Introduction
Breast cancer is a significant worldwide health
problem and the most commonly diagnosed neoplasm in women (1). Breast cancer accounts for 1.38 million
new cancer cases and more than 450,000 cancer deaths annually in
the world. Of the breast cancers up to 80% are invasive ductal
breast carcinoma (IDC) (2). IDC has
similar biological features or pathological types, but this
sporadic breast malignancy is triggered by a complex pathogenesis
and multiple gene mutations (3). To
date, the incidence rate of breast cancer has gradually reduced and
the survival rate has increased due to recent advancement in early
detection, prevention and targeted therapy. However, the mechanisms
responsible for breast cancer initiation, recurrence or metastasis
remain to be defined. Thus, new approaches and the discovery of
novel target-based molecules are urgently needed to effectively
control breast cancer and increase the survival of patients.
Interferon-induced transmembrane proteins (IFITM)
are a family of transmembrane proteins that contain two α helical
domains. To date, there are five IFITMs (IFITM1, IFITM2, IFITM3,
IFITM5 and IFITM10) (4–7) that are all clustered on chromosome 11
with less than 2 kb next to each fragment. The main function of
this protein family is to inhibit the invasion of many pathogenic
viruses, such as HIV (8). However,
more recently, they have been found to play a role in tumorigenesis
and cell adhesion and signaling, and are involved in early
embryonic development and the regulation of cell growth (9). Furthermore, they may act as tumor
markers (10,11). Indeed, in a previous study, Wang
et al(12) identified that
the expression of IFITM2 protein was significantly increased in
HER2-overexpressing mouse mammary tumors, while other investigators
proposed that IFITM3 protein may be involved in diverse cellular
processes (7,13,14),
including immune-cell regulation, somitogenesis, germ cell homing
and maturation, cell proliferation (15) and heart development (16). Previous studies demonstrated that
IFITM3 expression was significantly upregulated in colorectal
cancer (10) and played a critical
role in regulating tumor cell migration and invasion, which
contribute to the progression of gastric and head and neck cancers
(17,18).
However, up to date, only a few studies have
reported the role of IFITM3 in breast cancer. Thus, in the present
study, we first detected the expression of IFITM3 protein in
normal, premalignant and invasive breast tissues, and then
investigated the effects of IFITM3 knockdown with lentiviral shRNA
on the regulation of breast cancer cell growth and colony formation
and cell cycle distribution.
Materials and methods
Breast cancer tissue specimens and
clinicopathological features
In the present study, we collected 64 cases of
breast cancer tissue specimens from the Second Hospital of Jilin
University (Jilin, China) between March 2010 and April 2011. These
64 cases were formalin-fixed and paraffin-embedded specimens and
included 15 cases of ductal carcinoma in situ (DCIS) and 49
cases of IDC. Pathology diagnosis of each patient was made by at
least two pathologists and the patients underwent mastectomy at the
Second Hospital of Jilin University. Next, immunohistochemical
analysis of tumor tissues showed that there were 12 triple negative
breast cancers among the 49 IDC cases. Tumor staging for these
specimens was carried out according to the American Joint Committee
on Cancer Staging criteria (19).
The use of human specimens was approved by appropriate
institutional review boards.
Immunohistochemistry
The tissue samples were fixed in 10% neutral
formalin, embedded in paraffin and sectioned at 2 μm. After
de-paraffinization and rehydration, the sections were subjected to
antigen retrieval with a citrate buffer (pH 6.0) in a pressure
cooker. Immunohistochemistry was performed using an IFITM3
polyclonal antibody at a dilution of 1:200 from Proteintech Group,
Inc. (cat. #11714-1-AP; Chicago, IL, USA) at 4°C overnight or a
monoclonal antibody against estrogen receptor (ER), progesterone
receptor (PR) or Her2/neu protein (Dako, Glostrup, Denmark).
Negative controls were incubated with PBS instead of the primary
antibody.
The immunostained sections were independently
evaluated by two pathologists (W.S. and Q.S.S.). Both staining
intensity and percentage of staining were reviewed and scored using
a semi-quantitative grading system: negative and weak, moderate or
strongly positive. These three scores were summarized as negative,
low (weak staining) or high (moderate or strong staining)
expression of the gene.
Cells lines and cell culture
The human breast cancer MDA-MB-231 and MCF-7 cell
lines and the human renal epithelial 293T cell line (ATCC,
Manassas, VA, USA) were maintained at 37°C in a humidified
atmosphere of 95% air and 5% CO2. MDA-MB-231 cells were
cultured in monolayer with Dulbecco’s modified Eagle’s media (DMEM;
Gibco, New York, NY, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco) and 4 m ML-Glutamine (Sigma, St. Louis, MO, USA).
MCF-7 cells was cultured in a monolayer with DMEM containing 4 mM
L-Glutamine, 0.1 mM MEM Non-essential amino acids (Sigma) and 10%
FBS. The 293T cell line was cultured in DMEM supplemented with 10%
FBS, 100 units/ml penicillin and 0.1 mg/ml streptomycin
(Sigma).
Lentiviral plasmid construction and
infection
An shRNA sequence (5′-CCTGTTCAACACCCTCTTCAT-3′) was
used to target IFITM3 (GenBank code: NM_021034) and was purchased
by Neuron Biotech Co., Ltd. (Shanghai, China). IFITM3 RNAi was then
tested for the knockdown efficiency of IFITM3 in vitro. The
small hairpin RNA (shRNA) oligonucleotides were cloned into the
pLKD vector (Neuron Biotech Co.). Similarly, the widely used
negative control (NC) sequence (5′-TTCTCCGAACGTGTCACGT-3′) was also
synthesized and cloned into the pLKD vector as negative control.
These vectors were amplified, the DNA sequence was confirmed, and
then named as pLKD-shIFITM3 and pLKD-NC, respectively. Next, the
lentivirus vector, which contained a green fluorescent protein
(GFP) and a U6 terminator inserted between AgeI and
EcoRI, and pLKD-shIFITM3 or pLKD-NC were co-transfected into
293T cells. The 293T cells already carried the stably transfected
viral envelope and packaging vectors as previously described
(20). The lentiviral particles
were harvested from the media after 48-h transfection and purified
with ultracentrifugation. The virus titer (multiplicity of
infection; MOI) was determined using the plaque-forming assay as
previously described (21,22).
To knock down IFITM3 expression in breast cancer
cell lines, the lentiviruses were used to infect breast cancer
cells (si-CTRL group and si-IFITM3 group). In brief, MDA-MB-231 and
MCF-7 cells were seeded at 50,000 cells/well in 6-well plates with
a small volume of Opti-MEM, grown for 24 h, and then infected with
lentiviruses. The cells were then further incubated with normal
growth medium 24 h after infection, and after 72 h, the infection
was efficiency detected by the GFP in these MDA-MB-231 and MCF-7
cells. Our data showed an ~90% infection rate when a MOI of 15 and
20 was used, and these MOIs did not cause toxicity to the cells.
Finally, the cells were harvested for subsequent experiments.
Quantitative reverse transcription
polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cultured cells five days
after lentiviral infection using a TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s protocol. Two
micrograms of the RNA samples were then reversely transcribed into
cDNA using the protocol of the M-MLV Reverse transcriptase
(Promega, Madison, WI, USA). qPCR was performed with 20 μl of PCR
mixtures that contained 0.5 μl primer each, 10 μl 2X SYBR Premix Ex
Taq (Takara, Osaka, Japan) and 1 μl cDNA sample in TP800 (Takara).
The primer sequences were: IFITM3 (5′-TGTCCAAACCTTCTTCTC-3′ and
5′-CGTCGCCAACCATCTTCC-3′) and β-actin (5′-GGCGGCACCACCATGTACCCT-3′
and 5′-AGGGGCCGGACTCGTCATACT-3′) qPCR amplification conditions were
as follows: an initial denaturation at 95°C for 15 sec and then 45
cycles of 95°C for 5 sec and 60°C for 30 sec. The relative
expression of IFITM3 mRNA was calculated using the
2−ΔΔCT method (23). All
experiments were in duplicate and repeated at least three
times.
Protein extraction and western blot
analysis
After five days of viral infection, cells were
washed with phosphate-buffered saline (PBS; pH 7.4) and lysed with
a lysis buffer [10 mM Tris-HCl/pH 8.0, 85 mM KCl, 0.5% NP-40 plus
protease inhibitors (1 μM each of chymostatin, leupeptin, antipain
and pepstatin-A, and 1 mM each of phenylmethylsulfonyl fluoride and
benzamidine)] and then centrifuged at 4°C for 15 min. These cell
lysates were then separated on 12% sodium dodecyl sulfate
polyacrylamide gels and transferred onto PVDF membranes (Millipore,
Bedford, MA, USA). Next, the membranes were incubated with 5% skim
milk/PBS for 1 h at room temperature. Membranes were then incubated
with a rabbit polyclonal antibody against IFITM3 (1:200, cat.
#sc-130785; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a
mouse monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) antibody (1:1,000, cat. #sc-32233; Santa Cruz
Biotechnology) overnight at 4°C. The next day, the membranes were
washed with PBS-T and were further incubated with appropriate
horseradish peroxidase (HRP) conjugated secondary antibodies at
1:3,000 at room temperature for 2 h. The protein signals were
finally detected using an enhanced chemiluminescence system
(Amersham Life Sciences, Arlington Heights, IL, USA) according to
the protocol provided by the manufacturer.
Flow cytometry assay
Cells were grown in 6-well plates and infected with
lentiviruses for 24 h, and after medium changes, further grown for
4 days. For the flow cytometry assay, 1×106 cells from
each group were harvested by centrifugation at 1,200 rpm for 5 min,
and then pellets were washed with cold PBS, fixed with cold 70%
ethanol for 1 h, centrifuged at 1,500 rpm for 5 min to remove
ethanol and washed with PBS again. After that, DNA of these cells
was stained with propidium iodide (PI) at 4°C for 30 min in the
dark and then subjected to flow cytometric analysis by a flow
cytometer (BD Biosciences, San Jose, CA, USA). Each experiment was
performed in triplicate and repeated at least once.
Cell viability MTT assay
To monitor cell growth, 3 day-lentivirus-infected
cells were seeded into 96-well plates at a density of
2×103 cells in a final volume of 100 μl/well and
incubated at 37°C for up to 120 h. At the end of the experiments,
20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT, Sigma) was added to each well, and then the cells
were incubated for another 4 h. After discarding the growth media,
150 μl of dimethyl sulfoxide was added to each well of these cell
cultures to dissolve MTT. The optical density (OD) was measured at
490 nm using a spectrophotometer (SmartSpec Plus #170-2525; Bio-Rad
Laboratories, Hercules, CA, USA). Each experiment was performed
5-fold and repeated three times.
BrdU incorporation assay
Cells (2×104) were plated into 96-well
tissue culture plates and grown at 37°C for up to 72 h. At the end
of the experiments, bromodeoxyuridine (5-bromo-2′-deoxyuridine,
BrdU) from the BrdU Cell Proliferation Assay kit (cat. #2750;
Millipore) was added to the cell cultures and incubated for
additional 6 h. The cells were fixed with a fixing solution for 30
min and subjected to immunostaining with a monoclonal anti-BrdU
antibody and with the peroxidase conjugated goat anti-mouse IgG.
After adding the TMB (tetramethyl-benzidine) peroxidase substrate
on to the cells, the cells were incubated for 30 min in the dark
and the stop solution was added to terminate the reaction. The
plates were read at 490 nm using a spectrophotometer.
Colony formation assay
After 5 days of lentivirus infection, equal numbers
of breast cancer cells expressing either control shRNA or IFITM3
shRNA were trypsinized, resuspended, seeded at a density of 300/ml
into 35-mm culture plates and incubated at 37°C for 10 days. At the
end of the experiments, the cells were fixed with 4%
paraformaldehyde for 60 min and stained with Giemsa (Sigma) for 10
min. The positive colonies with >50 cells were counted under a
microscope.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
software (SPSS Inc., Chicago, IL, USA). The comparison between two
groups was analyzed by the Student’s t-test or Chi-square test. A
value of P<0.05 was defined to be of statistical significance,
and P<0.01 was defined to be highly statistically
significant.
Results
Overexpression of IFITM3 protein in
invasive breast ductal carcinoma tissue specimens
In the present study, we first detected the
expression of IFITM3 protein in 64 breast cancer tissue specimens,
which included 15 cases of DCIS and 49 cases of IDC, and in paired
adjacent normal breast specimens. Our data showed that IFITM3 was
positively stained in the cytoplasm of the majority of invasive
cancer specimens, whereas IFITM3 was negatively or weakly stained
in the adjacent normal mammary glands (Fig. 1). Of these 15 patients with DCIS,
only 5 (30.0%) were classified as high expression of IFITM3,
whereas 29 of 49 (59.1%) patients with invasive cancer highly
expressed IFITM3 protein, including 5 cases of grade I, 12 cases of
grade II and 12 cases of grade III. Furthermore, we associated the
trend of IFITM3 expression levels with clinicopathological features
from breast cancer patients and found that there was no association
of IFITM3 expression with age, tumor size and lymph node status
(Tables I and II). However, expression of IFITM3 protein
was significantly associated with ER (P<0.01, Table II) and PR status (P<0.05,
Table II).
 | Table IAssociations between IFITN3 expression
and clinicopathological characteristics of patients with breast
cancer in situ (n=15). |
Table I
Associations between IFITN3 expression
and clinicopathological characteristics of patients with breast
cancer in situ (n=15).
| | IFITM3 | | |
|---|
| |
| | |
|---|
| Clinicopathological
characteristics | N | Positive | Negative | χ2
value | P-value |
|---|
| Age at diagnosis
(years) | | | | 0.597 | >0.05 |
| <50 | 10 | 4 | 6 | | |
| ≥50 | 5 | 1 | 4 | | |
| Estrogen receptor
status | | | | 0.17 | >0.05 |
| Positive | 11 | 4 | 7 | | |
| Negative | 4 | 1 | 3 | | |
| Progesterone receptor
status | | | | 0.134 | >0.05 |
| Positive | 8 | 3 | 5 | | |
| Negative | 7 | 2 | 5 | | |
| HER-2 status | | | | 0.536 | >0.05 |
| Positive | 8 | 2 | 6 | | |
| Negative | 7 | 3 | 4 | | |
 | Table IIAssociations between IFITN3
expression and clinicopathological characteristics of patients with
breast invasive ductal carcinoma (n=49). |
Table II
Associations between IFITN3
expression and clinicopathological characteristics of patients with
breast invasive ductal carcinoma (n=49).
| | IFITM3 | | |
|---|
| |
| | |
|---|
| Clinicopathological
characteristics | N | Positive | Negative | χ2
value | P-value |
|---|
| Age at diagnosis
(years) | | | | 3.60 | >0.05 |
| <50 | 24 | 17 | 7 | | |
| ≥50 | 26 | 11 | 14 | | |
| Estrogen receptor
status | | | | 7.77 | <0.01 |
| Positive | 30 | 23 | 7 | | |
| Negative | 19 | 7 | 12 | | |
| Progesterone
receptor status | | | | 3.89 | <0.05 |
| Positive | 23 | 17 | 6 | | |
| Negative | 28 | 12 | 14 | | |
| HER-2 status | | | | 0.00 | >0.05 |
| Positive | 17 | 10 | 7 | | |
| Negative | 32 | 19 | 13 | | |
| Triple
negative | | | | 1.59 | >0.05 |
| Triple
negative | 12 | 7 | 5 | | |
| Non triple
negative | 37 | 12 | 25 | | |
| Lymph node
status | | | | 0.66 | >0.05 |
| Positive | 31 | 14 | 17 | | |
| Negative | 18 | 6 | 12 | | |
| Tumor size
(mm) |
| <20 | 13 | 7 | 6 | 0.21 | >0.05 |
| ≥20 | 36 | 22 | 14 | | |
| Grade | | | | 4.78 | >0.05 |
| I | 7 | 5 | 2 | | |
| II | 17 | 12 | 5 | | |
| III | 25 | 12 | 13 | | |
Lentivirus-delivered shRNA in silencing
IFITM3 expression in breast cancer cells
To determine the role of IFITM3 in breast cancer, we
constructed lentivirus-delivered shRNA vector to silence IFITM3
expression in breast cancer cells. First, we assessed the
efficiency of these lentiviruses in infecting breast cancer cells.
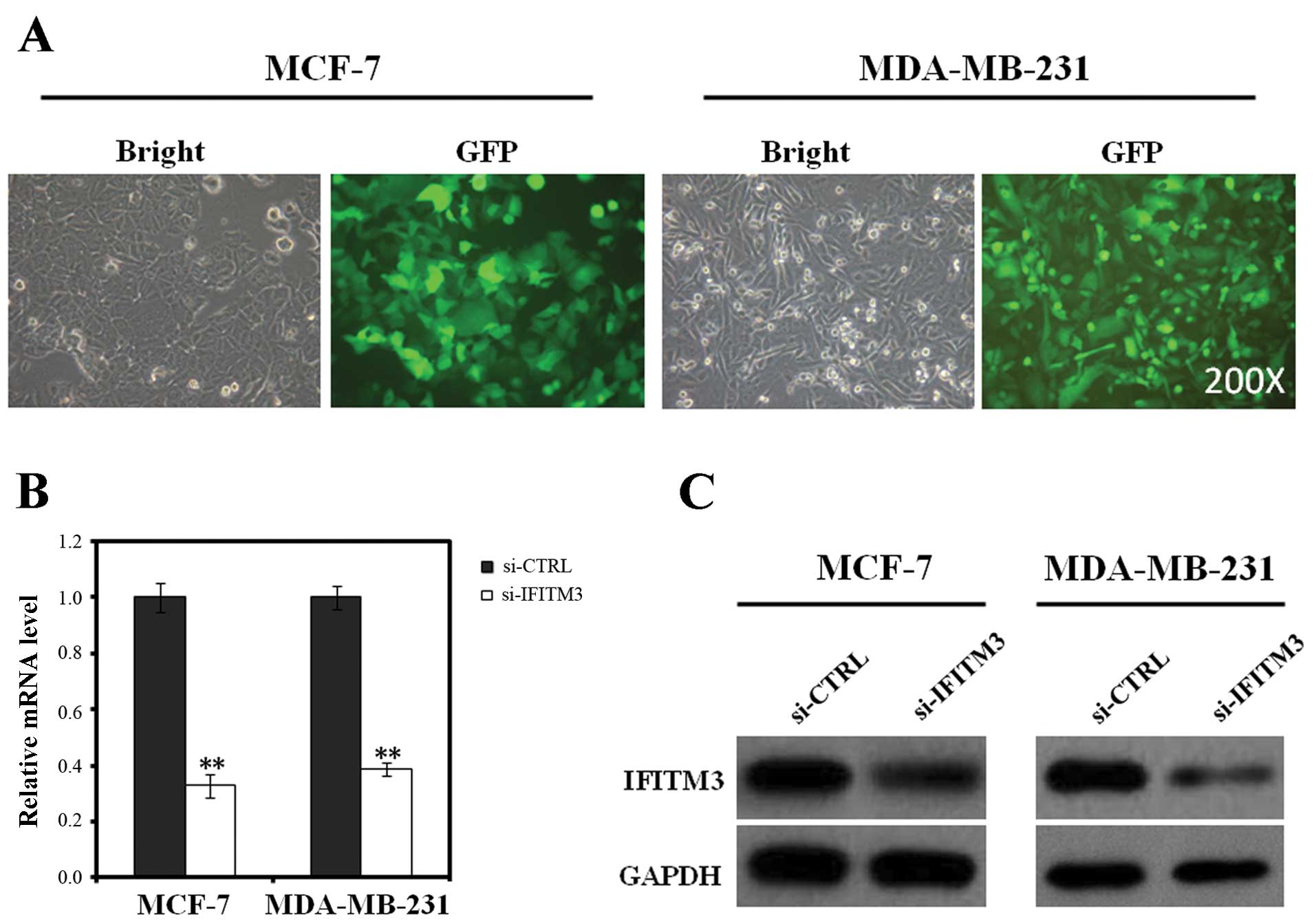
We found that >90% of MCF-7 and MDA-MB-231 cells expressed GFP
72 h after infection (Fig. 2A).
We then assessed the efficiency of lentiviral IFITM3
shRNA in knocking down IFITM3 expression in MCF-7 and MDA-MB-231
cells by using qRT-PCR and western blot analysis. The data showed
that levels of IFITM3 mRNA expression in the si-IFITM3-infected
MCF-7 and MDA-MB-231 cells was dramatically reduced by 67 and 61%,
respectively (P<0.01) compared to the 5-day si-CTRL-infected
cells (Fig. 2B). Similarly, western
blot analysis showed that expression of IFITM3 protein in
si-IFITM3-infected MDA-MB-231 and MCF-7 cells was also much lower
than that of si-CTRL-infected cells (Fig. 2C). These data confirmed that our
lentivirus carrying IFITM3 shRNA was able to efficiently repress
IFITM3 expression in breast cancer cells.
Effects of IFITM3 knockdown on the
regulation of breast cancer cell growth and cell cycle
distribution
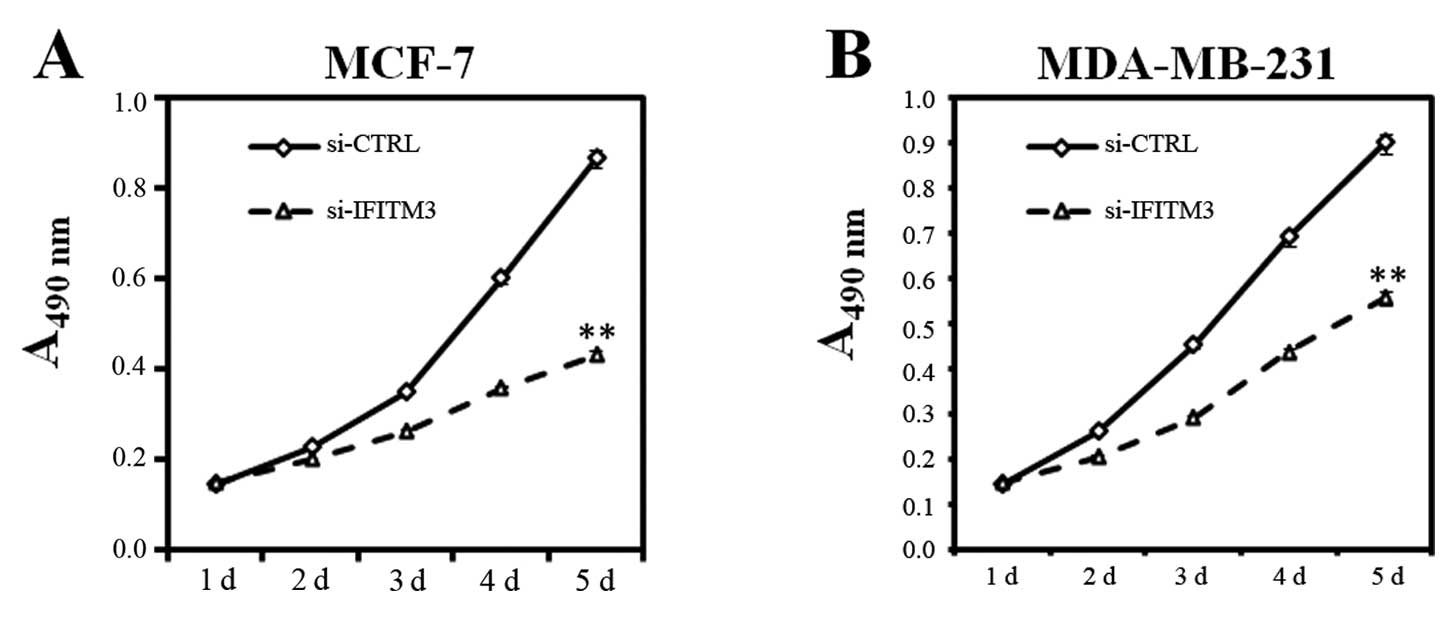
Next, we elucidated whether knockdown of IFITM3
expression exerted cell growth inhibition activity. We first
assayed for MTT, BrdU incorporation, and colony formation in
IFITM3-knocked down MCF-7 and MDA-MB-231 cells. In the MTT assay,
viability of MCF-7 and MDA-MB-231 cells in the si-IFITM3 group
dramatically reduced compared to the si-CTRL group for up to 5 days
(P<0.01, Fig. 3). Similarly, the
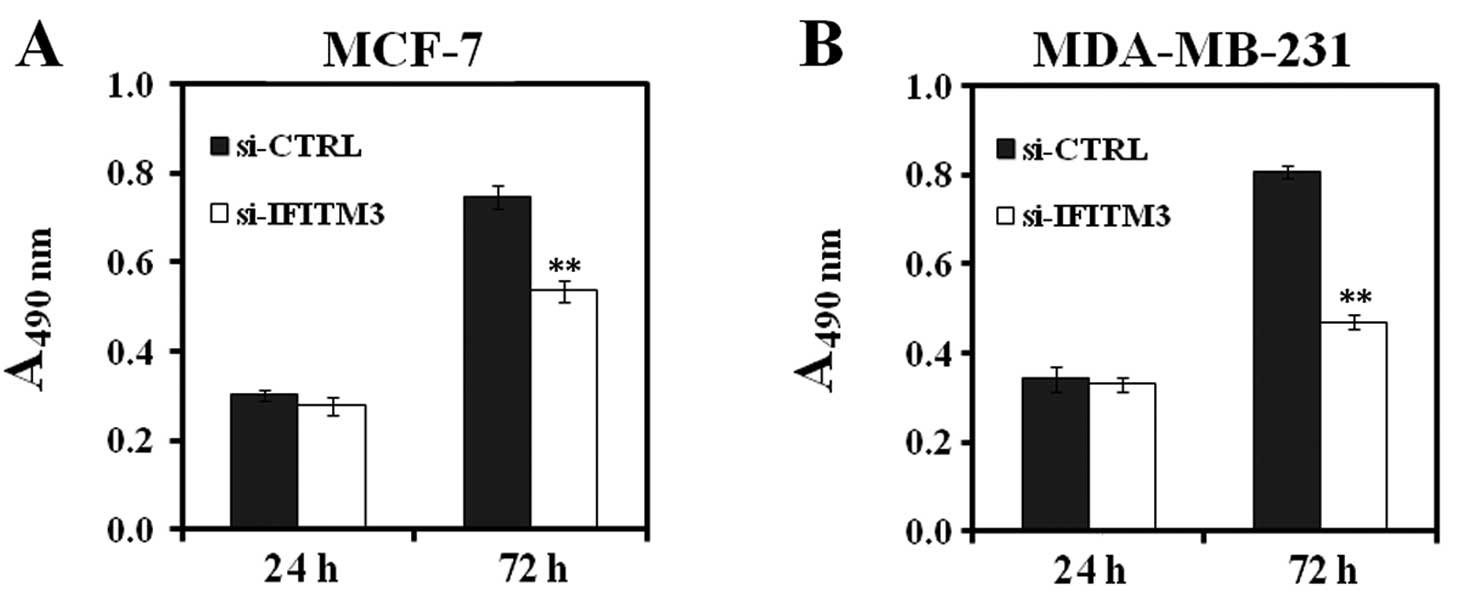
BrdU incorporation assay showed that reduced levels of DNA
synthesis in MCF-7 and MDA-MB-231 cells in the si-IFITM3 group was
detected and decreased by 28 and 41%, respectively, after 72-h
infection, compared to the si-CTRL-infected cells (P<0.01,
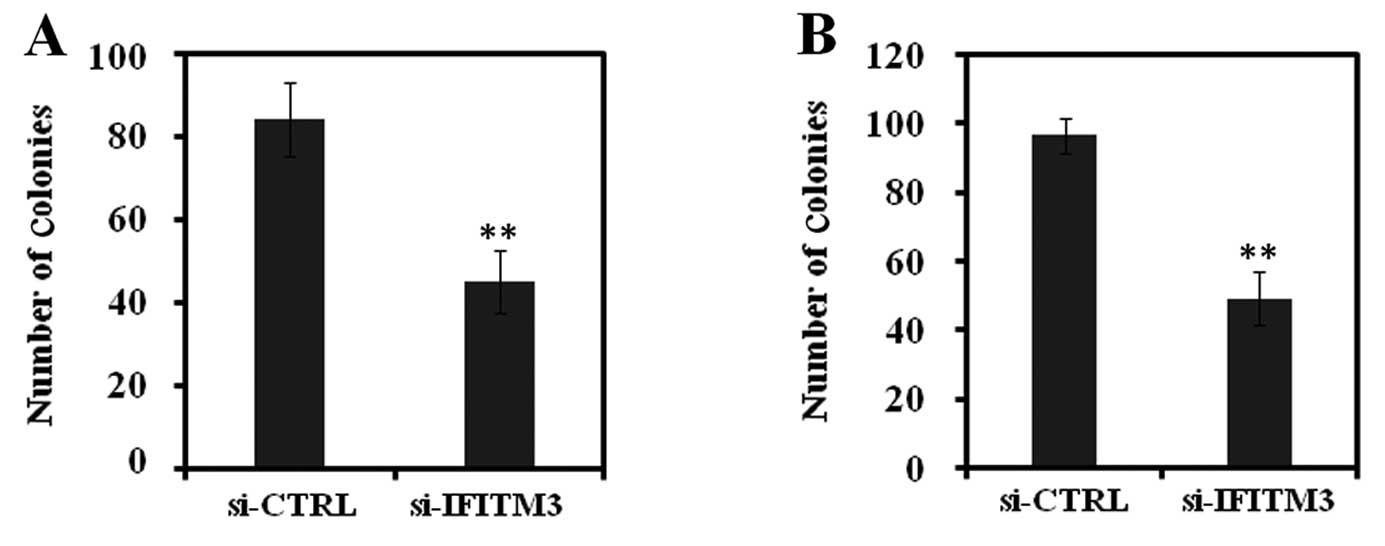
Fig. 4). Similarly, the colony
formation assay also showed that the colony-forming ability of
MCF-7 and MDA-MB-231 cells was pronouncedly suppressed by silencing
IFITM3 expression (Fig. 5). These
data clearly demonstrated that breast cancer cell growth after
si-IFITM3 infection was significantly inhibited compared with that
of the si-CTRL groups.
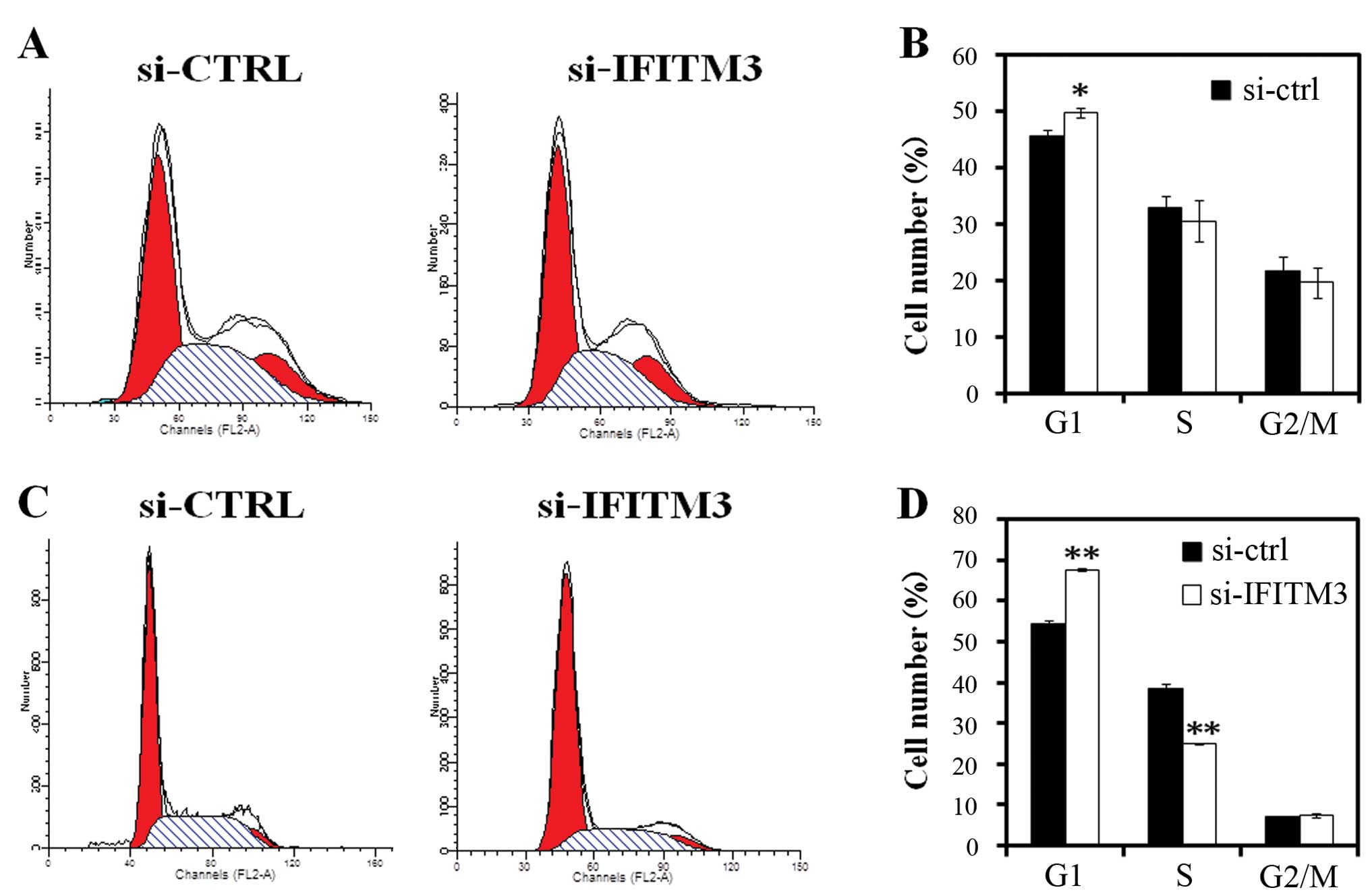
Furthermore, we analyzed the cell cycle distribution
in MCF-7 and MDA-MB-231 cells after knockdown of IFITM3 expression.
The flow cytometry data showed that 5 days after IFITM3 lentivirus
infection, the percentage of MCF-7 cells in the G0/G1 phase of the
cell cycle was higher than that of the si-CTRL group (P<0.05)
(Fig. 6A and B). The data are more
obvious shown in MDA-MB-231 cells (P<0.01) (Fig. 6C and D). Moreover, the number of
cells in the S phase of the cell cycle was lower in the si-IFITM3
group of MDA-MB-231 cells than that of the si-CTRL group cells
(p<0.01) (Fig. 6C and D).
Discussion
In the present study, we first detected the
expression of IFITM3 protein in normal, premalignant, and invasive
breast tissue specimens and found that IFITM3 protein was highly
expressed in invasive breast cancer tissues. We then constructed
and prepared lentiviral-carrying IFITM shRNA to knock down the
IFITM expression in breast cancer cells. Our data showed that
compared to the negative control lentivirus, IFITM shRNA lentivirus
effectively silenced expression of IFITM3 mRNA and protein in two
breast cancer cell lines. Knockdown of IFITM3 expression
significantly inhibited tumor cell viability, growth, and colony
formation and arrested tumor cells at the G1 phase of the cell
cycle, as well as reduced the number of cells in the S phase.
Although the present study is preliminary, the data clearly
indicate that IFITM3 protein plays a role in breast cancer
development and/or progression. Thus, IFITM3 may be a potential
target for the control of breast cancer.
IFITM3 is a protein that offers protection against
numerous viruses in the human body, including influenza A, Dengue
and HIV (8,24,25).
However, Abba et al(26)
recently reported that expression of IFITM3 mRNA was significantly
upregulated in invasive breast cancer tissues compared to DCIS.
Similarly, our present study showed that expression of IFITM3
protein was much higher in invasive breast cancer than in DCIS. It
has been postulated that infection caused by mouse mammary tumor
virus (MMTV) might play a role in the etiology of breast cancer
development. The molecular studies have shown that up to 40% of
sporadic breast cancer samples contain MMTV-like env gene sequences
(27,28), and Fernandez-Cobo et
al(29) found that 6
interferon-inducible proteins, including, IFI6, TRIM22, IFITM1,
IFITM2+IFITM3, IFI27 and IP-30, and the receptor IFNGR2 were
upregulated in MMTV-like env+ cells. However, it remains to
be determined whether the induced expression of IFITM3 protein in
invasive breast cancer is due to the mouse mammary tumor virus
infection.
Our present study showed that expression of IFITM3
protein was associated with the estrogen receptor and progesterone
receptor status in invasive breast cancer tissues, but these data
are contrary to that reported by Andreu et al(10). Estrogen exposure of normal breast
epithelium is a major risk factor for breast cancer development and
overexpression of estrogen receptor could promote cell cycle
progression and reduce apoptosis and DNA repair, in turn promoting
tumorigenesis (30,31). The estrogen receptor is expressed in
~60–80% of invasive breast cancer cases and the estrogen receptor
status is used to determine the sensitivity of breast cancer to
anti-estrogen therapy (32). Thus,
the positive association of IFITM3 expression with estrogen
receptor may indicate the same role of these two proteins in breast
cancer development, but may not have true relationship. However,
further investigation is needed to clarify this speculation.
We demonstrated that knockdown of IFITM3 expression
inhibited breast cancer cell viability, BrdU incorporation and
colony formation, as well as arrested breast cancer cells at the
G0/G1 phase of the cell cycle. These data are novel and indicate
the oncogenic activity of IFITM3 protein. However, our present
study is only a proof-of-principle and more studies are needed to
clarify the underlying molecular mechanisms. The limitations of
this study include: i) The number of breast cancer cases was small
and the data need to be verified in a large sample size to confirm
IFITM3 overexpression in breast cancer; and ii) we did not provide
any underlying molecular events to support IFITM3 actions in these
two breast cancer cell lines. Further studies are needed to address
these limitations and identify IFITM3 as a potential oncogene in
breast cancer.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Greenlee RT, Murray T, Bolden S and Wingo
PA: Cancer statistics. CA Cancer J Clin. 50:7–33. 2000.
|
|
3
|
Charpentier A and Aldaz CM: The molecular
basis of breast carcinogenesis. The Molecular Basis of Human
Cancer. Coleman WB, Tsongalis GJ and Totowa NJ: Human Press; New
Jersey: pp. 347–363. 2002, View Article : Google Scholar
|
|
4
|
Moffatt P, Gaumond MH, Salois P, et al:
Bril: a novel bone-specific modulator of mineralization. J Bone
Miner Res. 23:1497–1508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hickford D, Frankenberg S, Shaw G and
Renfree MB: Evolution of vertebrate interferon inducible
transmembrane proteins. BMC Genomics. 13:1552012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reid LE, Brasnett AH, Gilbert CS, et al: A
single DNA response element can confer inducibility by both α- and
γ-interferons. Proc Natl Acad Sci USA. 86:840–844. 1989.
|
|
7
|
Friedman RL, Manly SP, McMahon M, Kerr IM
and Stark GR: Transcriptional and post-transcriptional regulation
of interferon-induced gene expression in human cells. Cell.
38:745–755. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Pan Q, Rong L, He W, Liu SL and
Liang C: The IFITM proteins inhibit HIV-1 infection. J Virol.
85:2126–2137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saitou M, Payer B, Lange UC, Erhardt S,
Barton SC and Surani MA: Specification of germ cell fate in mice.
Philos Trans R Soc Lond B BiolSci. 358:1363–1370. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Andreu P, Colnot S, Godard C, et al:
Identification of the IFITM family as a new molecular marker in
human colorectal tumors. Cancer Res. 66:1949–1955. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegrist F, Ebeling M and Certa U: The
small interferon-induced transmembrane genes and proteins. J
Interferon Cytokine Res. 31:183–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Peng H, Zhong Y, et al:
Differential gene expression profiling of human epidermal growth
factor receptor 2-overexpressing mammary tumor. Acta Biochim
Biophys Sin. 40:397–405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tanaka SS, Nagamatsu G, Tokitake Y, Kasa
M, Tam PP and Matsui Y: Regulation of expression of mouse
interferon-induced transmembrane protein like gene-3, Ifitm3
(mil-1, fragilis), in germ cells. Dev Dyn. 230:651–659. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smith RA, Yong J, Weis JJ and Weis JH:
Expression of the mouse fragilis gene products in immune cells and
association with receptor signaling complexes. Genes Immun.
7:113–121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brem R, Oraszlan-Szovik K, Foser S,
Bohrmann B and Certa U: Inhibition of proliferation by 1-8U in
interferon-alpha-responsive and nonresponsive cell lines. Cell Mol
Life Sci. 60:1235–1248. 2003.PubMed/NCBI
|
|
16
|
Lau SL, Yuen ML, Kou CY, Au KW, Zhou J and
Tsui SK: Interferons induce the expression of IFITM1 and IFITM3 and
suppress the proliferation of rat neonatal cardiomyocytes. J Cell
Biochem. 113:841–847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Lee JH, Kim KY, et al: The
interferon-inducible 9-27 gene modulates the susceptibility to
natural killer cells and the invasiveness of gastric cancer cells.
Cancer Lett. 221:191–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hatano H, Kudo Y, Ogawa I, et al:
IFN-induced transmembrane protein 1 promotes invasion at early
stage of head and neck cancer progression. Clin Cancer Res.
14:6097–6105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singletary SE, Allred C, Ashley P, et al:
Revision of the American Joint Committee on cancer staging system
for breast cancer. J Clin Oncol. 20:3628–3636. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Federico Maurizio: Lentivirus Gene
Engineering Protocols (Methods in Molecular Biology). Humana Press;
1 edition. April 30–2003, View Article : Google Scholar
|
|
21
|
Sakoda T, Kasahara N, Hamamori Y and Kedes
L: A high-titer lentiviral production system mediates efficient
transduction of differentiated cells including beating cardiac
myocytes. J Mol Cell Cardiol. 31:2037–2047. 1999. View Article : Google Scholar
|
|
22
|
Soneoka Y, Cannon PM, Ramsdale EE, et al:
A transient three-plasmid expression system for the production of
high titer retroviral vectors. Nucleic Acids Res. 23:628–633. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30:e362002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brass AL, Huang IC, Benita Y, et al: IFITM
proteins mediate the innate immune response to influenza A H1N1
virus, West Nile virus and Dengue virus. Cell. 139:1243–1254. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang IC, Bailey CC, Weyer JL, et al:
Distinct patterns of IFITM-mediated restriction of filoviruses,
SARS coronavirus, and influenza A virus. PLoS Pathog.
7:e10012582011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abba MC, Drake JA, Hawkins KA, et al:
Transcriptomic changes in human breast cancer progression as
determined by serial analysis of gene expression. Breast Cancer
Res. 6:R499–R513. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Holland JF, Bleiweiss IK, Melana
SM, Xu D and Pogo BGT: Detection of mammary tumor virus env
gene-like sequences in human breast cancer. Cancer Res.
55:5173–5179. 1995.PubMed/NCBI
|
|
28
|
Ford CE, Tran D, Deng YM, Ta VT, Rawlinson
WD and Lawson JS: Mouse mammary tumor virus-like gene sequences in
breast tumors of Australian and Vietnamese women. Clin Cancer Res.
9:1118–1120. 2003.
|
|
29
|
Fernandez-Cobo M, Melana SM, Holland JF
and Pogo BG: Transcription profile of a human breast cancer cell
line expressing MMTV-like sequences. Infect Agent Cancer. 1:72006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khan SA, Rogers MA, Khurana KK, Meguid MM
and Numann PJ: Estrogen receptor expression in benign breast
epithelium and breast cancer risk. J Natl Cancer Inst. 90:37–42.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ricketts D, Turnbull L, Ryall G, et al:
Estrogen and progesterone receptors in the normal female breast.
Cancer Res. 51:1817–1822. 1991.PubMed/NCBI
|
|
32
|
Moy B and Goss PE: Estrogen receptor
pathway: resistance to endocrine therapy and new therapeutic
approaches. Clin Cancer Res. 12:4790–4793. 2006. View Article : Google Scholar : PubMed/NCBI
|