Introduction
Hepatocellular carcinoma (HCC) is a significant
worldwide health problem accounting for more than 750,000 new cases
and more than 690,000 cancer-related deaths in 2011 (1), in which half of these cases and deaths
were estimated to have occurred in China alone. HCC incidence rates
are increasing in many parts of the world, including the US and
Central Europe. To date, surgery is still the primary treatment for
HCC. However, there are not many patients who are eligible to
receive surgery due to a variety of reasons, such as unresectable
tumors, distant tumor metastasis, insufficient hepatic function and
poor health conditions. Moreover, HCC has a high recurrence rate
after resection. Thus, neoadjuvant therapy is usually used to treat
HCC patients. For example, transarterial chemoembolization (TACE),
which is an important neoadjuvant treatment against HCC, has been
extensively used to delay HCC progression in the clinic and to
improve the prognosis of HCC patients. Chemotherapeutical drugs,
such as adriamycin (ADM), mitomycin C (MMC), vincristine (VCR),
5-fluorouracil (5-FU), cisplatin (DDP) and carboplatin (CBP), are
frequently used in HCC treatment (2,3). Among
them, ADM is an antibiotic that eliminates tumor cells by
preventing RNA biosynthesis. MMC, also an antibiotic, is able to
directly destroy DNA and prevent DNA replication. Both DDP and CBP
confer antitumor effects through inhibition of DNA biosynthesis and
replication. In contrast, VCR targets the microtubules in cells,
interferes with protein metabolism and inhibits activity of RNA
polymerase and synthesis of plasmalemma adipoid. In the clinic, two
or three drugs are combined together with TACE. However, multidrug
resistance (MDR) prevents successful long-term use of chemotherapy.
MDR mechanisms are usually complicated and many factors, such as
P-glycoprotein (P-gp), resistance-associated protein (MRP), lung
resistance protein (LRP), glutathione S-transferase (GST),
cyclooxygenase 2 (COX-2), nuclear factor-κB (NF-κB) (4–9), have
been found to participate in MDR.
More recently, it has been shown that altered
expression of miRNAs plays an important role in MDR (10). miRNAs are small non-coding RNA
molecules of 20–23 nucleotides in length and regulate a variety of
biological processes. miRNAs inhibit mRNA translation of target
genes through imperfect base pairing with the 5′- or
3′-untranslated region of the target mRNAs. Previous studies have
underlined the involvement of miRNAs in drug resistance (11–18).
Different miRNAs have been shown to be important in the mediation
of chemosensitivity or chemoresistance in different cancer types,
which occurs through the regulation of MDR- or cell- growth-related
protein expression. Expression of miR-326 was found to downregulate
MRP-1 expression and sensitize tumor cells to VP-16 and doxorubicin
(13). Overexpression of miR-122
modulated HCC sensitivity to chemotherapeutic drugs through
downregulation of MDR-related genes MDR-1, GST-π, MRP,
antiapoptotic gene Bcl-w and cell cycle-related gene cyclin-B1
(15). Expression of miR-34a can
negatively regulate, at least in part, resistance of colorectal
cancer DLD-1 cells to 5-FU through targeting Sirt1 and E2F3 genes
(19). In contrast, stable
transfection of miR-21 induced drug resistance in K562 cells, while
suppression of miR-21 in K562/DNR cells enhanced DNR cytotoxicity
(20). miR-214 induces cell
survival and DDP resistance primarily through targeting the
PTEN/Akt pathway (21). miR-328
targets ABCG2 3′-UTR and, consequently, controls ABCG2 protein
expression and influences drug disposition in human breast cancer
cells (22). However, upregulation
of miR-138 reverses resistance of both P-gp-related and
P-gp-non-related drugs in HL-60/VCR cells and promotes
adriamycin-induced apoptosis (16).
Although these studies provide insightful information regarding
miRNA-mediated MDR in human cancers, a complete profile of
miRNA-mediated MDR is needed to systematically understand the role
of miRNAs in MDR in HCC. Therefore, in this study, we first
established five chemotherapeutical drug-resistant HCC cell
sublines, Huh-7/ADM, Huh-7/DDP, Huh-7/CBP, Huh-7/MMC and Huh-7/VCR.
We then subsequently profiled altered miRNA expression in these
sublines in comparison to the parental HCC cell line using miRCURY™
LNA array (v. 16.0).
Materials and methods
Cell line and culture
The human HCC Huh-7 cell line was obtained from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and
cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented
with 10% fetal bovine serum (FBS) (Gibco-BRL; Invitrogen Life
Technologies, Carlsbad, CA, USA) in a humidified incubator at 37°C
with 5% carbon dioxide.
Generation of drug-resistant HCC cell
sublines
The different drug-resistant HCC cell sublines were
established by exposing the parental Huh-7 cells to increased
concentrations of various chemotherapeutic drugs. Briefly, Huh-7
cells were inoculated in a 10-ml cell culture flask and cultivated
for 72 h in culture medium containing a low concentration of drugs
(0.02 μg/ml ADM, 0.02 μg/ml CBP, 0.0375 μg/ml DDP, 0.0015 μg/ml
MMC, or 0.01 μg/ml VCR). Subsequently, the cells were continuously
cultured without drug exposure for ~2 weeks. When cell growth was
in the logarithmic phase, the cells were collected and
re-inoculated in a 10-ml culture flask in culture medium containing
the above-mentioned drugs at an elevated concentration (1.5- to
2-fold of the previous dose) or at a previous concentration. This
procedure was repeated until the cells exhibited stable growth and
proliferation in a culture medium with 4.0 μg/ml ADM, 0.4 μg/ml
CBP, 0.6 μg/ml DDP, 0.1 μg/ml MMC or 4.0 μg/ml VCR. A period of
~10–15 months was required to establish these drug-resistant HCC
cell sublines. The level of drug resistance was determined using
the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT) assay.
Cell viability MTT assay
Exponentially growing cells were seeded at
6,000–10,000 cells/well in 96-well plates with 100 μl of culture
medium/well and incubated for 6 h. The cells were then exposed to
different concentrations of the drugs for 65 h. At the end of the
experiments, 20 μl of MTT (5 mg/ml in PBS) was added in each well,
and the cells were cultured for an additional 4 h for formation of
formazan crystals. Subsequently, 150 μl of DMSO was added to each
well to dissolve the crystals. The values of the optical density at
570 nm were then measured using a microplate ELISA reader. Each
experiment was performed in triplicate and repeated trice.
Resistance factors (RF) were calculated by dividing the
IC50 value (drug concentration resulting in 50%
reduction in absorbance compared with the control) of the
drug-resistant cells with that of the parental control cells.
miRNA microarray analysis
Total cellular RNA was isolated using TRIzol
(Invitrogen Life Technologies) and then cleaned using the miRNeasy
Mini kit (Qiagen) according to the manufacturer's instructions. RNA
quality and quantity were measured using a Nanodrop
spectrophotometer (ND-1000; Nanodrop Technologies, Wilmington, DE,
USA), and RNA integrity was determined using gel electrophoresis.
Subsequently, the miRCURY™ Hy3/Hy5 Power labeling kit (Exiqon,
Vedbaek, Denmark) was used to label the RNA as probes according to
the manufacturer's guideline. Next, the Hy3TM-labeled RNA samples
were hybridized on miRCURY™ LNA array (v. 16.0) (Exiqon) slides
according to the array manual. The slides were then washed three
times with wash buffer (Exiqon) and dried using centrifugation for
5 min at 400 rpm. Finally, the slides were scanned using the Axon
GenePix 4000B microarray scanner (Axon Instruments, Foster City,
CA, USA).
The scanned images were imported into GenePix Pro
6.0 software (Axon Instruments) for grid alignment and data
extraction. Replicated miRNAs were averaged and miRNAs with
intensities >50 in all the samples were chosen to calculate a
normalization factor. Expressed data were then normalized using the
median normalization. Differentially expressed miRNAs were then
identified according to the fold changes and intensities, and
hierarchical clustering was performed using MEV software (v. 4.6,
TIGR).
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
Total cellular RNA samples from the cells were
reversely transcribed into cDNA using a Universal cDNA synthesis
kit (Exiqon) according to the manufacturer's instructions.
Real-time PCR amplification was performed on an ABI 7500 Real-Time
PCR system with the DNA-binding dye technique (SYBR Green)
according to the manufacturer's instructions. The primers for the
different miRNAs and PCR reagents were purchased from Exiqon. U6
snRNA was used as the reference gene, and 2−ΔΔct was
used to calculate the expression levels of miRNAs in the
drug-resistant HCC cell sublines compared to the parental Huh-7
cell line.
Results
Establishment of different
chemotherapeutical drug-resistant HCC cell sublines
In this study, we first established drug-resistant
HCC cell sublines by treating HCC Huh-7 cells with increasing doses
of ADM, DDP, CBP, MMC and VCR for ~10–15 months. The drug-resistant
concentration of ADM was 4.0 μg/ml, CBP was 0.4 μg/ml, DDP was 0.6
μg/ml, MMC was 0.1 μg/ml and VCR was 4.0 μg/ml. The HCC cell
sublines were named Huh-7/ADM, Huh-7/DDP, Huh-7/CBP, Huh-7/MMC and
Huh-7/VCR, respectively. The IC50 values and RF, which
are shown in Table I, indicated
that these drug-resistant sublines were not only resistant to the
treatment drug, but also resistant to the other drugs. Under an
inverted microscope, although the drug-resistant cells were similar
to the parental Huh-7 cells, the drug-resistant cells grew slowly
as colonies and most of the cell lines had an enlarged cell body
(Fig. 1).
 | Table IIC50 values and resistance
factors (RF) in the HCC Huh-7 cell line and the five drug-resistant
sublines. |
Table I
IC50 values and resistance
factors (RF) in the HCC Huh-7 cell line and the five drug-resistant
sublines.
| Drug | Huh-7
IC50 | Huh-7/ADM
IC50 (RF) | Huh-7/CBP
IC50 (RF) | Huh-7/DDP
IC50 (RF) | Huh-7/MMC
IC50 (RF) | Huh-7/VCR
IC50 (RF) |
|---|
| ADM | 1.6±0.44 |
117.24±22.96a
(73.28) | 74.66±0.55b (46.66) | 11.87±0.81b (7.42) | 28.44±6.75a (17.78) |
148.73±13.52b
(92.96) |
| CBP | 19.79±1.86 | 27.43±1.31b (1.39) |
214.55±36.57a
(10.84) | 56.87±1.03b (2.87) | 74.71±3.93b (3.78) |
103.11±20.08a
(5.21) |
| DDP | 3.47±0.71 | 14.36±6.47
(4.14) | 15.87±1.57b (4.57) | 13.66±0.41b (3.94) | 4.37±0.71
(1.26) | 12.33±1.64b (3.55) |
| MMC | 1.98±0.12 | 6.9±0.84b (3.49) | 2.7±0.1b (1.36) | 1.72±0.394
(0.87) | 12.76±0.44b (6.44) | 9.77±0.84b (4.93) |
| VCR | 4.88±0.7 |
217.22±49.84a
(44.5) | 69.91±16.38a (14.33) | 46.52±4.43b (9.53) |
100.57±13.72b
(20.61) |
243.45±60.23a
(49.89) |
Differential expression of miRNAs in the
drug-resistant HCC cell sublines
To profile differential expression of various miRNAs
in the drug-resistant HCC sublines, we performed miRNA microarray
analysis in these sublines, as well as in the parental HCC Huh-7
cells. After normalizing the expression data with bioinformatical
methods, the differential miRNA expression profiles were plotted in
scatter-plots (Fig. 2). We
performed fold change filtering between the data for each subline
and the parental cells. The threshold for both the upregulated and
downregulated miRNAs was at least 2-fold and the intensity of the
hybridization signal was at least 500. Overall, compared to the
parental Huh-7 cell line, 53 upregulated miRNAs were found in
Huh-7/ADM, 56 in Huh-7/CBP, 58 in Huh-7/DDP, 58 in Huh-7/MMC and 49
in Huh-7/VCR, whereas there were 52 downregulated miRNAs in
Huh-7/ADM, 50 in Huh-7/CBP, 41 in Huh-7/DDP, 55 in Huh-7/MMC and 56
in Huh-7/VCR. Moreover, there were 26 upregulated and 25
downregulated miRNAs noted in the Huh-7/ADM, Huh-7/CBP, Huh-7/DDP
and Huh-7/MMC sublines. However, among these 51 upregulated and
downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were
downregulated in the Huh-7/VCR subline. We then performed a
hierarchical clustering analysis for these miRNAs (Fig. 3). Fig.
4 shows the normalized value of these differentially expressed
miRNAs between Huh-7 cells and the Huh-7/ADM, Huh-7/CBP, Huh-7/DDP,
Huh-7/MMC and Huh-7/VCR sublines.
 | Figure 2miRNA expression profiles between the
five drug-resistant cell sublines and their parental Huh-7 cell
line. The axes of the scatter-plot are the normalized signal values
(ratio scale). The variation in miRNA expression levels in the
drug-resistant sublines compared to Huh-7 cells were similar for
Huh-7/ADM, Huh-7/CBP, Huh-7/DDP and Huh-7/MMC. H0, Huh-7; A1,
Huh-7/ADM; C2, Huh-7/CBP; D3, Huh-7/DDP; M4, Huh-7/MMC; V5,
Huh-7/VCR. (A) RA1-H0=0.8277, (B)
RC2-H0=0.8451, (C) RD3-H0=0.9093, (D)
RM4-H0=0.8964, (E) RV5-H0=0.8647. |
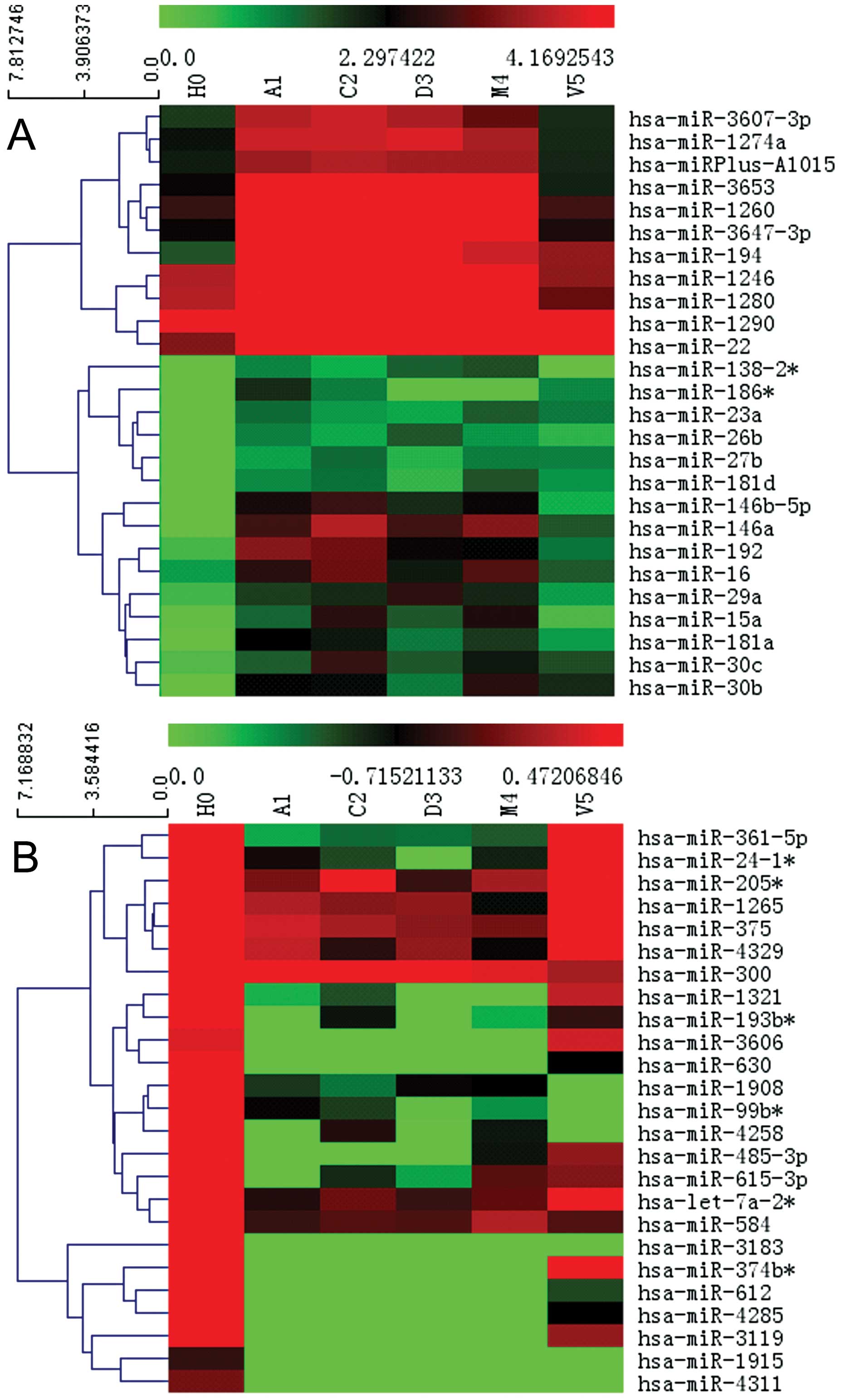 | Figure 3Hierarchical clustering analysis of
the miRNA microarray data. (A) Analyses of the 26 upregulated
miRNAs in Huh-7/ADM, Huh-7/CBP, Huh-7/DDP and Huh-7/MMC. (B)
Analyses of the 25 downregulated miRNAs in Huh-7/ADM, Huh-7/CBP,
Huh-7/DDP and Huh-7/MMC. The heat map diagram shows the results of
the two-way hierarchical clustering analysis of miRNA expression
levels and the cell lines. Each row represents an miRNA and each
column represents a cell line. The miRNA-clustering tree is shown
on the left and the cell line-clustering tree is at the top. The
color scale shown at the top illustrates the relative expression
level of an miRNA in a certain slide. A red color represents a high
relative expression level and a green color represents a low
relative expression level. H0, Huh-7; A1, Huh-7/ADM; C2, Huh-7/CBP;
D3, Huh-7/DDP; M4, Huh-7/MMC; V5, Huh-7/VCR. |
Validation of several differentially
expressed miRNAs in HCC cells
We validated the microarray data using real-time
RT-PCR analysis. We chose the five most significantly expressed
miRNAs (miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a) to
be verified in Huh-7, Huh-7/ADM, Huh-7/CBP, Huh-7/DDP, Huh-7/MMC
and Huh-7/VCR cells. Our qRT-PCR data confirmed the microarray data
(Table II). Table II also lists potential target genes
of these miRNAs.
 | Table IIConfirmation of miRNA expression
levels in the five drug-resistant sublines. |
Table II
Confirmation of miRNA expression
levels in the five drug-resistant sublines.
| Fold changes
(microarray, real-time PCRa) | |
|---|
|
| |
|---|
| A1/H0 | C2/H0 | D3/H0 | M4/H0 | |
|---|
|
|
|
|
| |
|---|
| miRNA | Array | qPCR | Array | qPCR | Array | qPCR | Array | qPCR | Target genes |
|---|
| miR-27b | 3.54 | 20.13 | 5.02 | 7.48 | 3.04 | 1.72 | 4.45 | 4.56 | BAX, P53, FoxO1,
KRAS |
| miR-181a | 9.98 | 22.32 | 8.99 | 12.41 | 4.77 | 2.17 | 7.02 | 6.45 | PTEN, KRAS,
RB1 |
| miR-146b-5p | 29.84 | 35.47 | 36.96 | 10.14 | 20.75 | 4.28 | 28.15 | 3.85 | TRAF6, PTGS2 |
| miR-181d | 7.08 | 6.97 | 8.20 | 3.86 | 4.97 | 2.02 | 9.80 | 3.19 | PTEN, KRAS,
RB1 |
| miR-146a | 24.44 | 441.13 | 42.60 | 96.45 | 24.57 | 12.95 | 33.69 | 50.66 | TRAF6, PTGS2 |
Discussion
In the present study, we first established five
different chemotherapeutic drug-resistant HCC cell sublines and
then profiled the altered miRNA expression in these sublines in
comparison to the parental HCC cell line using miRCURY™ LNA array
(v. 16.0). We found that cells that were resistant to one drug were
also resistant to the other drugs. miRNA microarray data revealed
53 upregulated miRNAs in Huh-7/ADM, 56 in Huh-7/CBP, 58 in
Huh-7/DDP, 58 in Huh-7/MMC and 49 in the Huh-7/VCR subline. In
contrast, there were 52 downregulated miRNAs in Huh-7/ADM, 50 in
Huh-7/CBP, 41 in Huh-7/DDP, 55 in Huh-7/MMC and 56 in Huh-7/VCR. In
total, there were 26 simultaneously upregulated and 25
simultaneously downregulated miRNAs in Huh-7/ADM, Huh-7/CBP,
Huh-7/DDP and Huh-7/MMC. Among these 51 altered miRNAs, 12 were
upregulated and 13 miRNAs were downregulated in the Huh-7/VCR
subline. We chose miR-27b, miR-181a, miR-146b-5p, miR-181d and
miR-146a to verify the results with real-time RT-PCR. This study
profiled altered miRNA expression in the five most commonly used
drug-induced HCC resistant sublines, which provides useful
information for further investigation of miRNA-mediated drug
resistance in HCC.
Generally, to study the mechanisms responsible for
tumor multidrug resistance, three methods are usually used to
establish a multidrug-resistant tumor cell line. These methods
include the induction of drug resistant cell lines in vitro,
MDR gene transfection, or the induction of drug resistance with a
nude mouse implanted tumor model. To induce tumor cell MDR in
vitro, two different cell culture methods are used: the drug
concentration incremental gradient method and the
high-concentration intermittent drug-induced method (23). In the present study, we established
drug-resistant cell sublines using the drug concentration
incremental gradient method. The drug-resistant cell sublines were
resistant not only to the drug used to induce the subline, but also
to the other drugs, which is characteristic of MDR. These data
provide the foundation to pursue subsequent research concerning the
altered expression of miRNAs in these sublines.
A large number of miRNAs with altered expression
were noted in these sublines compared to the parental HCC cell
line. Several of the miRNAs, such as the miR-15/16 family (14), miR-30c (24,25),
the miR-181 family (18,26,27),
miR-23a (28), miR-192 (29), miR-27b (12), miR-194 (12,29),
miR-22 (12), miR-29a (12,30),
miR-146a (29) and the let-7 family
(31,32), have been previously reported to be
related to chemoresistance in different types of cancers.
Specifically, miR-15b and miR-16 play a role in the development of
MDR in gastric cancer through modulation of apoptosis by targeting
Bcl-2 expression (14), while
miR-30c was found to be downregulated in various chemoresistant
tumor cell lines (23). Moreover,
miR-181d was upregulated in two docetaxel-induced head and neck
squamous cell cancer MDR cell lines (27), and miR-23a was shown to be an
upstream regulator of TOP2B that mediated cisplatin chemoresistance
in tongue squamous cell carcinoma cell lines (28). Furthermore, miR-192 was found to
target dihydrofolate reductase, a key enzyme in folate metabolism
that influences sensitivity in colorectal cancer cell lines when
treated with 5-FU (29). In
addition, breast cancer MCF-7/DOX cells resistant to DOX treatment
were associated with the increased expression of miR-22, miR-29a,
miR-194 and miR-132 (12).
miR-let-7a was downregulated in the NCI-60 cell line, which is
sensitive to cyclophosphamide (31). Our current data revealed 12
simultaneously upregulated and 13 downregulated miRNAs in the five
HCC drug-resistant cell sublines. Further investigation of these 25
miRNAs may help elucidate HCC cell MDR to these five drugs.
In addition, we used a SYBR-Green miRNA real-time
qPCR analysis to validate the expression of miR-27b, miR-181a,
miR-146b-5p, miR-181d and miR-146a in the drug-resistant HCC cell
sublines. miR-27b, which is localized at chromosome q22.32, has
been reported to play a role in cell growth, tumor progression and
the inflammatory response (33–36).
miRNA-146a/b has been shown to be involved in the modulation of
inflammatory responses (37),
immune responses (38) and
suppression of tumor metastasis (39–41).
miR-181a was reported to be associated with patient prognosis
(42,43). Finally, miR-181a and mir-181d
(44) have been shown to have
antitumor activity (43). However,
their role in HCC MDR will be further investigated in our next
study.
Acknowledgements
This study was supported in part by a grant from the
Department of Education, Fujian Province, China (#JA11114). We
would like to thank Ms. Ren-Nan of Fujian Medical University,
School of Public Health for her technical assistance in the qRT-PCR
analyses.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Lee JO, Kim DY, Lim JH, et al: Palliative
chemotherapy for patients with recurrent hepatocellular carcinoma
after liver transplantation. J Gastroenterol Hepatol. 24:800–805.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tomuleasa C, Soritau O, Fischer-Fodor E,
et al: Arsenic trioxide plus cisplatin/interferon
alpha-2b/doxorubicin/capecitabine combination chemotherapy for
unresectable hepatocellular carcinoma. Hematol Oncol Stem Cell
Ther. 4:60–66. 2011. View Article : Google Scholar
|
|
4
|
Walsh N, Larkin A, Kennedy S, et al:
Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and
ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 9:62009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi H, Lu D, Shu Y, Shi W, Lu S and Wang
K: Expression of multidrug-resistance-related proteins
P-glycoprotein, glutathione-S-transferases, topoisomerase-II and
lung resistance protein in primary gastric cardiac adenocarcinoma.
Cancer Invest. 26:344–351. 2008. View Article : Google Scholar
|
|
6
|
Mansilla S, Rojas M, Bataller M, Priebe W
and Portugal J: Circumvention of the multidrug-resistance protein
(MRP-1) by an antitumor drug through specific inhibition of gene
transcription in breast tumor cells. Biochem Pharmacol. 73:934–942.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filomeni G, Turella P, Dupuis ML, et al:
6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol, a specific
glutathione S-transferase inhibitor, overcomes the multidrug
resistance (MDR)-associated protein 1-mediated MDR in small cell
lung cancer. Mol Cancer Ther. 7:371–379. 2008. View Article : Google Scholar
|
|
8
|
Ronaldson PT, Ashraf T and Bendayan R:
Regulation of multidrug resistance protein 1 by tumor necrosis
factor alpha in cultured glial cells: involvement of nuclear
factor-kappaB and c-Jun N-terminal kinase signaling pathways. Mol
Pharmacol. 77:644–659. 2010. View Article : Google Scholar
|
|
9
|
Liu B, Qu L and Tao H: Cyclo-oxygenase 2
up-regulates the effect of multidrug resistance. Cell Biol Int.
34:21–25. 2010.PubMed/NCBI
|
|
10
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010. View Article : Google Scholar
|
|
11
|
Feng DD, Zhang H, Zhang P, et al:
Down-regulated miR-331-5p and miR-27a are associated with
chemotherapy resistance and relapse in leukaemia. J Cell Mol Med.
15:2164–2175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang Z, Wu H, Xia J, et al: Involvement
of miR-326 in chemotherapy resistance of breast cancer through
modulating expression of multidrug resistance-associated protein 1.
Biochem Pharmacol. 79:817–824. 2010. View Article : Google Scholar
|
|
14
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu Y, Xia F, Ma L, et al: MicroRNA-122
sensitizes HCC cancer cells to adriamycin and vincristine through
modulating expression of MDR and inducing cell cycle arrest. Cancer
Lett. 310:160–169. 2011.PubMed/NCBI
|
|
16
|
Zhao X, Yang L, Hu J and Ruan J: miR-138
might reverse multidrug resistance of leukemia cells. Leuk Res.
34:1078–1082. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao X, Yang L and Hu J: Down-regulation
of miR-27a might inhibit proliferation and drug resistance of
gastric cancer cells. J Exp Clin Cancer Res. 30:552011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu W, Shan X, Wang T, Shu Y and Liu P:
miR-181b modulates multidrug resistance by targeting BCL2 in human
cancer cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bai H, Xu R, Cao Z, Wei D and Wang C:
Involvement of miR-21 in resistance to daunorubicin by regulating
PTEN expression in the leukaemia K562 cell line. FEBS Lett.
585:402–408. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan YZ, Morris ME and Yu AM: MicroRNA-328
negatively regulates the expression of breast cancer resistance
protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol.
75:1374–1379. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong X, Xiong M, Meng X and Gong R:
Comparison of the multi-drug resistant human hepatocellular
carcinoma cell line Bel-7402/ADM model established by three
methods. J Exp Clin Cancer Res. 29:1152010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Galluzzi L, Morselli E, Vitale I, et al:
miR-181a and miR-630 regulate cisplatin-induced cancer cell death.
Cancer Res. 70:1793–1803. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai Y, Xie CH, Neis JP, Fan CY, Vural E
and Spring PM: MicroRNA expression profiles of head and neck
squamous cell carcinoma with docetaxel-induced multidrug
resistance. Head Neck. 33:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT
and Zhang CP: MicroRNAs contribute to the chemoresistance of
cisplatin in tongue squamous cell carcinoma lines. Oral Oncol.
46:317–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boni V, Bitarte N, Cristobal I, et al:
miR-192/miR-215 influence 5-fluorouracil resistance through cell
cycle-mediated mechanisms complementary to its post-transcriptional
thymidilate synthase regulation. Mol Cancer Ther. 9:2265–2275.
2010. View Article : Google Scholar
|
|
30
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang CJ, Hsu CC, Chang CH, et al: Let-7d
functions as novel regulator of epithelial-mesenchymal transition
and chemoresistant property in oral cancer. Oncol Rep.
26:1003–1010. 2011.PubMed/NCBI
|
|
32
|
Salter KH, Acharya CR, Walters KS, et al:
An integrated approach to the prediction of chemotherapeutic
response in patients with breast cancer. PLoS One. 3:e19082008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen L, Li H, Han L, et al: Expression and
function of miR-27b in human glioma. Oncol Rep. 26:1617–1621.
2011.PubMed/NCBI
|
|
34
|
Ji J, Zhang J, Huang G, Qian J, Wang X and
Mei S: Over-expressed microRNA-27a and 27b influence fat
accumulation and cell proliferation during rat hepatic stellate
cell activation. FEBS Lett. 583:759–766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JJ, Drakaki A, Iliopoulos D and Struhl
K: MiR-27b targets PPARgamma to inhibit growth, tumor progression
and the inflammatory response in neuroblastoma cells. Oncogene.
31:3818–3825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Rathinam R, Walch A and Alahari
SK: ST14 (suppression of tumorigenicity 14) gene is a target for
miR-27b, and the inhibitory effect of ST14 on cell growth is
independent of miR-27b regulation. J Biol Chem. 284:23094–23106.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhaumik D, Scott GK, Schokrpur S, et al:
MicroRNAs miR-146a/b negatively modulate the senescence-associated
inflammatory mediators IL-6 and IL-8. Aging (Albany, NY).
1:402–411. 2009.PubMed/NCBI
|
|
38
|
Chatzikyriakidou A, Voulgari PV, Georgiou
I and Drosos AA: The role of microRNA-146a (miR-146a) and its
target IL-1R-associated kinase (IRAK1) in psoriatic arthritis
susceptibility. Scand J Immunol. 71:382–385. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Campisi J and Benz CC: Expression of microRNA-146 suppresses
NF-kappaB activity with reduction of metastatic potential in breast
cancer cells. Oncogene. 27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical significance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–4284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gao W, Yu Y, Cao H, et al: Deregulated
expression of miR-21, miR-143 and miR-181a in non small cell lung
cancer is related to clinicopathologic characteristics or patient
prognosis. Biomed Pharmacother. 64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schwind S, Maharry K, Radmacher MD, et al:
Prognostic significance of expression of a single microRNA,
miR-181a, in cytogenetically normal acute myeloid leukemia: a
Cancer and Leukemia Group B study. J Clin Oncol. 28:5257–5264.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang XF, Shi ZM, Wang XR, et al: MiR-181d
acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2.
J Cancer Res Clin Oncol. 138:573–584. 2012. View Article : Google Scholar : PubMed/NCBI
|


















