Introduction
Tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL, also termed APO-2L) is a pro-apoptotic cytokine that
belongs to the tumor necrosis factor superfamily (1). A TRAIL homotrimer interacts with a
homotrimeric TRAIL-R1 [death receptor (DR)4] or TRAIL-R2 (DR5),
initiating extrinsic and indirectly intrinsic pathways of apoptosis
and nuclear facgtor-κB (NF-κB) activation (2). TRAIL also binds to two other membrane
receptors, TRAIL-R3 [decoy receptor (DcR)1] and TRAIL-R4 (DcR2)
that do not contain functional death domains and function as DcRs
(2). Another TRAIL receptor,
osteoprotegerin (OPG), acts as a soluble DcR for TRAIL (3). TRAIL is known to play critical roles
in immune surveillance and defense mechanisms against cancer cells,
as well as in normal hematopoiesis (4,5).
TRAIL is known to preferentially induce apoptosis in
cancer cells, with little to no toxicity to normal cells, which has
prompted research into its therapeutic application (6). Although TRAIL manifests fewer
side-effects than conventional chemotherapeutic reagents, there are
nevertheless considerable obstacles to the clinical application of
TRAIL. Since the majority of injected TRAIL is rapidly cleared, a
large dose of TRAIL may be required in clinical situations
(7). Additionally, sensitivity to
TRAIL-mediated apoptosis is widely divergent depending on the type
of cancer (8,9).
Cancer cells equipped with any type of
anti-apoptotic mechanisms can potentially evade TRAIL-mediated
apoptosis. The silencing of or loss-of-function mutations in DR4
and DR5 via genetic and epigenetic changes in cancer cells can
render the cancer cells resistant to TRAIL (10–13).
The increased expression of DcRs is also involved in the TRAIL
resistance of cancer cells (14).
In addition, cancer cells that overexpress anti-apoptotic
molecules, including cellular FLICE-inhibitory protein (c-FLIP),
X-linked inhibitor of apoptosis (XIAP) and Bcl-2, are resistant to
TRAIL-mediated apoptosis (15–17).
Since TRAIL-mediated apoptosis can be regulated by the expression
status of various anti- and pro-apoptotic molecules, the focus of
research on the therapeutic application of TRAIL has shifted toward
the discovery of indicators and enhancers of the TRAIL sensitivity
of cancer cells (8,18,19).
Gastric cancer was estimated to be the fourth most
common malignancy and the second leading cause of cancer-related
mortality worldwide in 2008 (20).
The incidence of gastric cancer is reported to be particularly high
in East Asian countries, including Japan, China and Korea. Gastric
cancer is treated by the surgical resection of the operable tumor,
accompanied by localized radiotherapy and chemotherapy with
conventional chemotherapeutics (21). However, the non-specific toxicity of
the drugs necessitates the development of novel therapeutic
measures to treat the disease, which has prompted a search for
novel therapeutics (22).
TRAIL has been tested as a candidate drug for the
treatment of gastric cancer (23).
Although TRAIL induces the apoptosis of many gastric cancer cells,
considerable resistance has also been reported in a few gastric
cancer cells (16,24). To optimize TRAIL utility as an
effective therapeutic strategy for gastric cancer, it is necessary
to identify critical indicators of TRAIL treatment outcomes and
potential enhancers of TRAIL efficacy (11,23,25,26).
In the present study, we investigated the response of various
gastric cancer cell lines to TRAIL, the mechanism of TRAIL-mediated
apoptosis and the molecular basis of the differential sensitivity
to TRAIL. Four gastric cancer cells grown in suspension manifested
differential susceptibility to TRAIL treatment. The expression of
signaling components involved in TRAIL-mediated apoptosis,
including TRAIL receptors, caspases and apoptosis-modulating
proteins, was examined to investigate the mechanism of
TRAIL-mediated apoptosis and the molecular basis of the divergent
responses to TRAIL treatment.
Materials and methods
Cell culture
The gastric cancer cell lines, SNU-1, SNU-5, SNU-16
and SNU-620, were cultured in RPMI-1640 (HyClone, South Logan, UT,
USA) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml
penicillin and 100 μg/ml streptomycin (HyClone) at 37°C in a
humidified 5% CO2 incubator. They were grown in
suspension and subcultured twice a week by splitting 1:10 after
collection by centrifugation.
MTT assay
Methylthiazolyldiphenyl-tetrazolium bromide (MTT;
Sigma-Aldrich, St. Louis, MO, USA) assay was carried out as
previously described in the study by Huynh et al(27), with some modifications for cell
suspension. A total of 2,000 cells/well plated in a 96-well plate
on the previous day were treated with either human TRAIL APO-II
Ligand (PeproTech Inc., Rocky Hill, NJ, USA) or the vehicle, as
specified in the figure legends. At 24, 48 or 72 h after treatment,
20 μl/well MTT solution (2.5 mg/ml PBS) were added to the cells and
incubated for 3 h. Solubilizer (80 μl/well, 10% SDS with 0.01 N
HCl) was added, and the plate was incubated at 37°C overnight to
dissolve the MTT formazan. The absorbance at 570 nm, with reference
absorbance at 650 nm, was measured with a Multiskan GO
spectrophotometer (Thermo Scientific, Rockland, IL, USA). The
percentage viability was calculated by [(absorbance of experimental
well)/(absorbance of control well)] ×100%. The IC50 was
obtained by plotting log[percentage viability] against TRAIL
concentration and determining the TRAIL concentration at 50%
viability.
Flow cytometric analysis
Annexin V binding and the expression of TRAIL
receptors on the cell surface were analyzed by flow cytometry. For
Annexin V binding analysis, cells seeded at 1.2×105/well
in 6-well plates on the previous day were treated with 50 ng/ml
TRAIL for 16 h. The cells were collected by centrifugation (520 ×
g, 2 min) and were incubated in PBS containing 2 mM EDTA and 0.5%
FBS for 2 min. The cells were washed once with Annexin V binding
buffer and then incubated in buffer containing FITC-conjugated
Annexin V (100 μl, 0.05 μg/ml; Annexin V-FITC detection kit,
eBioscience, San Diego, CA, USA) and propidium iodide (PI, 5 μg/ml)
at room temperature in the dark for 30 min. Annexin V binding and
PI infiltration were measured by flow cytometry using a
FACSCalibur™ (BD Biosciences, Sparks, MD, USA), and the data were
analyzed with CellQuest Pro™ software (BD Biosciences). To detect
cell surface receptor expression, cells, plated as described above,
were allowed to grow for 48 h. The pelleted cells (520 × g, 2 min)
were dispersed by incubation in PBS containing 2 mM EDTA and 0.5%
FBS for 2 min, and the cells were then incubated in 100 μl PBS
containing phycoerythrin (PE)-conjugated α-TRAIL, α-DcR1 (CD263),
α-DR4 (CD261), α-DR5 (all from eBioscience) or α-DcR2 antibody
(B-R27, Santa Cruz Biotechnology) for 30 min in the dark at room
temperature. A PE-conjugated mouse IgG isotype control
(eBioscience) was used as the negative control. The fluorescence
signal from the bound antibodies was measured by flow cytometry, as
described above.
RNA isolation and RT-PCR
The total RNA was isolated, and an aliquot (5 μg)
was reverse-transcribed, as previously described (27). A total of 2 μl of each RT reaction
mixture was then employed for real-time PCR using the
SYBR®-Green PCR kit (Qiagen, Hilden, Germany) with
LightCycler 2.0 (Roche, Basel, Switzerland) as directed by the
manufacturer. The primers used in the experiments were as follows:
TRAIL (GGAACCCAAGGTGGGTAGAT/TCTCACCACACTGCAACCTC), DR4
(CTGAGCAACGCAGACTCGCTGTCCAC/TCCAAGGACACGGCAGAGCCTGTGCCAT), DR5
(GCCTCATGGACAATGAGATAAAGGTGGCT/CCAAATCTCAAAGTACGCACAAACGG), DcR1
(GAAGAATTTGGTGCCAATGCCACTG/CTCTTGGACTTGGCTGGGAGATGTG), DcR2
(GGCTGCTGGTTCCAGTGAATGACGCT/GTTTCTTCCAGGCTGCTTCCCTTTGTAG) and
GAPDH (TGATGACATCAAGAAGGTGG/TCCTTGGAGGCCATGTGGGCCAT). The
PCR reactions consisted of one cycle of 95°C for 10 min, followed
by 45 cycles of 95°C for 10 sec, 62°C for 5 sec and 72°C for 20
sec. The input cDNA levels were monitored by measuring GAPDH in a
parallel reaction. The specific mRNA levels were calculated by
(1/2)ΔΔCT. ΔΔCT is defined as
CT of targets-CT of GAPDH.
Protein extraction and western blot
analysis
Total cell lysates were prepared and analyzed by
western blot analysis, as previously described (27). The antibodies used in the
experiments were α-caspase-9, α-caspase-3, α-XIAP (Cell Signaling
Technology, Danvers, MA, USA), α-BID, α-caspase-8 (Chemicon,
Temecula, CA, USA), α-DR5, α-FLIP (eBioscience), α-DR4 (Novus
Biologicals, Littleton, CO, USA) and α-β-actin (Bethyl Laboratories
Inc., Montgomery, TX, USA).
Results
Response of gastric cancer cells to
TRAIL
The responsiveness of the four gastric cancer cell
lines, SNU-1, SNU-5, SNU-16 and SNU-620, to TRAIL was determined by
MTT assay. TRAIL induced cell death in a concentration-dependent
manner in all four cell lines. However, the IC50 of
TRAIL at each time-point demonstrated the differential TRAIL
sensitivity of the cell lines (Fig.
1A). The IC50 values at 48 h were 12.7 ng/ml for
SNU-16, 14.0 ng/ml for SNU-620, 30.3 ng/ml for SNU-5 and 78.7 ng/ml
for SNU-1. In general, the IC50 values of the SNU-16 and
SNU-620 cells were 4- to 6-fold lower than those of the SNU-1 cells
at each measured time-point. The TRAIL-mediated apoptosis of the
gastric cancer cells was confirmed by flow cytometric analysis of
the Annexin V-bound cells. In the SNU-16 and SNU-620 cells, TRAIL
significantly increased the number of both Annexin V- and
PI-positive cells, whereas there was no significant increase in
these populations in the SNU-1 cells (Fig. 1B). The fraction of Annexin V- and/or
PI-positive cells was displayed in the following order: SNU-16 ≈
SNU-620 > SNU-5 >> SNU-1. Both the MTT assay and flow
cytometry analysis results indicated that the SNU-16 and SNU-620
cells were highly sensitive to TRAIL-mediated cell death, whereas
the SNU-1 cells were relatively resistant and the SNU-5 cells
showed intermediate sensitivity to TRAIL.
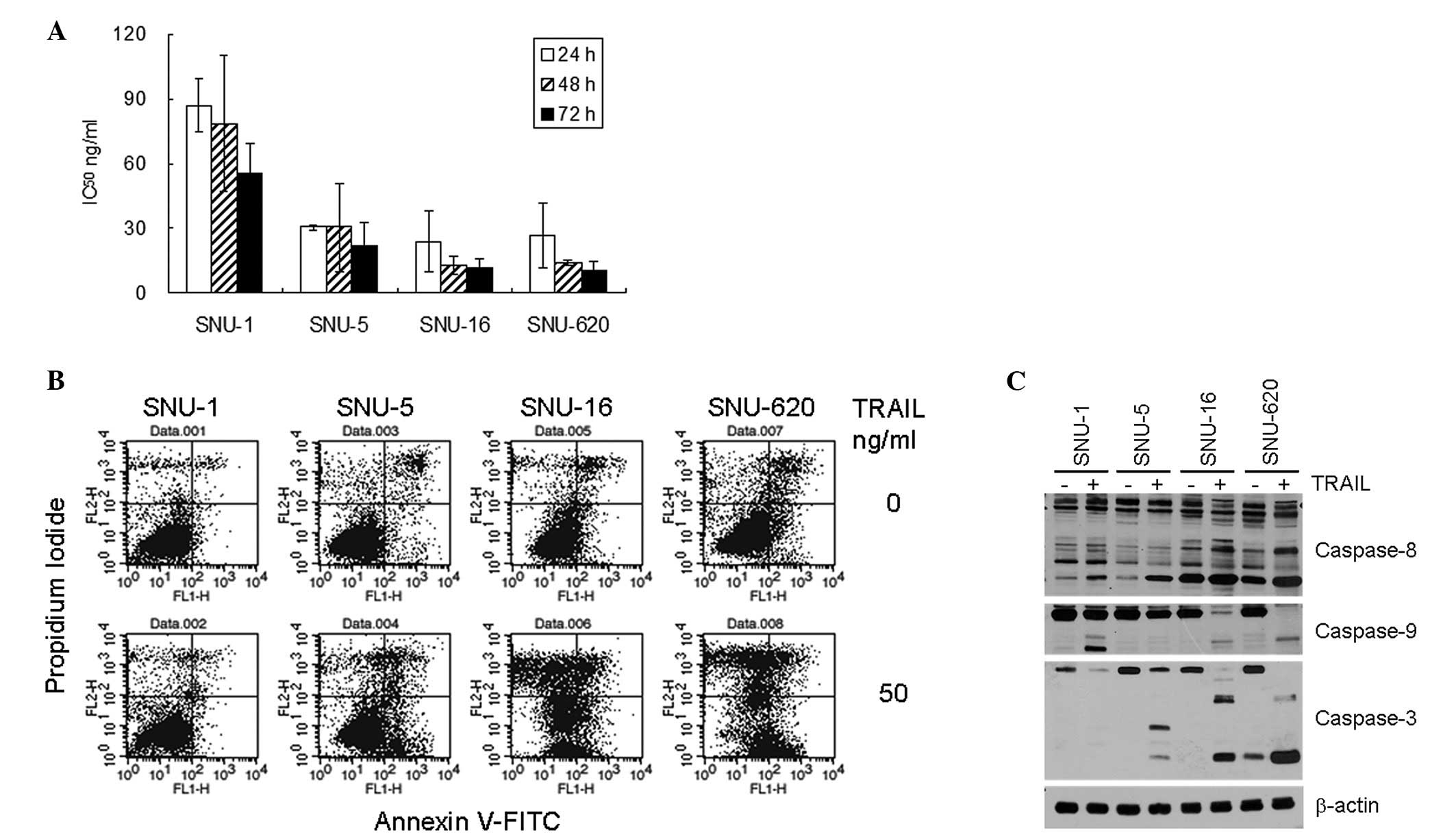 | Figure 1TRAIL-mediated apoptosis of the four
gastric cancer cell lines. (A) Daily IC50 of
TRAIL-induced cell death of the four gastric cancer cells. The
cells were plated one day prior to TRAIL treatment (0, 6.25, 12.5,
25, 50 or 100 ng/ml), and MTT assay was carried out at 24, 48 or 72
h after treatment. The percentage cell viability and
IC50 were calculated as described in Materials and
methods. The results shown are the average ± standard deviation of
three independent experiments. (B) Flow cytometric analysis of
Annexin V binding. The cells were plated one day prior to TRAIL
treatment (50 ng/ml), and 16 h after treatment, Annexin V
binding/PI infiltration of the cells was measured by flow
cytometry. The results shown are representatives of three
independent experiments. (C) Activation of caspases upon TRAIL
treatment. The activating fragmentation of caspase-3, -8 and -9 was
detected by western blot analysis of cell lysates that were
untreated or treated with 50 ng/ml TRAIL for 16 h. Cell lysates
were prepared and analyzed by western blot analysis, as described
in Materials and methods. β-actin was used to monitor protein
content on the western blots. The data shown are representatives of
three independent experiments. |
The activation of caspase-3, -8 and -9 upon TRAIL
treatment was measured by western blot analysis (Fig. 1C). The expression levels of
procaspase-3, -8 and -9 were comparable between the gastric cancer
cells. Although TRAIL treatment increased the amount of active
caspase-8 fragments in all four cell lines, the quantity of the
fragments in the control and TRAIL-treated cells was much higher in
the TRAIL-sensitive SNU-16 and SNU-620 cells compared to the SNU-1
and SNU-5 cells. Similarly, the quantity of active caspase-3
fragments was significantly increased in the SNU-16 and SNU-620
cells and slightly increased in the SNU-5 cells, although
procaspase-3 expression diminished upon TRAIL treatment in all four
cell lines. By contrast, the activation of caspase-9 was apparent
in the SNU-1, SNU-16 and SNU-620 cells. Overall, the levels of
caspase activation, particulrly those of caspase-3 and -8
positively correlated with the differential TRAIL sensitivity of
the gastric cancer cells.
Expression of TRAIL receptors in the
gastric cancer cells
Since TRAIL receptor expression is associated with
the differential susceptibility to TRAIL (11,28),
we examined the mRNA levels of TRAIL receptors and TRAIL in the
gastric cancer cells by real-time RT-PCR (Fig. 2A). The mRNA expression of DcR1 and
DcR2 was the lowest in the TRAIL-sensitive SNU-16 cells among the
four gastric cancer cells. The SNU-5 cells showed a ~10-fold higher
level of TRAIL mRNA compared to SNU-1 and a 2- to 3-fold higher
level compared to the SNU-16 and SNU-620 cells. The DR4 mRNA level
in the SNU-1 cells was ~1,000-fold lower than that in the SNU-5,
SNU-16 and SNU-620 cells and was almost negligible, while the SNU-1
cells demonstrated the highest level of DR5 mRNA expression among
the tested cell lines. Western blot analysis of whole cell DR4
protein levels confirmed the absence of its expression in the SNU-1
cells (Fig. 2B). In addition, DR4
expressoin was almost completely diminished upon TRAIL treatment in
the TRAIL-sensitive SNU-16 and SNU-620 cells (Fig. 2C). By contrast, the DR5 protein
level was highest in the SNU-5 cells but was comparable among the
other cell lines (Fig. 2B). DR5
expression was reduced upon TRAIL treatment only in the SNU-16
cells (Fig. 2C).
Since TRAIL binds to its receptors on the cell
surface, the expression of TRAIL receptor proteins on the cell
surface was also measured by flow cytometry (Fig. 3). DR5 expression on the cell surface
was apparent and comparable among the four gastric cancer cell
lines. By contrast, the surface level expression of DR4 was obvious
in the SNU-5, SNU-16 and SNU-620 cells but not in the SNU-1 cells,
which correlated with the mRNA results. In contrast to the mRNA
levels, the expression of DcR1 on the cell surface was higher in
the SNU-5 and SNU-620 cells than in the other cell lines, while
surface DcR2 expression levels were noticeably higher in the SNU-1
cells. The expression of TRAIL was comparable among the four cell
lines. Taken together, these results show that the expression of
DR4 at the mRNA and protein levels and that of DcR2 on the cell
surface positively correlated with the differential TRAIL
sensitivity of the gastric cancer cells, whereas the expression of
DR5 and DcR1 did not correlate with TRAIL sensitivity.
Expression of apoptosis modulators
Several pro- and anti-apoptotic proteins, including
the proteins of the Bcl-2 and inhibitors of apoptosis (IAP)
families, either stimulate or inhibit TRAIL-induced apoptosis
(15–17). Therefore, to further understand the
mechanism underlying the differential susceptibility to
TRAIL-mediated apoptosis, we examined the expression of apoptosis
modulators in the gastric cancer cell lines by western blot
analysis. The fragmentation of Bid into tBid upon TRAIL treatment
directly reflected the activation of caspase-8 and TRAIL
sensitivity (Fig. 4, upper panel).
In addition, although the basal level of XIAP was similar in all
four cell lines, XIAP was almost completely degraded in the
TRAIL-sensitive SNU-16 and SNU-620 cells (Fig. 4, second panel from the top).
However, the expression of the long and short forms of FLIP was
relatively low in the SNU-1 cells and the short form completely
disappeared upon TRAIL treatment in all cell lines (Fig. 4, third panel from the top). In
conclusion, the fragmentation of Bid to tBid and XIAP degradation
corresponded well to the TRAIL sensitivity of the gastric cancer
cells, whereas the expression level of FLIP in the cells that were
untreated or treated with TRAIL did not correlate with TRAIL
sensitivity.
Since XIAP and cellular IAPs (cIAPs) are known to
inhibit TRAIL-mediated apoptosis through interaction with caspases,
XIAP cleavage may be required to promote TRAIL-mediated cell death
(15). Otherwise, caspases
activated by TRAIL cleave XIAP, thus facilitating apoptosis by a
feed forward amplification loop (29). Therefore, we determined whether XIAP
was cleaved by caspases activated by TRAIL treatment in the
TRAIL-sensitive SNU-620 cells (Fig.
5). The amount of XIAP protein gradually decreased with time in
the SNU-620 cells treated with TRAIL (Fig. 5A). However, the inhibition of
caspase activity following pre-treatment with the pan-caspase
inhibitor, Z-VAD, prevented the degradation of XIAP upon TRAIL
treatment (Fig. 5B). These results
suggest that XIAP may be cleaved by TRAIL-activated caspases, which
accelerates the apoptosis induced by TRAIL.
Discussion
The differential susceptibility of cancer cells
toward TRAIL-induced apoptosis is an obstacle to the wide
application of TRAIL as a cancer therapeutics. The effective
application of TRAIL in cancer treatment necessitates the
identification of potential indicators of TRAIL efficacy in cancer
cells, prompting intense investigation into the molecular
mechanisms of the TRAIL resistance of various cancer cells
(8,19). In this study, we aimed to identify
potential indicators of TRAIL response through quantitative
measurement of TRAIL cytotoxicity and perfomed a detailed analysis
of the molecular mechanisms involved in the differential TRAIL
sensitivity of four gastric cancer cell lines.
The efficacy of TRAIL decreased in the four gastric
cancer cell lines in the following order: SNU-16 ≈ SNU-620 >
SNU-5 >> SNU-1. This was further supported by an Annexin V
binding assay and the caspase activation profile. TRAIL
IC50 of the gastric cancer cells ranged between
23.8–87.1 ng/ml after 24 h of treatment, which was comparable to
effective concentration of TRAIL (10–100 ng/ml) in the
TRAIL-sensitive cancer cells. Of note, there was a positive
correlation between TRAIL sensitivity and the location of the
gastric cancer cell source. SNU-1 cells which were relatively
resistant to TRAIL, were isolated from a solid tumor, while the
other cell lines were established from ascites (30,31).
While the silencing of DR4 expression was implicated in the
tempered TRAIL response of the SNU-1 cells in this study, no
significant difference in the expression of DR4 was found between
primary gastric carcinomas and metastasized ones from ascites
(32). Thus, although tumor
metastasis from ascites is different from ascites per se,
the question of whether tumor cells in ascites are more sensitive
to TRAIL than those in solid tumors remains to resolved.
The expression level of death-inducing and decoy
TRAIL receptors has often been associated with TRAIL responsiveness
(9). The downmodulation or loss of
DR expression resulting from gene loss, mutations, epigenetic
control and/or post-translational regulation has been implicated in
gastric cancer cells with TRAIL resistance (13,33–35).
The expression of DR4 mRNA and protein was ~1,000-fold lower in the
SNU-1 cells which were relatively resistant to TRAIL, than in the
other tested cell lines, while the expression of DR5 and DcR1 was
comparable between the cell lines. DR4 expression has been reported
to correlate with TRAIL-induced apoptosis in various cancer cells
despite the presence of functional DR5 (11,36).
By contrast, DR5 has been shown to play a critical role in the
TRAIL-mediated apoptosis of certain bladder cancer cells (37), suggesting that TRAIL preferentially
exploits distinct DRs in different cells. In the gastric cancer
cells, the silenced DR4 expression is the most critical component
that determines TRAIL-mediated apoptosis. The downregulation of DR4
by promoter methylation has been reported in gastric carcinoma,
which could abate the effectiveness of TRAIL (33,34).
However, since azacytidine, a DNA methylation inhibitor, did not
restore DR4 expression (data not shown), its reduced expression in
the SNU-1 cells was not attributed to downregulation by
methylation.
Bid truncation into tBid by active caspase-8 relays
an extrinsic apoptotic signal triggered by TRAIL to the intrinsic
apoptotic pathway (38). Akt
activation renders ovarian cancer cells resistant to TRAIL by the
downregulation of Bid, suggesting that, in addition to the
extrinsic pathway, the intrinsic pathway significantly contributes
to TRAIL-induced apoptosis (39).
While the expression level of Bid was similar among the gastric
cancer cell lines, the full cleavage of Bid, as well as the
appearance of active caspase-9 and -3 fragments were obvious in the
TRAIL-sensitive gastric cancer cells. In addition, although cleaved
caspase-9 fragments were also observed in the SNU-1 cells, a
significant amount of procaspase-9 still remained intact upon TRAIL
treatment, including a significant amount of Bid. Thus, TRAIL
sensitivity better correlates with the activation of caspases via
the extrinsic pathway, which accentuates the importance of DRs and
the subsequent activation of caspase-8 in the gastric cancer
cells.
The sensitivity of cancer cells to TRAIL-mediated
cell death also correlates with intracellular levels of pro- and
anti-apoptotic proteins (9). Two
forms of FLIP, a long form and a short form, inhibit TRAIL-mediated
apoptosis by displacing caspase-8 from the death-inducing signaling
complex (DISC). The upregulation of FLIP by Akt has been suggested
to be an inhibitory mechanism of TRAIL-induced apoptosis in SNU-216
gastric cancer cells (16). TRAIL
response may also be influenced by the modulation of FLIP
expression (40). However, the
level of FLIP is not always indicative of the sensitivity to TRAIL
(41). In the current study, there
was no significant difference in the expression of both forms of
FLIP among the gastric cancer cells, and, in all of the cell lines
tested, the short form of FLIP disappeared upon TRAIL treatment
(Fig. 4). Thus, the TRAIL
susceptibility of the gastric cancer cells is likely independent of
the FLIP expression and degradation level.
IAPs are inhibitors of apoptosis that interact with
caspases and inhibit their activities (38). The expression of IAPs, particulary
XIAP, has been reported to be inversely correlated with TRAIL
sensitivity, and the modulation of XIAP expression influences TRAIL
sensitivity in a number of cancer cell lines (15). Whereas a substantial amount of XIAP
was detected in the gastric cancer cells, regardless of their
susceptibility to TRAIL, XIAP expression diminished upon TRAIL
treatment only in the TRAIL sensitive SNU-16 and SNU-620 cells. The
inhibition of caspase activity by the caspase inhibitor, Z-VAD,
fully rescued XIAP in the gastric cancer cells, suggesting that
XIAP was cleaved by activated caspases in the TRAIL-treated gastric
cancer cells (Fig. 5). XIAP
degradation by activated caspases upon TRAIL treatment can further
propagate apoptosis, which exerts positive feedback to enhance
TRAIL-mediated apoptosis (29).
TRAIL is an emerging candidate for gastric cancer
therapeutics and our results clearly support this possibility
(23). However, the differential
sensitivity to TRAIL-mediated apoptosis was also apparent among the
gastric cancer cells tested. Our results demonstrated that the
amounts of DR4 mRNA and protein coincided well with the TRAIL
sensitivity of the gastric cancer cells. Therefore, the expression
of DRs, particularly DR4, may serve as an indicator of TRAIL
response in gastric cancer cells, although it would be too hasty to
rule out other contributing factors. By contrast, since the
expression of Bid, FLIP and XIAP was comparable between the gastric
cancer cells, and since the cleavage of Bid to tBid and the
degradation of XIAP were apparent only in the TRAIL-sensitive
cells, these phenomena therefore play a limited role in enhancing
TRAIL-induced apoptosis by a positive feedback loop in the gastric
cancer cells.
Acknowledgements
The present study was supported by a grant from the
National Research Foundation of Korea (no. 2010-0013381 to
D.K.).
References
|
1
|
Wiley SR, Schooley K, Smolak PJ, et al:
Identification and characterization of a new member of the TNF
family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bouralexis S, Findlay DM and Evdokiou A:
Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis.
10:35–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simonet WS, Lacey DL, Dunstan CR, et al:
Osteoprotegerin: a novel secreted protein involved in the
regulation of bone density. Cell. 89:309–319. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Almasan A and Ashkenazi A: Apo2L/TRAIL:
apoptosis signaling, biology, and potential for cancer therapy.
Cytokine Growth Factor Rev. 14:337–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zauli G and Secchiero P: The role of the
TRAIL/TRAIL receptors system in hematopoiesis and endothelial cell
biology. Cytokine Growth Factor Rev. 17:245–257. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ashkenazi A, Pai RC, Fong S, et al: Safety
and antitumor activity of recombinant soluble Apo2 ligand. J Clin
Invest. 104:155–162. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiang H, nguyen CB, Kelley SK, Dybdal N
and Escandon E: Tissue distribution, stability, and
pharmacokinetics of Apo2 ligand/tumor necrosis factor-related
apoptosis-inducing ligand in human colon carcinoma COLO205
tumor-bearing nude mice. Drug Metab Dispos. 32:1230–1238. 2004.
View Article : Google Scholar
|
|
8
|
Johnstone RW, Frew AJ and Smyth MJ: The
TRAIL apoptotic pathway in cancer onset, progression and therapy.
Nat Rev Cancer. 8:782–798. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L and Fang B: Mechanisms of
resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther.
12:228–237. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hopkins-Donaldson S, Ziegler A, Kurtz S,
et al: Silencing of death receptor and caspase-8 expression in
small cell lung carcinoma cell lines and tumors by DNA methylation.
Cell Death Differ. 10:356–364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim K, Fisher MJ, Xu SQ and el-Deiry WS:
Molecular determinants of response to TRAIL in killing of normal
and cancer cells. Clin Cancer Res. 6:335–346. 2000.PubMed/NCBI
|
|
12
|
Ozoren N, Fisher MJ, Kim K, et al:
Homozygous deletion of the death receptor DR4 gene in a
nasopharyngeal cancer cell line is associated with TRAIL
resistance. Int J Oncol. 16:917–925. 2000.PubMed/NCBI
|
|
13
|
Wajant H, Pfizenmaier K and Scheurich P:
TNF-related apoptosis inducing ligand (TRAIL) and its receptors in
tumor surveillance and cancer therapy. Apoptosis. 7:449–459. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sanlioglu AD, Dirice E, Aydin C, Erin N,
Koksoy S and Sanlioglu S: Surface TRAIL decoy receptor-4 expression
is correlated with TRAIL resistance in MCF7 breast cancer cells.
BMC Cancer. 5:542005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cummins JM, Kohli M, Rago C, Kinzler KW,
Vogelstein B and Bunz F: X-linked inhibitor of apoptosis protein
(XIAP) is a nonredundant modulator of tumor necrosis factor-related
apoptosis-inducing ligand (TRAIL)-mediated apoptosis in human
cancer cells. Cancer Res. 64:3006–3008. 2004. View Article : Google Scholar
|
|
16
|
Nam SY, Jung GA, Hur GC, et al:
Upregulation of FLIP(S) by Akt, a possible inhibition mechanism of
TRAIL-induced apoptosis in human gastric cancers. Cancer Sci.
94:1066–1073. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun SY, Yue P, Zhou JY, et al:
Overexpression of Bcl2 blocks TNF-related apoptosis-inducing ligand
(TRAIL)-induced apoptosis in human lung cancer cells. Biochem
Biophys Res Commun. 280:788–797. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JJ, Knudsen S, Mazin W, Dahlgaard J
and Zhang B: A 71-gene signature of TRAIL sensitivity in cancer
cells. Mol Cancer Ther. 11:34–44. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mahalingam D, Szegezdi E, Keane M, de Jong
S and Samali A: TRAIL receptor signalling and modulation: are we on
the right TRAIL? Cancer Treat Rev. 35:280–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cervantes A, Rosello S, Roda D and
Rodriguez-Braun E: The treatment of advanced gastric cancer:
current strategies and future perspectives. Ann Oncol. 19(Suppl 5):
v103–v107. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoong J, Michael M and Leong T: Targeted
therapies for gastric cancer: current status. Drugs. 71:1367–1384.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qiao L and Wong BC: Targeting apoptosis as
an approach for gastrointestinal cancer therapy. Drug Resist Updat.
12:55–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Belkhiri A, Zhu S, Chen Z, Soutto M and
El-Rifai W: Resistance to TRAIL Is Mediated by DARPP-32 in Gastric
Cancer. Clin Cancer Res. 18:3889–3900. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu J, Zhao M, Teng Y, et al:
Interferon-alpha sensitizes human gastric cancer cells to
TRAIL-induced apoptosis via activation of the c-CBL-dependent
MAPK/ERK pathway. Cancer Biol Ther. 12:494–502. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu J, Qu XJ, Xu L, et al: Bortezomib
synergizes TRAIL-induced apoptosis in gastric cancer cells. Dig Dis
Sci. 55:3361–3368. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huynh KM, Soh JW, Dash R, Sarkar D, Fisher
PB and Kang D: FOXM1 expression mediates growth suppression during
terminal differentiation of HO-1 human metastatic melanoma cells. J
Cell Physiol. 226:194–204. 2011. View Article : Google Scholar
|
|
28
|
Uno K, Inukai T, Kayagaki N, et al:
TNF-related apoptosis-inducing ligand (TRAIL) frequently induces
apoptosis in Philadelphia chromosome-positive leukemia cells.
Blood. 101:3658–3667. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hornle M, Peters N, Thayaparasingham B,
Vorsmann H, Kashkar H and Kulms D: Caspase-3 cleaves XIAP in a
positive feedback loop to sensitize melanoma cells to TRAIL-induced
apoptosis. Oncogene. 30:575–587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park JG, Frucht H, LaRocca RV, et al:
Characteristics of cell lines established from human gastric
carcinoma. Cancer Res. 50:2773–2780. 1990.PubMed/NCBI
|
|
31
|
Park JG, Yang HK, Kim WH, et al:
Establishment and characterization of human gastric carcinoma cell
lines. Int J Cancer. 70:443–449. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Koyama S, Koike N and Adachi S: Expression
of TNF-related apoptosis-inducing ligand (TRAIL) and its receptors
in gastric carcinoma and tumor-infiltrating lymphocytes: a possible
mechanism of immune evasion of the tumor. J Cancer Res Clin Oncol.
128:73–79. 2002. View Article : Google Scholar
|
|
33
|
Lee KH, Lim SW, Kim HG, et al: Lack of
death receptor 4 (DR4) expression through gene promoter methylation
in gastric carcinoma. Langenbecks Arch Surg. 394:661–670. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park WS, Lee JH, Shin MS, et al:
Inactivating mutations of KILLER/DR5 gene in gastric cancers.
Gastroenterology. 121:1219–1225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fisher MJ, Virmani AK, Wu L, et al:
Nucleotide substitution in the ectodomain of trail receptor DR4 is
associated with lung cancer and head and neck cancer. Clin Cancer
Res. 7:1688–1697. 2001.PubMed/NCBI
|
|
36
|
Lemke J, Noack A, Adam D, et al: TRAIL
signaling is mediated by DR4 in pancreatic tumor cells despite the
expression of functional DR5. J Mol Med. 88:729–740. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Szliszka E, Mazur B, Zydowicz G, Czuba ZP
and Krol W: TRAIL-induced apoptosis and expression of death
receptor TRAIL-R1 and TRAIL-R2 in bladder cancer cells. Folia
Histochem Cytobiol. 47:579–585. 2009.PubMed/NCBI
|
|
38
|
Aggarwal BB, Bhardwaj U and Takada Y:
Regulation of TRAIL-induced apoptosis by ectopic expression of
antiapoptotic factors. Vitam Horm. 67:453–483. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goncharenko-Khaider N, Lane D, Matte I,
Rancourt C and Piche A: The inhibition of Bid expression by Akt
leads to resistance to TRAIL-induced apoptosis in ovarian cancer
cells. Oncogene. 29:5523–5536. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seal S, Hockenbery DM, Spaulding EY, Kiem
HP, Abbassi N and Deeg HJ: Differential responses of FLIPLong and
FLIPShort-overexpressing human myeloid leukemia cells to TNF-alpha
and TRAIL-initiated apoptotic signals. Exp Hematol. 36:1660–1672.
2008. View Article : Google Scholar
|
|
41
|
Zhang XD, Franco A, Myers K, Gray C,
Nguyen T and Hersey P: Relation of TNF-related apoptosis-inducing
ligand (TRAIL) receptor and FLICE-inhibitory protein expression to
TRAIL-induced apoptosis of melanoma. Cancer Res. 59:2747–2753.
1999.PubMed/NCBI
|



















