNA gene of H. pylori.
Materials and methods
DNA extraction
DNA was extracted from paraffin sections of 51
resected gastric adenocarcinomas including 21 intestinal, 24
diffuse and 6 mixed types. Briefly, 50–100 μl of DNA extraction
buffer solution [50 mM Tris buffer (pH 8.3), 1 mM EDTA (pH 8.0), 5%
Tween-20 and 100 μg/ml proteinase K] with 10% resin was added to
scraped tissue and incubated at 56°C for a minimum of 1 h.
Following incubation, the tubes were heated at 100°C for 10 min.
Tubes were centrifuged to pellet the debris, and 5 μl of the
supernatant was used in the PCR reaction.
PCR amplification for Helicobacter pylori
identification
To identify H. pylori the primers used were:
forward, 5′-biotin-AGGGGTAAAATCCGTAGAGAT-3′ and reverse,
5′-CGTTTAGGGCGTGGACTA-3′. The latter primer amplifies a 133-bp DNA
fragment from the ‘16S rRNA’ region of H. pylori. Briefly, 5
μl of DNA was added to reach 50 μl of PCR solution mix, containing
0.2 mmol each of dNTP, 1.5 mmol/l MgCl2, 1X PCR buffer,
1.5 units of Immolase DNA Taq polymerase (Bioline, London, UK) and
20 pmol of each primer. PCR was performed for 5 min at 95°C, 50
cycles (30 sec at 95°C, 30 sec at 52°C and 30 sec at 72°C) and 10
min at 72°C using a PTC-220 thermal cycler (Bio-Rad, USA). The PCR
products were electrophoresed in an agarose gel to confirm
successful amplification of the PCR product.
Pyrosequencing analysis for Helicobacter
pylori identification
Biotinylated PCR products were immobilized to
streptavidin-coated beads (Amersham Pharmacia Biotech AB, Sweden)
using solution from the PSQ™ 96 Sample Preparation kit
(Pyrosequencing AB, UK), following a standard protocol. Beads (10
μl) were diluted in binding buffer with biotinylated PCR products
and incubated for 10 min at room temperature. The beads were
transferred to a filter probe, and liquid was removed by vacuum
filtration. DNA was separated in denaturation solution for 2 min.
The templates were washed with washing buffer, transferred to a PSQ
96 SQA plate and annealed with the sequencing primer, reverse,
5′-CTCCCCA CGCTTT-3′ in annealing buffer at room temperature.
Samples were analyzed using the PyroMark ID system (Biotage, UK)
with SQA software and the SQA reagent kit (Biotage) for sequence
analysis.
Results
DNA was extracted from the paraffin-embedded tissues
of 51 resected gastric adenocarcinomas. PCR primers were designed
to amplify the 133-bp PCR fragment in highly conserved regions of
the 16S rRNA gene. The sequence of the PCR products was analyzed
using the PyroMark ID system with SQA software and the SQA reagent
kit. Sequence analysis for the identification of H. pylori
by sequencing a section of the 16S rRNA gene covering the H.
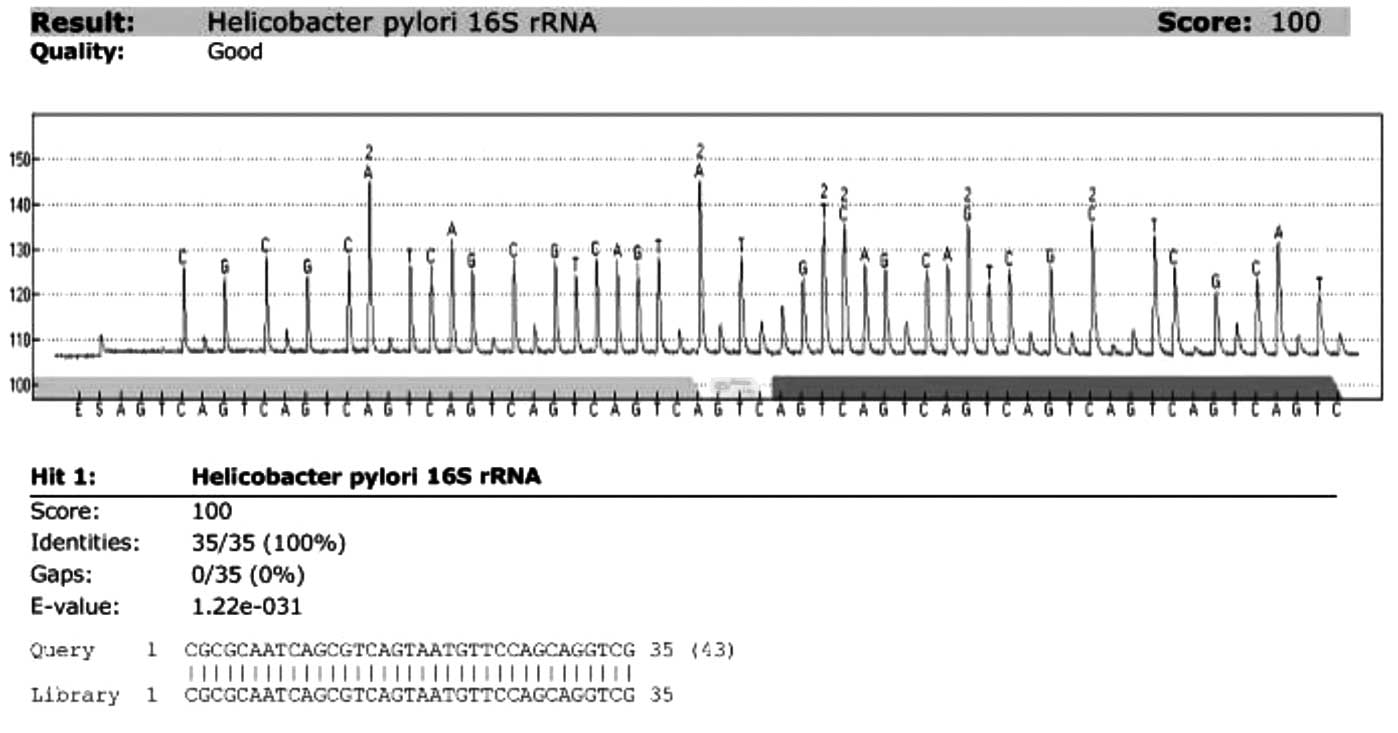
pylori signature sequence was carried out. Fig. 1 shows the representative results
from the analysis of the paraffin-embedded tissues of 51 resected
gastric adenocarcinomas. Pyrosequencing analysis of 16S rRNA showed
that H. pylori was present in 47 (92.2%) of the 51 gastric
adenocarcinomas: 18 of the 21 intestinal-, 23 of the 24 diffuse-
and all of the 6 mixed-type. In the 4 H. pylori-negative
cases, Helicobacter cinaedi (H. cinaedi) (2 cases),
Helicobacter mustelae (H. mustelae) (1 case) and
Campylobacter hyointestinalis (C. hyointestinalis) (1
case) were detected. Two H. cinaedi- and 1 C.
hyointestinalis-positive cases were detected in the
intestinal-type, and 1 H. mustelae case was detected in the
diffuse-type adenocarcinomas.
Discussion
A variety of diagnostic procedures are used to
identify H. pylori in clinical samples. No single test is
optimal due to the length of time required to perform the test,
lack of sensitivity or irreproducibility (5). Culture and histological examination of
biopsy specimens using different stains and assaying for urease
activity have the disadvantages of lack of sensitivity and long
incubation periods. Assays based on the use of PCR to detect the
presence of H. pylori DNA using several different gene
targets showed that PCR is feasibile for the rapid, sensitive and
specific detection of H. pylori (2–5,7–10,12).
Using the PyroMark ID system with SQA software and the SQA reagent
kit to amplify a section of the 16S rRNA gene it was possible to
analyze bacterial genetic targets in DNA extracted directly from
human gastric tissues without the prolonged culturing of bacteria.
Subsequently, the organism was differentiated from a set of other
bacterial species (6).
H. pylori has been classified as a Group I
carcinogen. Previous epidermiologic studies established a strong
causal relationship between H. pylori infection and gastric
cancer (13–16). A nationwide survey conducted in
South Korea in 1998 on the serologic prevalence of H. pylori
infection concluded that the prevalence of H. pylori was
66.9% among adults (≥16 years), a percentage that decreased to
59.6% in 2005 (17). In this study,
H. pylori was present in 47 (92.2%) of 51 gastric
adenocarcinoma tissues from Korean patients. Persistent infection
of the gastric mucosa by H. pylori initiates an inflammatory
cascade that progresses into atrophic gastritis, a condition
associated with a reduced capacity for the secretion of gastric
acid and an increased risk of developing gastric cancer (18).
Two cases of H. cinaedi infection, 1 case of
C. hyointestinalis infection and 1 case of H.
mustelae infection in the 4 H. pylori-negative cases
were noted in the present study. Two H. cinaedi- and 1 C.
hyointestinalis-positive cases were detected in the
intestinal-type and 1 H. mustelae in the diffuse-type
adenocarcinomas. Molecular evidence of H. cinaedi organisms
in 2 of 126 urease-negative human gastric biopsy specimens was
previously reported (19). H.
cinaedi was found to cause gastroenteritis (20) and extraintestinal infection,
particularly in immunocompromised patients (21). C. hyointestinalis was
initially described by Gebhart et al as a possible cause of
porcine proliferative enteritis (22). The organism has subsequently been
isolated from the feces of humans with gastroenteritis and, in a
few cases, from the blood of patients with bacteremia (23). Although H. cinaedi and C.
hyointestinalis were previously associated with
gastroenteritis, the incidence and roles of these organisms in
gastric carcinogenesis remain unclear. H. mustelae is a
gastric pathogen that has many biochemical, molecular and
phenotypic characteristics similar to those of H. pylori
(24). H. mustelae infection
was found to increase gastric epithelial proliferation, as noted in
H. pylori-infected humans, presumably due to a chronic
inflammatory response (25). A
previous study suggested that the high tumor incidence reported in
MNNG-treated ferrets reflected the involvement of H.
mustelae infection in the carcinogenic process in these animals
(26). A previously reported case
linking H. mustelae and gastric adenocarcinoma supports the
hypothesis that H. mustelae, similar to H. pylori in
humans, may be a gastric co-carcinogen in ferrets (27). However, this hypothesis has yet to
be confirmed in humans.
Pyrosequencing technology is useful in the
identification and differentiation of H. pylori from other
species by analyzing the gene encoding 16S rRNA. Gastric
adenocarcinoma tissues contain bacteria and the majority are H.
pylori. H. cinaedi, H. mustelae and C.
hyointestinalis rarely occur. The roles of these organisms in
the pathogenesis of gastric adenocarcinoma remain unclear.
Acknowledgements
This study was supported by Konkuk University.
References
|
1
|
Blaser MJ and Parsonnet J: Parasitism by
the ‘slow’ bacterium Helicobacter pylori leads to altered
gastric homeostasis and neoplasia. J Clin Invest. 94:4–8. 1994.
|
|
2
|
Weiss J, Mecca J, da Silva E and Gassner
D: Comparison of PCR and other diagnostic techniques for detection
of Helicobacter pylori infection in dyspeptic patients. J
Clin Microbiol. 32:1663–1668. 1994.PubMed/NCBI
|
|
3
|
Clayton CL, Kleanthous H, Coates PJ,
Morgan DD and Tabaqchali S: Sensitive detection of Helicobacter
pylori by using polymerase chain reaction. J Clin Microbiol.
30:192–200. 1992.
|
|
4
|
Engstrand L, Nguyen AM, Graham DY and
el-Zaatari FA: Reverse transcription and polymerase chain reaction
amplification of rRNA for detection of Helicobacter species.
J Clin Microbiol. 30:2295–2301. 1992.PubMed/NCBI
|
|
5
|
Hammar M, Tyszkiewicz T, Wadstrom T and
O’Toole PW: Rapid detection of Helicobacter pylori in
gastric biopsy material by polymerase chain reaction. J Clin
Microbiol. 30:54–58. 1992.
|
|
6
|
Hjalmarsson S, Alderborn A, Fock C, Muldin
I, Kling H, Uhlen M and Engstrand L: Rapid combined
characterization of microorganism and host genotypes using a single
technology. Helicobacter. 9:138–145. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoshina S, Kahn SM, Jiang W, et al: Direct
detection and amplification of Helicobacter pylori ribosomal
16S gene segments from gastric endoscopic biopsies. Diagn Microbiol
Infect Dis. 13:473–479. 1990.PubMed/NCBI
|
|
8
|
Valentine JL, Arthur RR, Mobley HL and
Dick JD: Detection of Helicobacter pylori by using the
polymerase chain reaction. J Clin Microbiol. 29:689–695. 1991.
|
|
9
|
Van Zwet AA, Thijs JC, Kooistra-Smid AM,
Schirm J and Snijder JA: Sensitivity of culture compared with that
of polymerase chain reaction for detection of Helicobacter
pylori from antral biopsy samples. J Clin Microbiol.
31:1918–1920. 1993.PubMed/NCBI
|
|
10
|
Wang JT, Lin JT, Sheu JC, Yang JC, Chen DS
and Wang TH: Detection of Helicobacter pylori in gastric
biopsy tissue by polymerase chain reaction. Eur J Clin Microbiol
Infect Dis. 12:367–371. 1993.
|
|
11
|
Eckloff BW, Podzorski RP, Kline BC and
Cockerill FR III: A comparison of 16S ribosomal DNA sequences from
five isolates of Helicobacter pylori. Int J Syst Bacteriol.
44:320–323. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ho SA, Hoyle JA, Lewis FA, et al: Direct
polymerase chain reaction test for detection of Helicobacter
pylori in humans and animals. J Clin Microbiol. 29:2543–2549.
1991.PubMed/NCBI
|
|
13
|
El-Omar EM, Oien K, Murray LS, et al:
Increased prevalence of precancerous changes in relatives of
gastric cancer patients: critical role of H. pylori.
Gastroenterology. 118:22–30. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eslick GD, Lim LL, Byles JE, Xia HH and
Talley NJ: Association of Helicobacter pylori infection with
gastric carcinoma: a meta-analysis. Am J Gastroenterol.
94:2373–2379. 1999.
|
|
15
|
Limburg P, Qiao Y, Mark S, et al:
Helicobacter pylori seropositivity and subsite-specific
gastric cancer risks in Linxian, China. J Natl Cancer Inst.
93:226–233. 2001. View Article : Google Scholar
|
|
16
|
Uemura N, Okamoto S, Yamamoto S, et al:
Helicobacter pylori infection and the development of gastric
cancer. N Engl J Med. 345:784–789. 2001. View Article : Google Scholar
|
|
17
|
Kim N, Park RY, Cho SI, et al:
Helicobacter pylori infection and development of gastric
cancer in Korea: long-term follow-up. J Clin Gastroenterol.
42:448–454. 2008. View Article : Google Scholar
|
|
18
|
Dicksved J, Lindberg M, Rosenquist M,
Enroth H, Jansson JK and Engstrand L: Molecular characterization of
the stomach microbiota in patients with gastric cancer and in
controls. J Med Microbiol. 58:509–516. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pena JA, McNeil K, Fox JG and Versalovic
J: Molecular evidence of Helicobacter cinaedi organisms in
human gastric biopsy specimens. J Clin Microbiol. 40:1511–1513.
2002.
|
|
20
|
Quinn TC, Goodell SE, Fennell C, Wang SP,
Schuffler MD, Holmes KK and Stamm WE: Infections with
Campylobacter jejuni and Campylobacter-like organisms
in homosexual men. Ann Intern Med. 101:187–192. 1984.
|
|
21
|
Burman WJ, Cohn DL, Reves RR and Wilson
ML: Multifocal cellulitis and monoarticular arthritis as
manifestations of Helicobacter cinaedi bacteremia. Clin
Infect Dis. 20:564–570. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gebhart CJ, Ward GE, Chang K and Kurtz HJ:
Campylobacter hyointestinalis (new species) isolated from
swine with lesions of proliferative enteritis. Am J Vet Res.
44:361–367. 1983.
|
|
23
|
Lastovica AJ:
Campylobacter/Helicobacter bacteremia in Cape Town,
South Africa, 1977–1995. Campylobacters, Helicobacters and Related
Organisms. Newell DG, Ketley JM and Feldman RA: Plenum Press; New
York: pp. 475–479. 1996, View Article : Google Scholar
|
|
24
|
Fox JG, Edrise BM, Cabot EB, Beaucage C,
Murphy JC and Prostak KS: Campylobacter-like organisms
isolated from gastric mucosa of ferrets. Am J Vet Res. 47:236–239.
1986.
|
|
25
|
Yu J, Russell RM, Salomon RN, Murphy JC,
Palley LS and Fox JG: Effect of Helicobacter mustelae
infection on ferret gastric epithelial cell proliferation.
Carcinogenesis. 16:1927–1931. 1995.
|
|
26
|
Fox JG, Correa P, Taylor NS, Lee A, Otto
G, Murphy JC and Rose R: Helicobacter mustelae associated
gastritis in ferrets: an animal model of Helicobacter pylori
gastritis in humans. Gastroenterology. 99:352–361. 1990.
|
|
27
|
Fox JG, Dangler CA, Sager W, Borkowski R
and Gliatto JM: Helicobacter mustelae-associated gastric
adenocarcinoma in ferrets (Mustela putorius furo). Vet
Pathol. 34:225–229. 1997. View Article : Google Scholar
|