Introduction
Colorectal cancer is the third most commonly
diagnosed cancer and the second leading cause of cancer-related
death in the Western world (1,2).
Patients with early stage disease generally have an excellent
prognosis after curative resection, while the prognosis for
patients with distant metastases remains poor (3). Research has demonstrated that
epithelial-mesenchymal transition (EMT) plays a key role in the
early process of the metastasis of cancer cells. EMT has been
implicated in the development of invasive and metastatic tumor
cells during tumor progression (4–6). This
process involves the acquisition of the expression of mesenchymal
molecules, such as vimentin and fibronectin, together with the loss
of epithelial cell adhesion molecules such as E-cadherin (7).
CD44 is a cell adhesion molecule, which belong to a
family of hyaluronan binding proteins. The smallest and most
abundant isoform of CD44 is the so-called standard form (CD44s).
The different isoforms are mainly generated by alternative splicing
of 10 variant exons that account for sequences located in the
extracellular part of the CD44 protein (8). Recently, CD44s has been considered to
be one of the cancer stem cell markers in many solid tumors,
including colorectal cancer (9,10).
CD44s can contribute to the activation of stem cell regulatory
genes and can be a target of these genes (11). However, clinically, the value of
CD44v6 rather than CD44s has been reported by many investigators as
an immunohistochemical prognostic markers in colorectal cancer.
Increased expression of CD44v6 has been reported in lymph node
metastases, linked to adverse prognosis independent of Dukes’ stage
and UICC stage (12–16).
HGF-mediated activation of the c-Met tyrosine kinase
pathway induces the proliferation, motility, adhesion and invasion
of colon cancer cells (17–21). The HGF/c-Met pathway can contribute
to EMT, including malignant tumor progression (22,23).
The activation of c-Met and the downstream signaling pathway to
extracellular signal-regulated kinase (Erk) in response to HGF
requires the presence of the CD44v6-containing isoform (24,25).
A recent study reported that the hyaluronan-CD44
interaction plays a key role in EMT-associated fibrotic disorder
(26). With regard to cancer
progression and metastasis, the relevance of CD44s and/or CD44v6 to
EMT is still poorly understood. In this study, we investigated
whether CD44s and CD44v6 expression is associated with the EMT
phenomenon in patients with colorectal cancer using
immunohistochemistry and in vitro analysis.
Materials and methods
Patients and tissue samples
Among 166 patients with colorectal cancer who
underwent curative surgery between 2000 and 2007 at the Department
of Gastroenterological Surgery of the Kumamoto University Hospital,
Kumamoto, Japan, we selected 113 patients diagnosed with stage II
and III disease, according to the 7th edition of the UICC
classification (27). None of the
patients had undergone preoperative chemotherapy. Tissue specimens
were collected from the patients after informed consent had been
obtained, in accordance with the institutional guidelines of our
hospital.
Cell lines and reagents
The human colon carcinoma cell line, LoVo, was
purchased from Riken Bioresource Center (Osaka, Japan) and was
cultured in Ham’s F12 medium (Wako, Osaka, Japan). The HCT116 cell
line was purchased from the ATCC (Manassas, VA, USA) and cultured
in RPMI medium (Wako). All media were supplemented with 10% fetal
bovine serum (Gibco, Tokyo, Japan), penicillin (100 units/ml), and
streptomycin (100 μg/ml). All cells were incubated at 37°C in a
humidified chamber supplemented with 5% CO2. The
anti-CD44s (clone, SFF-304) and CD44v6 (clone, VFF-18) antibodies
were purchased from Bender MedSystems (Vienna, Austria). The
antibodies against E-cadherin and fibronectin were purchased from
BD-Biosciences (San Jose, CA, USA). The anti-vimentin antibody was
purchased from Santa Cruz Biotechnology (Heidelberg, Germany). The
anti-β-actin antibody was purchased from Cell Signaling Technology
(Tokyo, Japan).
Immunohistochemical staining
The immunohistochemical procedure was performed as
previously described (28). In
brief, after deparaffinization and rehydration of 4-μm sections,
the endogenous peroxidase activity of the specimens was blocked
with a methanol solution containing 3% hydrogen peroxide for 10 min
at room temperature. Heat-induced antigen retrieval by microwave
pretreatment in citrate buffer solution at pH 6.0 (for E-cadherin)
or pH 9.0 (for CD44s, CD44v6 and vimentin) for 5–20 min was
performed. Samples were incubated with the primary antibodies
overnight at 4°C at the dilutions noted below. Anti-CD44s was used
at a 1:100 dilution; anti-CD44v6 was used at 1:500; anti-E-cadherin
was used at 1:1,200 and anti-vimentin was used at 1:50. A
subsequent reaction was performed with the EnVision Plus detection
system (Dako Co., Tokyo, Japan). A positive reaction was visualized
with a diaminobenzidine solution, followed by counterstaining with
Mayer’s hematoxylin.
Evaluation and scoring
We randomly selected 5 fields within the tumor
invasive front under high power magnification (x400) for
evaluation. Each molecule expressed on cancer cells was quantified
as a percentage of the total number of stained cells. For the
expression of CD44s and CD44v6, we applied a three-grade scoring
system of: i) strong, ii) moderate, or iii) weak/none. Strong
staining was defined as staining in >25% of the tumor cells,
moderate staining was indicated when <25% of the cells were
stained, and staining in <10% of the tumor cells or an absence
of staining was scored as weak or none (13,15).
Both CD44s and CD44v6 were considered to be highly expressed when
the staining was strong. For the expression of E-cadherin and
vimentin, the median value of staining was determined to be the
cut-off value. The immunostaining results were independently
evaluated by two investigators who were blinded with respect to the
clinical and histopathologic features.
Invasion assay
Cell invasion was assessed using the Matrigel
invasion chamber (BD Biosciences, San Jose, CA, USA) as previously
described (28). Cells
(1×105/well) transfected with either negative-control
siRNA (200 nM) or CD44v6 siRNA (200 nM) were plated on Transwell
chambers precoated with Matrigel in a 24-well plate. After the
cells were incubated for 24 h at 37°C in a humidified incubator
with 5% CO2, the non-invading cells were removed with
cotton swabs. The cells that had invaded through the membrane were
fixed in 100% methanol and stained with toluidine blue. In five
randomly selected fields, the number of invading cells was counted
under a light microscope. Each experiment was performed in
triplicate.
Wound healing assay
The migration activity was determined using the
wound healing assay. A suspension of LoVo cells
(2.5×105/well) was poured into each well using a 12-well
plate. After 24 h of incubation, the cells were transfected with
siRNA and grown until subconfluence. The confluent cell layer was
scratched with a pipette tip, followed by medium replacement with
or without 50 ng/ml recombinant HGF (R&D Systems, Minneapolis,
MN, USA). The wound distances were measured and averaged from 5
points per wound area as a baseline width. After 24 h, the width of
the mean wound distance was calculated. To evaluate the ‘wound
closure’, five randomly selected points along each wound were
marked, and the horizontal distance the migrating cells traveled
into the wound was measured. The data are reported as the means ±
SD.
RNA extraction, cDNA synthesis and
quantitative RT-PCR assay
Total RNA was obtained from the cell lines using the
RNeasy Mini kit (Qiagen, Tokyo, Japan), according to the
manufacturer’s instructions. cDNA was synthesized with the
SuperScript III Transcriptor First Strand cDNA Synthesis system for
RT-PCR (Invitrogen, Tokyo, Japan), according to the manufacturer’s
instructions. Quantitative reverse transcription PCR (qRT-PCR) was
performed using a LightCycler 480 II system (Roche Diagnostics,
Tokyo, Japan) as previously described (29). To perform qPCR, primers were
designed using the Roche Webpage and the Universal Probe Library
following the manufacturer’s recommendations. The primers used were
as follows: CD44v6 forward, 5′-AACAGCTACCCA GAAGGAACAG-3′; CD44v6
reverse, 5′-CTTTGGGTGTTT GGCGATA-3′; and universal probe #145;
GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′; GAPDH reverse, 5′-GC
CCAATACGACCAAATCC-3′; and universal probe #60. For amplification,
an initial denaturation at 95°C for 10 min was followed by 15 sec
at 95°C, 15 sec at 60°C, and 13 sec at 72°C. All experiments were
performed two times to confirm their reproducibility.
Western blot analysis
For isolating the proteins, cells harvested in
6-well plates were washed once in PBS and lysed in lysis buffer
[Tris-HCl (pH 7.4), 25 mmol/l; NaCl, 100 mmol/l; EDTA, 2 mmol/l;
Triton X-100, 1%; with 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1
mmol/l Na3VO4, 1 mmol/l phenylmethylsulfonyl
fluoride]. Equal amounts of proteins were loaded onto 10% gels and
were separated by SDS-PAGE. The resolved proteins were
electrophoretically transferred to polyvinylidene fluoride (PVDF)
membranes (Bio-Rad Laboratories, Hercules, CA, USA). The membranes
were blocked with 5% low-fat dry milk in TBS-T [25 mM Tris (pH
7.4), 125 mM NaCl, 0.4% Tween 20] for 90 min at room temperature,
followed by incubation with the primary antibody at 4°C overnight.
The primary antibodies for CD44v6 (1:1,000), E-cadherin (1:2,000),
and vimentin (1:1,000) were the same as used for
immunohistochemistry. Primary antibody for fibronectin was used at
1:1,000. The membranes were extensively washed and incubated with a
1:2,000 dilution of HRP-conjugated secondary antibody (Santa Cruz
Biotechnology) for 1 h at room temperature. The membranes were
washed and visualized using the chemiluminescence detection reagent
kit (ECL Plus; GE Healthcare Corp., Tokyo, Japan).
Small interfering RNA transfection
Two different sequences of small interfering RNA
(siRNA) targeting human CD44v6 and negative control siRNA were
purchased from Qiagen (Tokyo, Japan). The sequences of CD44v6 were
as follows: CD44v6#1 sense, 5′-GGCAACUCCUAGUAGUACATT-3′ and
antisense, 5′-UGUACUACUAGGAGUUGCCTG-3′; CD44v6#2 sense,
5′-GAAGACUCCCAUUCGACAATT-3′ and antisense,
5′-UUGUCGAAUGGGAGUCUUCTT-3′. The cells were transfected with the
CD44v6-targeting siRNA or non-silencing siRNA using Lipofectamine
2000 reagent (Qiagen) according to the manufacturer’s protocol.
Statistical analyses
Statistical analyses were performed using the
StatView 5.0 software program (SAS Institute, Inc., Cary, NC, USA).
The progression-free survival (PFS) and overall survival (OS) were
calculated using the Kaplan-Meier method and the log-rank test was
used to determine the statistical significance. A Cox
proportional-hazards model was used to assess the risk ratio with
simultaneous contribution from several covariates. Associations
among discrete variables were assessed using the χ2
test. Mean values were compared using the Student’s t-test. We
analyzed the data by an ANOVA, followed by a Tukey-Kramer post-hoc
test to compare multiple samples. P-values <0.05 were considered
to indicate a statistically significant result.
Results
Relevance of CD44v6 and CD44s to EMT
markers in patients with colorectal cancer
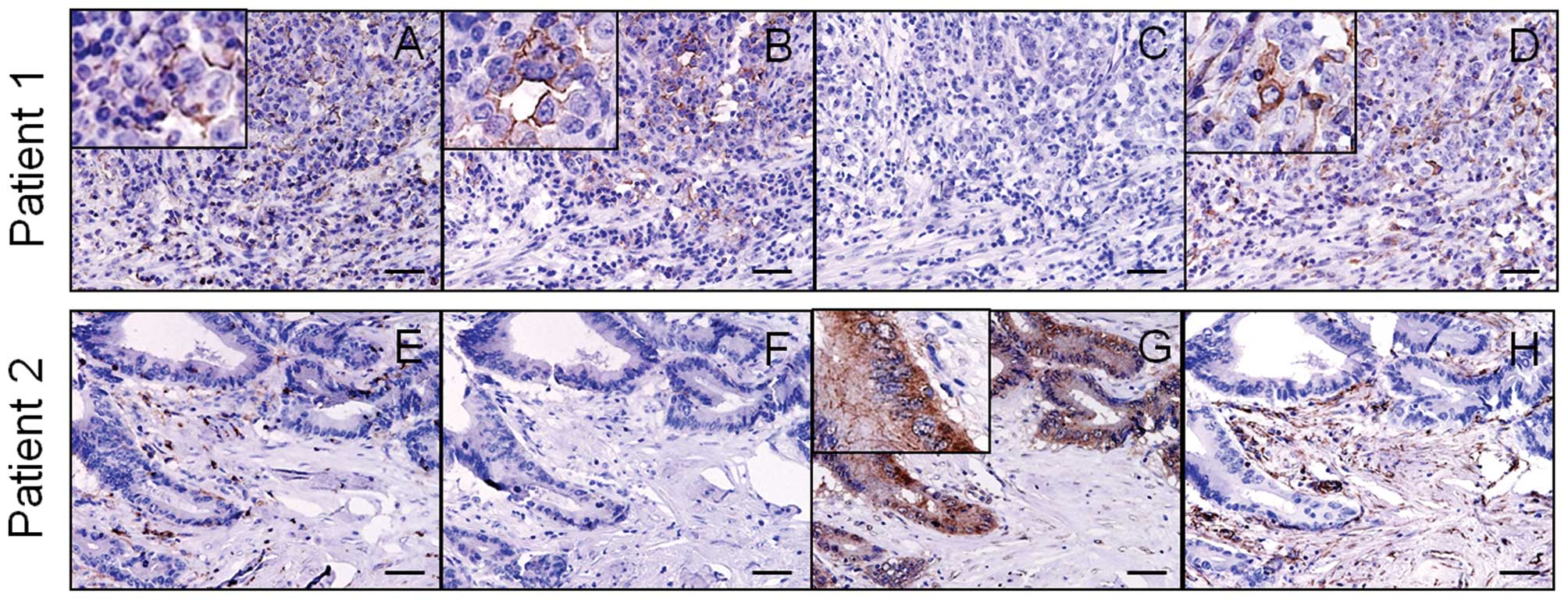
The expression of CD44s, CD44v6 and E-cadherin was
observed on the membrane of cancer cells, while vimentin expression
was observed in the cytoplasm. Although CD44s expression was noted
in both cancer and stromal cells, such as fibroblasts and immune
cells, CD44v6 expression was localized to the cancer cells. CD44v6
expression showed a significant inverse correlation with E-cadherin
expression (P=0.0007) and a positive correlation with vimentin
expression (P=0.0096) (Table I),
and the results from two representative patients are shown in
Fig. 1A–H. In contrast, CD44s
expression showed an inverse correlation with vimentin expression
(P=0.031), and no correlation with E-cadherin expression (data not
shown). High CD44s expression was not associated with a poor
prognosis (data not shown). Based on these results, we examined the
role of CD44v6 in EMT of colon cancer.
 | Table IExpression levels of E-cadherin and
vimentin compared to that of CD44v6. |
Table I
Expression levels of E-cadherin and
vimentin compared to that of CD44v6.
| | CD44v6 | |
|---|
| |
| |
|---|
| Molecule | Total (n=113) | High (n=38) | Low (n=75) | P-value |
|---|
| E-cadherin |
| High | 55 | 10 | 45 | 0.0007 |
| Low | 58 | 28 | 30 | |
| Vimentin |
| High | 55 | 25 | 30 | 0.0096 |
| Low | 58 | 13 | 45 | |
Clinicopathological features of patients
and the impact of CD44v6 expression on the prognosis of stage II or
III colorectal cancer
Table II shows the
association between CD44v6 expression and the clinicopathological
features of the 113 patients. A high level of CD44v6 expression was
inversely correlated with histological differentiation of the tumor
(P=0.048). To examine the prognostic value of CD44v6 expression,
univariate and multivariate analyses were carried out (Table III). High CD44v6 expression was
found to be an independent poor prognostic factor in disease-free
survival (DFS) and overall survival (OS) (data not shown).
Kaplan-Meier curves of the DFS and OS determined based on CD44v6
expression are shown in Fig. 1I and
J (P=0.03 and P=0.047, respectively).
 | Table IICorrelation between the CD44v6
expression pattern and clinicopathological factors of the
colorectal cancer patients. |
Table II
Correlation between the CD44v6
expression pattern and clinicopathological factors of the
colorectal cancer patients.
| | CD44v6 | |
|---|
| |
| |
|---|
| Clinicopathological
factors | Total (n=113) | High (n=38) | Low (n=75) | P-value |
|---|
| Age (years)a |
| ≤68 | 54 | 14 | 40 | 0.10 |
| >68 | 59 | 24 | 35 | |
| Gender |
| Male | 62 | 20 | 42 | 0.73 |
| Female | 51 | 18 | 33 | |
| Location of primary
tumor |
| Colon | 78 | 27 | 51 | 0.74 |
| Rectum | 35 | 11 | 24 | |
| Histological
type |
| Well | 68 | 18 | 50 | 0.048 |
| Other | 45 | 20 | 25 | |
| Tumor size
(mm)a |
| ≤50 | 54 | 16 | 38 | 0.39 |
| >50 | 59 | 22 | 37 | |
| Lymphatic
invasionb |
| Negative | 90 | 28 | 62 | 0.26 |
| Positive | 23 | 10 | 13 | |
| Venous
invasionb |
| Negative | 49 | 19 | 30 | 0.31 |
| Positive | 64 | 19 | 45 | |
| pT stage |
| T1/T2 | 4 | 1 | 3 | 0.71 |
| T3/T4 | 109 | 37 | 72 | |
| pN stage |
| N0 | 68 | 21 | 47 | 0.45 |
| N1/N2 | 45 | 17 | 28 | |
 | Table IIIResults of the univariate and
multivariate analyses for disease-free survival. |
Table III
Results of the univariate and
multivariate analyses for disease-free survival.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
| Clinicopathological
factors | Total (n=113) | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years)a |
| ≤68 | 54 | 1.00 | 0.55–2.52 | 0.67 | 1.00 | 0.74–4.11 | 0.21 |
| >68 | 59 | 1.18 | | | 1.74 | | |
| Gender |
| Male | 62 | 1.00 | 0.49–2.21 | 0.92 | 1.52 | 0.61–3.78 | 0.37 |
| Female | 51 | 1.04 | | | 1.00 | | |
| Location of primary
tumor |
| Colon | 78 | 1.00 | 0.90–4.11 | 0.09 | 1.00 | 0.60–3.36 | 0.43 |
| Rectum | 35 | 1.92 | | | 1.41 | | |
| Tumor size
(mm)a |
| ≤50 | 54 | 1.00 | 0.83–3.98 | 0.13 | 1.00 | 0.68–3.57 | 0.30 |
| >50 | 59 | 1.82 | | | 1.56 | | |
| Histological
type |
| Well | 68 | 1.27 | 0.57–2.82 | 0.56 | 3.03 | 1.09–8.46 | 0.03 |
| Other | 45 | 1.00 | | | 1.00 | | |
| Lymphatic
invasionb |
| Negative | 23 | 1.00 | 0.97–4.81 | 0.06 | 1.00 | 0.98–8.98 | 0.05 |
| Positive | 90 | 2.15 | | | 2.97 | | |
| Venous
invasionb |
| Negative | 64 | 1.00 | 0.63–3.04 | 0.41 | 1.00 | 0.44–2.72 | 0.85 |
| Positive | 49 | 1.39 | | | 1.09 | | |
| Invasion of primary
tumor |
| Negative | 98 | 1.00 | 0.62–4.32 | 0.32 | 1.00 | 0.78–7.16 | 0.13 |
| Positive | 15 | 1.63 | | | 2.36 | | |
| Lymph node
metastasis |
| Negative | 68 | 1.00 | 1.19–5.55 | 0.02 | 1.00 | 1.26–7.54 | 0.01 |
| Positive | 45 | 2.57 | | | 3.08 | | |
| CD44v6 |
| Low | 75 | 1.00 | 1.11–5.04 | 0.03 | 1.00 | 1.31–7.07 | 0.01 |
| High | 38 | 2.37 | | | 3.05 | | |
Knockdown of CD44v6 results in the
downregulation of mesenchymal markers
Since we hypothesized that CD44v6 has a role in the
EMT phenomenon of colon cancer, we assessed whether CD44v6
knockdown impairs EMT in colon cancer cells. We transfected HCT116
and LoVo cells with two different siRNAs against CD44v6, and both
reduced CD44v6 mRNA and protein expression compared with the
control siRNA (Fig. 2A and B). We
used siCD44v6#1 in the subsequent experiments. Knockdown of CD44v6
downregulated the protein expression of vimentin and fibronectin
when compared with the control, while little change was noted in
E-cadherin expression (Fig. 2B).
Vimentin expression in HCT116 cells and fibronectin expression in
LoVo cells were too small to compare.
Downregulation of CD44v6 decreases cancer
cell invasion, migration and HGF-induced cell scattering
The Matrigel invasion assay revealed that the
knockdown of CD44v6 decreased the invasive activity of HCT116 and
LoVo cells (fold-changes were 0.34 and 0.50; P=0.0003 and 0.0001)
(Fig. 2C). In addition, knockdown
of CD44v6 inhibited the migration of HCT116 and LoVo cells
(fold-changes, 0.54 and 0.34; P=0.0085 and 0.0094, respectively)
(Fig. 2D). The knockdown of CD44v6
did not cause any marked change in cell proliferation in either
cell line (data not shown).
Based on previous reports indicating that CD44v6
induces cell scattering through HGF-cMet signaling in certain
cancer cell cultures (24,30), we examined whether CD44v6 mediates
this phenomenon. HGF stimulation of the cancer cells for 48 h led
to an increase in cell migration (4.4-fold, P<0.0001) and CD44v6
knockdown decreased the HGF-mediated cell migration (0.55-fold,
P<0.05) (Fig. 2E) in HCT116
cells. A similar effect was noted in LoVo cells, but the difference
was not significant (data not shown). Taken together, these results
indicate that CD44v6 supports HGF-induced cell scattering.
Discussion
The present study demonstrated that expression of
CD44v6, but not CD44s, was highly correlated with the
downregulation of E-cadherin and upregulation of vimentin
expression in human samples, and CD44v6 supports the mesenchymal
phenotype, such as cellular invasion, migration, HGF-induced cell
scattering, and expression of mesenchymal markers, in colon cancer
cells in vitro. Clinical research using immunohistochemistry
also showed that a high level of CD44v6 expression was an
independent prognostic factor for DFS and OS of stage II/III
colorectal cancer patients following curative resection.
The levels of CD44s and CD44v6 expression in
patients with colorectal cancer using immunohistochemical analysis
remain controversial. Concerning CD44s, several studies have
reported that CD44s expression is associated with an advanced stage
of disease and a poor prognosis (31–34),
whereas other studies found no significant correlation between
CD44s expression and the progression of colorectal cancer patients
(35,36). Furthermore, the absence of CD44
expression in the stromal matrix was reported to be associated with
a poor prognosis (37). Concerning
CD44v6, its increased expression has been associated with poor
prognosis, linked to adverse prognosis independent of Dukes’ and
UICC stages (12–16). However, others have reported that
CD44v6 expression is associated with a favorable prognosis
(38–40). Various clones of antibody, CD44s and
CD44v6, appeared to affect the outcome of their clinical
significance in patients with colorectal cancer. The clinical
outcome and information regarding the antibodies used in previous
reports are listed in Table
IV.
 | Table IVSummary of immunohistochemical-based
studies for CD44v6 in colorectal cancer. |
Table IV
Summary of immunohistochemical-based
studies for CD44v6 in colorectal cancer.
| Authors (ref.) | Year of study | No. of
patients | Stage | Clone no. | Cut-off | Effect of high
CD44v6 expression on survival |
|---|
| Mulder et
al(13) | 1994 | 68 | Dukes’ A–D | VFF4, 7 | (−) <10%, (+)
10–50%, (++) >50% | Adverse effect |
| Wielenga et
al(15) | 1993 | 70 | Dukes’ A–D | VFF4, 7 | 10% | Dukes’ stage |
| Finke et
al(12) | 1995 | 102 | I–IV | VFF7 | (−) 0, (+) ≤20%,
(+) 20–70%, (++) ≥70% | UICC stage |
| Koretz et
al(43) | 1995 | 180 | Dukes’ A–D | VFF7 | 10% | No effect |
| Gotley et
al(42) | 1996 | 109 | Dukes’ A–D | 2F10, 2G9 | | Not done |
| Nanashima et
al(39) | 1999 | 113 | Dukes’ B–D,
metastatic tumor | VFF7 | (−), (+) | Favorable
effect |
| Neumayer et
al(44) | 1999 | 81 | Adenoma,
carcinoma-in-adenoma, T1 | VFF7 | 10% | Not done |
| Nanashima et
al(38) | 2001 | 62 | Liver
metastases | VFF7 | 10% | Favorable
effect |
| Günther et
al(41) | 2002 | 116 | I–III | 2F10 | (−) <10%, (+)
10–50%, (++) >50% | No effect |
| Köbel et
al(16) | 2004 | 145 | I–IV | VFF7 | 10% | Adverse effect |
| Kuhn et
al(45) | 2007 | 170 | I–IV | VFF18 | | No effect |
| Peng et
al(14) | 2008 | 179 | II/III | VFF7 | 10% | Adverse effect |
| Zlobec et
al(40) | 2009 | 1,279 | I–IV, metastatic
tumor | VFF18 | 30% | Favorable
effect |
Although many previous studies concerning CD44v6 in
colon cancer have reported the significance of CD44v6 expression
using immunohistochemistry-based prognostic studies (12–16,38–45),
the effect of CD44v6 expression on EMT is unclear. This study
suggests a molecular mechanism for how CD44v6 expression is linked
to the malignant phenotype. The relationship of CD44v6 and EMT
markers was particularly clear in the invasive front of colorectal
cancer. In addition, we observed that a high level of CD44v6
expression was inversely correlated with cell differentiation. This
result is consistent with previous studies (46,47),
and CD44v6 may be related to the phenotype of poorly differentiated
cancer cells that is defined by a lack of cellular polarity and
regularity.
To confirm these clinical observations, we examined
the role of CD44v6 in colon cancer cells using RNA interference.
The knockdown of CD44v6 decreased cell invasive and migratory
capabilities, and also decreased vimentin expression, but had no
obvious effect on E-cadherin expression in colon cancer cells.
HGF/c-Met signaling and cell scattering were previously reported to
be regulated by CD44v6 expression (24,48),
and we herein showed that CD44v6 expression was related to
HGF-induced cell scattering. When this phenotypic change occurred,
the E-cadherin expression level did not decrease after HGF
treatment (data not shown).
This study was limited with regard to how
CD44v6-expressing cells functionally differ from CD44s-expressing
cells. A recent study indicated that the functional role of CD44s
expression on breast cancer cells differed from that of a variant
isoform in EMT regulation (49).
Variant-specific functional roles have been identified not only for
CD44v6, but also for CD44v3 in colorectal cancer (21,50,51).
Although our clinical outcome showed the relevance of CD44v6 to
tumor progression and EMT phenomenon, further investigation is
required to determine how the cellular phenotype is regulated under
various microenvironmental conditions.
In conclusion, the clinical outcome suggests that
CD44v6 but not CD44s is associated with E-cadherin downregulation
and vimentin upregulation and thereby relates to tumor progression.
In vitro analysis supported that CD44v6 may affect the
mesenchymal phenotype of colon cancer cells.
Acknowledgements
We thank Keisuke Miyake, Naoko Yokoyama and Yuko
Taniguchi for their technical assistance.
Abbreviations:
|
EMT
|
epithelial-mesenchymal transition
|
|
siRNA
|
small interfering RNA
|
|
RT-PCR
|
reverse transcription polymerase chain
reaction
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
HGF
|
hepatocyte growth factor
|
|
TGF-β
|
transforming growth factor-β
|
|
TNFα
|
tumor necrosis factor α
|
References
|
1
|
Ferlay J, Autier P, Boniol M, Heanue M,
Colombet M and Boyle P: Estimates of the cancer incidence and
mortality in Europe in 2006. Ann Oncol. 18:581–592. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar
|
|
3
|
Dhar DK, Yoshimura H, Kinukawa N, et al:
Metastatic lymph node size and colorectal cancer prognosis. J Am
Coll Surg. 200:20–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kalluri R: EMT: when epithelial cells
decide to become mesenchymal-like cells. J Clin Invest.
119:1417–1419. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponta H, Sherman L and Herrlich PA: CD44:
from adhesion molecules to signalling regulators. Nat Rev Mol Cell
Biol. 4:33–45. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dalerba P, Dylla SJ, Park IK, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zoller M: CD44: can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Finke LH, Terpe HJ, Zorb C, Haensch W and
Schlag PM: Colorectal cancer prognosis and expression of
exon-v6-containing CD44 proteins. Lancet. 345:5831995. View Article : Google Scholar
|
|
13
|
Mulder JW, Kruyt PM, Sewnath M, et al:
Colorectal cancer prognosis and expression of exon-v6-containing
CD44 proteins. Lancet. 344:1470–1472. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng J, Lu JJ, Zhu J, et al: Prediction of
treatment outcome by CD44v6 after total mesorectal excision in
locally advanced rectal cancer. Cancer J. 14:54–61. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wielenga VJ, Heider KH, Offerhaus GJ, et
al: Expression of CD44 variant proteins in human colorectal cancer
is related to tumor progression. Cancer Res. 53:4754–4756.
1993.PubMed/NCBI
|
|
16
|
Kobel M, Weichert W, Cruwell K, Schmitt
WD, Lautenschlager C and Hauptmann S: Epithelial hyaluronic acid
and CD44v6 are mutually involved in invasion of colorectal
adenocarcinomas and linked to patient prognosis. Virchows Arch.
445:456–464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kammula US, Kuntz EJ, Francone TD, et al:
Molecular co-expression of the c-Met oncogene and hepatocyte growth
factor in primary colon cancer predicts tumor stage and clinical
outcome. Cancer Lett. 248:219–228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang WG, Lloyds D, Puntis MC, Nakamura T
and Hallett MB: Regulation of spreading and growth of colon cancer
cells by hepatocyte growth factor. Clin Exp Metastasis. 11:235–242.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uchiyama A, Essner R, Doi F, et al:
Interleukin 4 inhibits hepatocyte growth factor-induced invasion
and migration of colon carcinomas. J Cell Biochem. 62:443–453.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Renzo MF, Olivero M, Giacomini A, et
al: Overexpression and amplification of the met/HGF receptor gene
during the progression of colorectal cancer. Clin Cancer Res.
1:147–154. 1995.PubMed/NCBI
|
|
21
|
Wielenga VJ, van der Voort R, Taher TE, et
al: Expression of c-Met and heparan-sulfate proteoglycan forms of
CD44 in colorectal cancer. Am J Pathol. 157:1563–1573. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weidner KM, Sachs M and Birchmeier W: The
Met receptor tyrosine kinase transduces motility, proliferation,
and morphogenic signals of scatter factor/hepatocyte growth factor
in epithelial cells. J Cell Biol. 121:145–154. 1993. View Article : Google Scholar
|
|
23
|
Bladt F, Riethmacher D, Isenmann S, Aguzzi
A and Birchmeier C: Essential role for the c-met receptor in the
migration of myogenic precursor cells into the limb bud. Nature.
376:768–771. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Orian-Rousseau V, Chen L, Sleeman JP,
Herrlich P and Ponta H: CD44 is required for two consecutive steps
in HGF/c-Met signaling. Genes Dev. 16:3074–3086. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van der Voort R, Taher TE, Wielenga VJ, et
al: Heparan sulfate-modified CD44 promotes hepatocyte growth
factor/scatter factor-induced signal transduction through the
receptor tyrosine kinase c-Met. J Biol Chem. 274:6499–6506.
1999.
|
|
26
|
Takahashi E, Nagano O, Ishimoto T, et al:
Tumor necrosis factor-alpha regulates transforming growth
factor-beta-dependent epithelial-mesenchymal transition by
promoting hyaluronan-CD44-moesin interaction. J Biol Chem.
285:4060–4073. 2010. View Article : Google Scholar
|
|
27
|
Edge SB, Byrd BD and Compton CC: AJCC
Cancer Staging Manual. 7th edition. Springer; New York, NY:
2010
|
|
28
|
Okabe H, Beppu T, Hayashi H, et al:
Hepatic stellate cells may relate to progression of intrahepatic
cholangiocarcinoma. Ann Surg Oncol. 16:2555–2564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Okabe H, Beppu T, Ueda M, Hayashi H,
Ishiko T, Masuda T, Otao R, Horlad H, Mima K, Miyake K, Iwatsuki M,
Baba Y, Takamori H, Jono H, Shinriki S, Ando Y and Baba H:
Identification of CXCL5 involved in the interaction between
cholangiocarcinoma and cancer associated fibroblasts. Int J Cancer.
131:2234–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grotegut S, von Schweinitz D, Christofori
G and Lehembre F: Hepatocyte growth factor induces cell scattering
through MAPK/Egr-1-mediated upregulation of Snail. EMBO J.
25:3534–3545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huh JW, Kim HR, Kim YJ, et al: Expression
of standard CD44 in human colorectal carcinoma: association with
prognosis. Pathol Int. 59:241–246. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ichikawa W: Positive relationship between
expression of CD44 and hepatic metastases in colorectal cancer.
Pathobiology. 62:172–179. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Visca P, Del Nonno F, Botti C, et al: Role
and prognostic significance of CD44s expression in colorectal
cancer. Anticancer Res. 22:2671–2675. 2002.PubMed/NCBI
|
|
34
|
Fernandez JC, Vizoso FJ, Corte MD, et al:
CD44s expression in resectable colorectal carcinomas and
surrounding mucosa. Cancer Invest. 22:878–885. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Heider KH, Hofmann M, Hors E, et al: A
human homologue of the rat metastasis-associated variant of CD44 is
expressed in colorectal carcinomas and adenomatous polyps. J Cell
Biol. 120:227–233. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ylagan LR, Scholes J and Demopoulos R:
CD44: a marker of squamous differentiation in adenosquamous
neoplasms. Arch Pathol Lab Med. 124:212–215. 2000.PubMed/NCBI
|
|
37
|
Furuta K, Zahurak M, Goodman SN, Hamilton
SR and August JT: CD44 expression in the stromal matrix of
colorectal cancer: association with prognosis. Clin Cancer Res.
4:21–29. 1998.PubMed/NCBI
|
|
38
|
Nanashima A, Yamaguchi H, Sawai T, et al:
Prognostic factors in hepatic metastases of colorectal carcinoma:
immunohistochemical analysis of tumor biological factors. Dig Dis
Sci. 46:1623–1628. 2001. View Article : Google Scholar
|
|
39
|
Nanashima A, Yamaguchi H, Sawai T, et al:
Expression of adhesion molecules in hepatic metastases of
colorectal carcinoma: relationship to primary tumours and prognosis
after hepatic resection. J Gastroenterol Hepatol. 14:1004–1009.
1999. View Article : Google Scholar
|
|
40
|
Zlobec I, Gunthert U, Tornillo L, et al:
Systematic assessment of the prognostic impact of membranous CD44v6
protein expression in colorectal cancer. Histopathology.
55:564–575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gunther K, Dworak O, Remke S, et al:
Prediction of distant metastases after curative surgery for rectal
cancer. J Surg Res. 103:68–78. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gotley DC, Fawcett J, Walsh MD, Reeder JA,
Simmons DL and Antalis TM: Alternatively spliced variants of the
cell adhesion molecule CD44 and tumour progression in colorectal
cancer. Br J Cancer. 74:342–351. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koretz K, Moller P, Lehnert T, Hinz U,
Otto HF and Herfarth C: Effect of CD44v6 on survival in colorectal
carcinoma. Lancet. 345:327–328. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Neumayer R, Rosen HR, Reiner A, et al:
CD44 expression in benign and malignant colorectal polyps. Dis
Colon Rectum. 42:50–55. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kuhn S, Koch M, Nubel T, et al: A complex
of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins
promotes colorectal cancer progression. Mol Cancer Res. 5:553–567.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu YJ, Yan PS, Li J and Jia JF:
Expression and significance of CD44s, CD44v6, and nm23 mRNA in
human cancer. World J Gastroenterol. 11:6601–6606. 2005.PubMed/NCBI
|
|
47
|
Bendardaf R, Elzagheid A, Lamlum H,
Ristamaki R, Collan Y and Pyrhonen S: E-cadherin, CD44s and CD44v6
correlate with tumour differentiation in colorectal cancer. Oncol
Rep. 13:831–835. 2005.PubMed/NCBI
|
|
48
|
Tremmel M, Matzke A, Albrecht I, et al: A
CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and
angiogenesis. Blood. 114:5236–5244. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brown RL, Reinke LM, Damerow MS, et al:
CD44 splice isoform switching in human and mouse epithelium is
essential for epithelial-mesenchymal transition and breast cancer
progression. J Clin Invest. 121:1064–1074. 2011. View Article : Google Scholar
|
|
50
|
Kuniyasu H, Oue N, Tsutsumi M, Tahara E
and Yasui W: Heparan sulfate enhances invasion by human colon
carcinoma cell lines through expression of CD44 variant exon 3.
Clin Cancer Res. 7:4067–4072. 2001.PubMed/NCBI
|
|
51
|
Yamaguchi A, Urano T, Goi T, et al:
Expression of a CD44 variant containing exons 8 to 10 is a useful
independent factor for the prediction of prognosis in colorectal
cancer patients. J Clin Oncol. 14:1122–1127. 1996.PubMed/NCBI
|