Introduction
Gliomas are the most common and most aggressive
types of primary brain tumor in humans. Despite recent advances in
treatment by a combination of surgery and chemotherapy and/or
radiotherapy, the prognosis of malignant glioma remains extremely
poor. Glioblastoma (GBM) is the most common and malignant glioma,
and has a median survival time of approximately one year (1–7). In
the United States, 75% of glioma patients die within 5 years of
diagnosis (8). There is, therefore,
an urgent need for the development of a prognostic biomarker for
this disease.
To date, a considerable amount of research has
discovered that epigenetic mechanisms play an important role in
tumors. Aberrant epigenetic mechanisms, such as promoter
hyper/hypomethylation, histone modifications, or non-coding RNA
expression, are key in tumor formation (9). Moreover, DNA methylation is typically
a more stable and inheritable epigenetic pattern that can persist
for several cell generations, which potentially broadens its
practical applicability in the clinic (10). Changes in DNA methylation patterns
are an important hallmark of tumor development and progression
(11). Although the role of
hypermethylation in the silencing of tumor suppressor genes is now
well-documented (12), a reduction
in the level of methylation also contributes to neoplastic
progression in numerous types of human cancer, including gliomas
(13,14).
KAZALD1 is also known as IGFBP-rP10, BONO1, FKSG28
or FKSG40. This gene encodes a secreted member of the insulin
growth factor-binding protein (IGFBP) superfamily. It contains an
N-terminal insulin growth factor-binding domain, a central
Kazal-type serine protease inhibitor and follistatin-like domain,
and a C-terminal immunoglobulin-like domain. Studies of the mouse
ortholog suggest that this gene product may have a function in bone
development and bone regeneration. The insulin-like factor binding
domain has high affinity for insulin-like growth factors, while the
Kazal domain contains sequence and structural homology to serine
proteases and follistatins (15).
The immunoglobulin-like domain appears to have a molecular binding
function perhaps regulating cell adhesion and extracellular ligand
binding (16), and is associated
with regions of matrix mineralization in bone and dentin matrices
(17). Invasion and proliferation
may be the way KAZALD1 promotes tumor progression.
In the present study, promoter methylation status of
KAZALD1 and its correlation with clinicopathological parameters
were evaluated in our methylation microarray and in 91 independent
samples. The promoter methylation status of KAZALD1 in glioma was
investigated to determine any correlation between tumors and DNA
methylation. Correlation of KAZALD1 methylation status with
clinical data was investigated to assess its predictive and
prognostic value for glioma patients. Expression of KAZALD1 in
these tumors was also assessed to determine whether promoter
hypomethylation correlated with protein overexpression in gliomas.
Regular follow-up of these patients and correlation of molecular
findings with disease outcome emphasized the prognostic relevance
of KAZALD1 methylation and expression in glioma patients.
Functional assays in glioma cell lines were also performed.
Materials and methods
Patients and samples
All patients were from the Chinese Glioma Genome
Atlas (CGGA). The patients selected in the present study underwent
surgical resection between January 2006 and December 2010 and
subsequently received concomitant and adjuvant temozolomide and
radiotherapy. Clinical data, including patient age at diagnosis,
gender and preoperative Karnofsky performance status (KPS) score
were obtained from the medical records. Overall survival (OS) time,
defined as the period from operation to mortality, was calculated
mainly when patients visited the clinics or by phone interview with
patients and/or their relatives. Patients who succumbed to
non-primary diseases were excluded (18). Tumor tissue samples were obtained by
surgical resection before the treatment with radiation and
chemotherapy. Respective specimens were snap-frozen and stored in
liquid nitrogen until nucleic acid extraction. The present study
was approved by the Ethics Committee of the Capital Medical
University, and written informed consent was obtained from all
patients. Only samples with >80% tumor cells were selected.
DNA extraction
All tissue samples were immediately snap-frozen in
liquid nitrogen after surgery. A hematoxylin and eosin-stained
frozen section was prepared for assessment of the percentage of
tumor cells before DNA extraction. Genomic DNA was isolated from
frozen tumor tissues using the QIAamp DNA Mini kit (Qiagen)
according to the manufacturer’s protocol. DNA concentration and
quality were measured using the NanoDrop ND-1000 spectrophotometer
(NanoDrop Technologies, Houston, TX, USA).
Genome-wide DNA methylation
profiling
A series of 119 glioma samples (63 low-grade
gliomas, 33 anaplastic gliomas and 23 glioblastomas) were measured
by methylation microarray. We used the Illumina Infinium Human
Methylation 27 BeadChips (Illumina Inc.), as previously described
(19). The BeadChip contains 27,578
highly informative CpG sites covering more than 14,000 human RefSeq
genes. This allows researchers to interrogate all these sites per
sample at a single nucleotide resolution. Bisulfite modification of
DNA, chip processing and data analysis were performed following the
manufacturer’s manual at the Wellcome Trust Centre for Human
Genetics Genomics Laboratory (Oxford, UK). The array results were
analyzed with the BeadStudio software (Illumina).
Immunohistochemistry
Immunohistochemistry (IHC) was performed as
previously described (20).
Briefly, surgical biopsies from the patients above were fixed in
formalin, routinely processed and paraffin-embedded. Five
micron-thick sections were prepared, and immunohistochemical
staining with streptavidin-biotin immunoperoxidase assay was
performed using goat polyclonal antibody to KAZALD1 (Santa Cruz
Biotechnology). The degree of immunostaining of sections was viewed
and scored separately by two independent investigators. The scores
were determined by combining the proportion of positively stained
tumor cells and the intensity of staining. The proportion of
positively stained tumor cells was graded as follows: 0, no
positive tumor cells; 1, <5% positive tumor cells; 2, 5–20%
positive tumor cells; and 3, >20% positive tumor cells (5). The intensity of staining was recorded
on a scale of 0 (no staining), 1 (weak staining, light yellow), 2
(moderate staining, yellowish brown) and 3 (strong staining,
brown). The staining index was calculated as follows: staining
index = staining intensity × proportion of positively stained tumor
cells. High KAZALD1 expression was defined as a staining index
score >6, while low expression was defined as a staining index
≤6.
Cell lines and cell culture
The human glioblastoma cell lines LN229 and U251
were obtained from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Science, Shanghai, China. The cells were
maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS)
and incubated at 37ºC in 5% CO2. Upon 80% confluency,
cells were starved in DMEM with 1% FBS for 24 h and maintained in
this low-serum condition for the course of the treatment.
KAZALD1 gene knockdown by siRNA
Specific oligos targeting the KAZALD1 gene were
selected (OriGene Technologies, Inc., Rockville, MD, USA). siRNA1,
the most efficient one, screened from three siRNAs, was used to
knockdown KAZALD1 (sequence, 5′-GCUAGGCACCAAUAAAC AUUUCUAA-3′).
Logarithmically growing cells were seeded at a density of
105 cells per 6-cm dish and transfected with 5 μmol
KAZALD1 siRNA using Lipofectamine® 2000 (Invitrogen),
according to the manufacturer’s instructions. Forty-eight hours
later, cells were used for in vitro functional assays as
described below.
Western blot analysis
Following the cell treatment, cell lysates were
prepared via lysis buffer, electrophoresed onto SDS-polyacrylamide
gels and transferred to polyvinylidene difluoride membranes.
Membranes were probed with rabbit antibodies against KAZALD1 (Santa
Cruz Biotechnology) and glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) (A-3; Santa Cruz Biotechnology) at dilutions of 1:1,000.
Blots were detected with horseradish peroxidase-labeled anti-goat
antibodies (1:5,000 dilution), developed using enhanced
chemiluminescence (ECL) reagents (Amersham Pharmacia,
Buckinghamshire, UK).
Transwell invasion assay
The transwell invasion assay was carried out in
24-well cell culture chambers using transwell inserts (Corning Life
Sciences, Corning, NY, USA) with 8 m pore membrane precoated with
matrigel (BD Biosciense, San Jose, CA, USA). LN229 and U251 cells
were plated at the density of 1×104 per upper well in
200 μl culture medium (DMEM, no FBS), control group and siRNA
group, respectively. The lower chamber was filled with 500 μl
medium (DMEM, 12% FBS). The cells were allowed to invade for 24 h,
after which time the non-invading cells with Matrigel matrix were
removed from the upper surface of the membrane by scrubbing with a
cotton-tipped swab. Cells on the lower surface of the filter were
fixed for 30 min in methanol and glacial acetic acid mixture (3:1),
air-dried briefly and stained with crystal violet. The mean number
of invaded cells was counted from five preselected microscopic
fields at ×200 magnification. All experiments were performed in
triplicate.
Colony formation assay
LN229 and U251 cells were transfected with siRNA or
a negative control for 48 h and were then plated into six orifice
plates (1,000 per orifice) and transfected with siRNA one more time
on the 6th day. On the 12th day, plates were washed with PBS and
stained with crystal violet. The number of colonies with >30
cells was counted. The colonies were counted manually using a
microscope.
Nude mouse tumor xenograft model and
siRNA of KAZALD1 treatment
U251 glioma cells were subcutaneously injected into
5-week-old female nude mice (Cancer Institute of the Chinese
Academy of Medical Science). When the tumor volume reached 50
mm3, mice were randomly divided into two groups (six
mice per group). Each group was treated with siRNA of KAZALD1 or
negative control oligo in 10 μl Lipofectamine through local
injection of the xenograft tumor at multiple sites. The treatment
was performed once every 3 days for 15 days. The tumor volume was
measured with a caliper twice a week, using the following formula:
volume = length × width2/2.
Statistical analysis
Significance analysis of microarrays (SAM) was used
for genes significantly differently methylated between high-grade
and low-grade gliomas. Cox-regression was performed using Matleb
software. A two-sided χ2 test was performed using SPSS
13.0. Kaplan-Meier survival curves were obtained and differences in
the OS were tested using the log-rank test (GraphPad Prism 5).
Differences of tumor cell invasion and colony formation number
between treated and control groups were analyzed by t-test.
P<0.05 was considered to indicate a statistically significant
result.
Results
Gene screening
The methylation microarray from CGGA contains 27,578
highly informative CpG sites covering more than 14,475 human RefSeq
genes. Following SAM analysis between high- and low-grade gliomas,
with FDR<0.2, 306 probes were screened out. Then, by survival
Cox-regression analysis using Metlab software, 25 probes (25 genes)
were left. For univariate analysis validation, we performed t-test
and Kaplan-Meier survival curve and log-rank test, after which, 12
probes (12 genes) remained. We further searched for the CpG islands
upstream of the promoter, 6 genes were in the list in the end.
These 6 genes were ADCY1, KAZALD1, KLF4, SLMAP, TETRAN and
TP53INP1. We selected the oncogene KAZALD1 (Fig. 1).
High grade and poor prognosis association
of KAZALD1 hypomethylation in CGGA data
The methylation level of KAZALD1 was measured in a
series of 119 glioma samples (63 low-grade gliomas, 33 anaplastic
gliomas and 23 glioblastomas) via microarray. KAZALD1 was
significantly hypomethylated in high-grade gliomas compared to
low-grade gliomas (P<0.001). The correlation between KAZALD1
methylation and OS was measured through Kaplan-Meier survival curve
analysis and log-rank test. KAZALD1 methylation was inversely
correlated with OS in the high-grade glioma samples (P=0.002)
(Fig. 2B).
Patients with low-methylation level had a median OS
of 544 days compared to 958 days in patients with high methylation
level (Table I). Preoperative KPS
score was also correlated with OS (P=0.004). There was no
significant association with age, gender and OS. The multivariate
Cox proportional hazards model, after adjusting for age, KPS score,
identified hypomethylation of KAZALD1 as an independent unfavorable
prognostic factor for OS (P=0.009).
 | Table IVariables related to OS in 56
high-grade gliomas in methylation microarray: univariate and
multivariate analysis. |
Table I
Variables related to OS in 56
high-grade gliomas in methylation microarray: univariate and
multivariate analysis.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
| Variables | No. of patients | Median OS (days) | 95% CI (days) | P-value | Relative risk | 95% CI | P-value |
|---|
| Gender |
| Male | 23 | 591 | 249–745 | | | | |
| Female | 33 | 497 | 317–865 | 0.528 | | | |
| Age (years) |
| ≤41.5 | 28 | 878 | 304–1452 | | | | |
| >41.5 | 28 | 530 | 293–767 | 0.178 | 1.248 | 0.605–2.573 | 0.549 |
| KPS |
| <80 | 19 | 290 | 135–445 | | | | |
| ≥80 | 37 | 878 | NR | 0.004 | 0.426 | 0.208–0.871 | 0.019 |
|
KAZALD1-methylation |
| Low | 28 | 544 | 400–688 | | | | |
| High | 28 | 958 | 768–1150 | 0.002 | 0.367 | 0.172–0.782 | 0.009 |
Expression of KAZALD1 is relative to
glioma grade progression and confers a poor prognosis of high
KAZALD1 expression in high-grade glioma patients
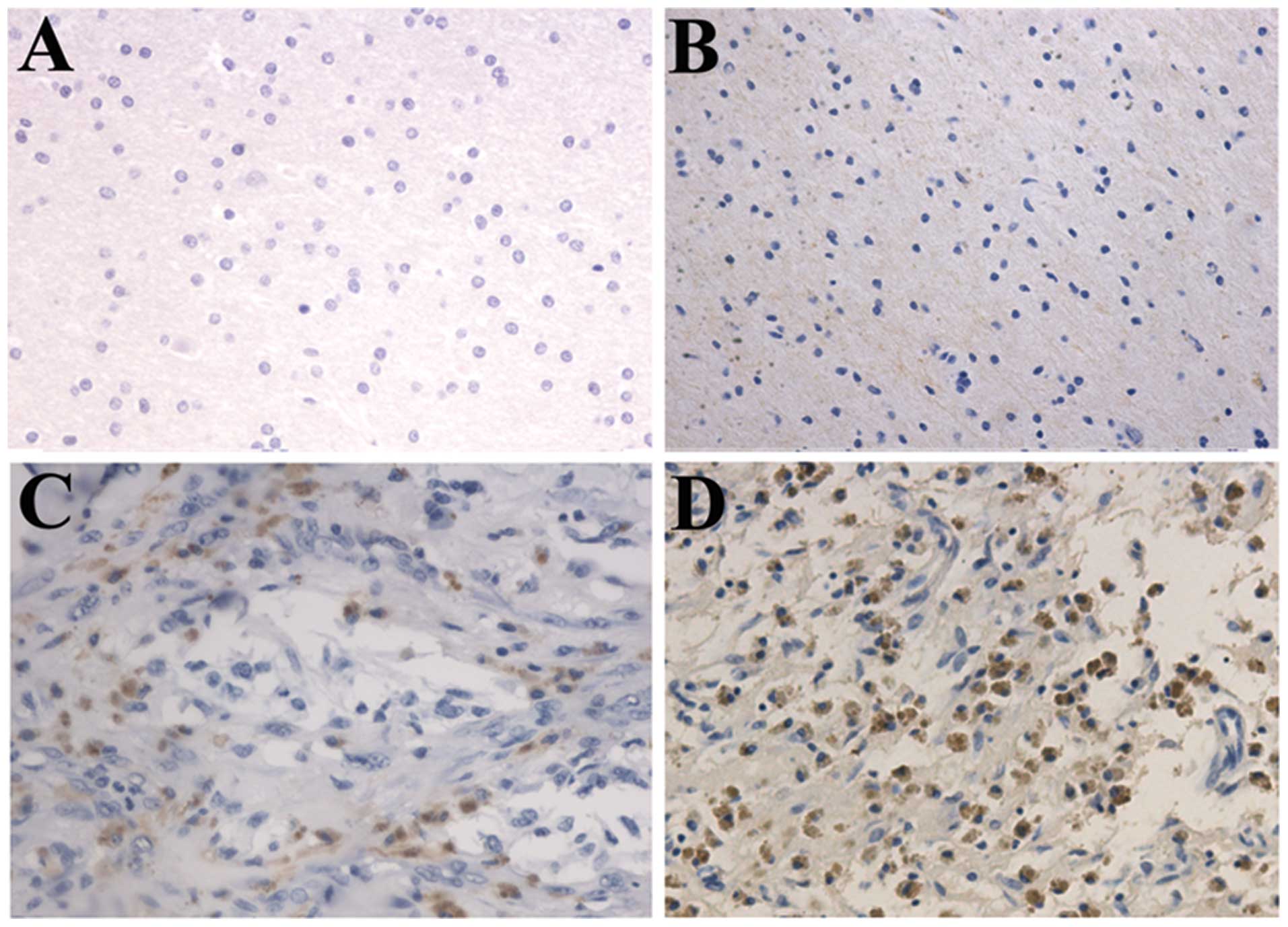
We performed immunohistochemical staining in 91
glioma samples from mainland Han Chinese glioma patients. The 91
patients consisted of 30 patients with astrocytoma (A, WHO grade
II), 20 with anaplastic astrocytoma (AA, WHO grade III) and 41 with
glioblastoma multiforme (GBM, WHO grade IV). We analyzed the
correlation between KAZALD1 protein expression and histological
staging of gliomas, which showed KAZALD1 expression ranged from low
to high along with grade progression of gliomas (P<0.01;
Fig. 3A). Survival analysis showed
that patients with high KAZALD1 expression had significantly
shorter OS (P<0.001) than those with low expression in
high-grade glioma patients (Fig.
3B). We also confirmed there was no KAZALD1 expression in
normal brain tissue by IHC (Fig.
4).
Patients with high KAZALD1 expression had median OS
time of 295 days compared to 660 days in patients with low KAZALD1
expression (Table II).
Preoperative KPS score was also correlated with OS (P=0.022). There
were no significant associations with age, gender and OS. The
multivariate Cox proportional hazards model, after adjusting for
age, KPS score, identified high KAZALD1 expression as an
independent unfavorable prognostic factor for OS (HR, 4.096;
P=0.001).
 | Table IIVariables related to OS in 61
high-grade glioma validated samples (58 survivals were available):
univariate and multivariate analysis. |
Table II
Variables related to OS in 61
high-grade glioma validated samples (58 survivals were available):
univariate and multivariate analysis.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
| Variables | No. of
patients | Median OS
(days) | 95% CI (days) | P-value | Relative risk | 95% CI | P-value |
|---|
| Gender |
| Male | 38 | 493 | 339–647 | | | | |
| Female | 20 | 362 | 200–523 | 0.410 | | | |
| Age (years) |
| ≤50 | 29 | 570 | 466–674 | | | | |
| >50 | 29 | 354 | 332–376 | 0.192 | 1.207 | 0.541–2.689 | 0.646 |
| KPS |
| <80 | 37 | 376 | 245–507 | | | | |
| ≥80 | 21 | 572 | 461–683 | 0.022 | 0.519 | 0.207–1.300 | 0.161 |
|
KAZALD1-expression |
| Low | 36 | 660 | 467–853 | | | | |
| High | 22 | 295 | 184–406 | 0.000 | 4.096 | 1.843–9.106 | 0.001 |
KAZALD1 promotes invasion and
proliferation of LN229 and U251 cell lines
Following siRNA knockdown of KAZALD1 in two cell
lines, LN229 and U251, we validated its expression level (Fig. 5A). The cells were further used for
transwell invasion assay and colony formation assay which showed
that siRNA significantly attenuated the effect of KAZALD1 on cell
invasion (P<0.05; Fig. 5B) as
well as proliferation (P<0.05; Fig.
5C), respectively.
KAZALD1 promotes glioma growth in
vivo
To investigate the potential impact of KAZALD1
expression in vivo, a U251 xenograft model was utilized. The
KAZALD1-treated group displayed a significant growth and tumor
reduction, whereas tumor growth was not impacted by negative
control (Fig. 6C).
Discussion
In the present study, we investigated the
methylation and expression level of KAZALD1 in two independent
cohorts, totaling 210 glioma patients. We found that methylation
level of KAZALD1 promoter was associated with glioma grade
progression and overall survival (OS) in high-grade gliomas using
genome-wide DNA methylation profiling. We further compared the
expression level of KAZALD1 in low-grade (astrocytoma,
oligodendrocytoma and oligoastrocytoma) and high-grade (anaplastic
gliomas and glioblastomas) human gliomas, and demonstrated a
significant increase in KAZALD1 expression from low- to high-grade
gliomas. KAZALD1 was an independent prognostic factor predicting OS
in high-grade gliomas, indicating a significant correlation between
KAZALD1 expression and clinical outcome in glioma patients. KAZALD1
functional analyses were performed in LN229 and U251 cell lines,
which showed KAZALD1 promoted invasion and proliferation of glioma
both in vitro and in vivo. To the best of our
knowledge, this is the first report on the methylation status,
expression difference and function of KAZALD1 in glioma.
DNA methylation of promoter CpG islands has been
recognized as an important mechanism for the regulation of gene
expression and transcriptional modification in mammals. DNA
methylation status is more stable and can be detected using a
number of high-throughput and sensitive techniques with little
patient material (21). Aberrations
in DNA methylation patterns may have critical effects on tumor
initiation and progression. Although considerable research has been
conducted on the epigenetic control of tumor suppressor genes,
little is known about the potential role of promoter CpG
demethylation in the activation of oncogenes. In the present study,
we performed genome-wide DNA methylation profiling of 119 glioma
tissues via microarrays. We set a series of screening conditions
including grade correlation, survival correlation and CpG island in
upstream of gene promoter (Fig. 1).
KAZALD1 was one of six genes conformed to appeal conditions. We
also found that methylation level of KAZALD1 in high-grade glioma
was significantly lower than that in low-grade glioma (Fig. 2A). Kaplan-Meier survival curve
analysis showed that low methylation level of KAZALD1 had shorter
OS compared to the high methylation group in high-grade glioma
patients (Fig. 2B). Multivariate
analysis identified hypomethylation of KAZALD1 was an independent
unfavorable prognostic factor for OS (Table I; P=0.009; low methylated group, 544
days; high methylated group, 958 days).
To explore whether the expression level of KAZALD1
correlated with the status of methylation in glioma, we detected
the KAZALD1 expression in 91 independent cohorts including 30
low-grade and 61 high-grade glioma patients using
immunohistochemistry. We found that high-grade glioma patients had
significantly higher KAZALD1 expression than low-grade glioma
patients (Figs. 3A and 4). According to our original hypothesis,
there was a negative correlation between KAZALD1 expression and
methylation status in low- and high-grade gliomas. In 61 high-grade
glioma patients, patients with high KAZALD1 expression had shorter
OS than those with low expression (Fig.
3B, Table II). These
observations indicated that glioma with KAZALD1 hypomethylation and
higher protein expression might be a more biologically aggressive
phenotype than those without it. These results showed that
increased expression of KAZALD1 in gliomas with a poor prognosis
was at least partially due to hypomethylation of the KAZALD1
promoter region.
To date, no information has been provided regarding
the regulation of the KAZALD1 gene in gliomas. However, rather
limited data are available on the relationship between the KAZALD1
gene and its precise mechanism of action on tumors, and their
clinical effect on outcome for patients with glioma remains
unclear. The function of KAZALD1 in dental development and bone
regeneration was investigated only recently and, thus far, its
biological function is unknown (17). The KAZALD1 insulin-like factor
binding domain has high affinity for insulin-like growth factors,
while the Kazal domain contains sequence and structural homology to
serine proteases and follistatins (15). The immunoglobulin-like domain
appears to have a molecular binding function perhaps regulating
cell adhesion and extracellular ligand binding (16), and is associated with regions of
matrix mineralization (17).
KAZALD1 and MMP-20 are coexpressed in secretory odontoblasts at
developmental stages (22). Based
on the above findings, we hypothesize that KAZALD1 promoted the
glioma progression through invasion and proliferation. We performed
functional assays in LN229 and U251 cell lines and found that
invasion and colony formation number decreased when KAZALD1 was
knocked down by siRNA, and the same results were founded in
vivo. We consider that KAZALD1 could combine with IGF, to
activate the IGF pathway. A series of downstream genes of the IGF
pathway promote glioma grade progression and shorten survival time.
However, this requires further assays.
In summary, promoter hypomethylation of KAZALD1 was
associated with high KAZALD1 expression. Patients harboring
hypomethylation and high expression KAZALD1 had shorter OS in
high-grade glioma. On the basis of these observations and the
results from subset analysis, it is reasonable to conclude that
KAZALD1 promoter methylation status is an important prognostic
biomarker in glioma. KAZALD1 promoted glioma malignant progression
through invasion and proliferation.
Acknowledgements
The authors thank Dr Susan Furness for her critical
reading and Professor Chen (Beijing Sanbo Brain Hospital) for IHC
technical support. This study was supported by grants from the
National High Technology Research and Development Program (no.
2012AA02A508), the International Science and Technology Cooperation
Program (no. 2012DFA30470), the National 973 program (no.
2011CB707804) and the National Natural Science Foundation of China
(no. 81201993).
References
|
1
|
Zhang W, Yan W, You G, Bao Z, Wang Y, et
al: Genome-wide DNA methylation profiling identifies ALDH1A3
promoter methylation as a prognostic predictor in G-CIMP-primary
glioblastoma. Cancer Lett. 328:120–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang J, Huang K, Shi Z, Zou J, Wang Y, et
al: High β-catenin/Tcf-4 activity confers glioma progression via
direct regulation of AKT2 gene expression. Neuro Oncol. 13:600–609.
2011.
|
|
3
|
Chen L, Han L, Zhang K, Shi Z, Zhang J, et
al: VHL regulates the effects of miR-23b on glioma survival and
invasion via suppression of HIF-1α/VEGF and β-catenin/Tcf-4
signaling. Neuro Oncol. 14:1026–1036. 2012.PubMed/NCBI
|
|
4
|
Yan W, Zhang W, You G, Zhang J, Han L, et
al: Molecular classification of gliomas based on whole genome gene
expression: a systematic report of 225 samples from the Chinese
Glioma Cooperative Group. Neuro Oncol. 14:1432–1440. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Li S, Chen L, You G, Bao Z, et al:
Glioblastoma with an oligodendroglioma component: distinct clinical
behavior, genetic alterations, and outcome. Neuro Oncol.
14:518–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Zhang J, Yan W, You G, Bao Z, et
al: Whole-genome microRNA expression profiling identifies a
5-microRNA signature as a prognostic biomarker in Chinese patients
with primary glioblastoma multiforme. Cancer. 119:814–824. 2013.
View Article : Google Scholar
|
|
7
|
Zhang W, Zhang J, Hoadley K, Kushwaha D,
Ramakrishnan V, et al: miR-181d: a predictive glioblastoma
biomarker that downregulates MGMT expression. Neuro Oncol.
14:712–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Starkweather AR, Sherwood P, Lyon DE,
McCain NL, Bovbjerg DH, et al: A biobehavioral perspective on
depressive symptoms in patients with cerebral astrocytoma. J
Neurosci Nurs. 43:17–28. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gaspar-Maia A, Alajem A, Meshorer E and
Ramalho-Santos M: Open chromatin in pluripotency and reprogramming.
Nat Rev Mol Cell Biol. 12:36–47. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feinberg AP and Tycko B: The history of
cancer epigenetics. Nat Rev Cancer. 4:143–153. 2004. View Article : Google Scholar
|
|
12
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2(Suppl 1): S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ehrlich M: DNA methylation in cancer: too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cadieux B, Ching TT, VandenBerg SR and
Costello JF: Genome-wide hypomethylation in human glioblastomas
associated with specific copy number alteration,
methylenetetrahydrofolate reductase allele status, and increased
proliferation. Cancer Res. 66:8469–8476. 2006. View Article : Google Scholar
|
|
15
|
Forbes B, Szabo L, Baxter RC, Ballard FJ
and Wallace JC: Classification of the insulin-like growth factor
binding proteins into three distinct categories according to their
binding specificities. Biochem Biophys Res Commun. 157:196–202.
1988. View Article : Google Scholar
|
|
16
|
Bork P, Holm L and Sander C: The
immunoglobulin fold. Structural classification, sequence patterns
and common core. J Mol Biol. 242:309–320. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
James MJ, Jarvinen E and Thesleff I:
Bono1: a gene associated with regions of deposition of bone and
dentine. Gene Expr Patterns. 4:595–599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li S, Yan C, Huang L, Qiu X, Wang Z, et
al: Molecular prognostic factors of anaplastic oligodendroglial
tumors and its relationship: a single institutional review of 77
patients from China. Neuro Oncol. 14:109–116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hill VK, Ricketts C, Bieche I, Vacher S,
Gentle D, et al: Genome-wide DNA methylation profiling of CpG
islands in breast cancer identifies novel genes associated with
tumorigenicity. Cancer Res. 71:2988–2999. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Qiu XG, Chen BS, Li SW, Cui Y, et
al: Antiangiogenic therapy with bevacizumab in recurrent malignant
gliomas: analysis of the response and core pathway aberrations.
Chin Med J. 122:1250–1254. 2009.PubMed/NCBI
|
|
21
|
Schatz P, Distler J, Berlin K and Schuster
M: Novel method for high throughput DNA methylation marker
evaluation using PNA-probe library hybridization and MALDI-TOF
detection. Nucleic Acids Res. 34:e592006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kiukkonen A, Sahlberg C, Lukinmaa PL,
Alaluusua S, Peltonen E, et al: 2,3,7,8-tetrachlorodibenzo-p-dioxin
specifically reduces mRNA for the mineralization-related dentin
sialophosphoprotein in cultured mouse embryonic molar teeth.
Toxicol Appl Pharmacol. 216:399–406. 2006. View Article : Google Scholar
|