Introduction
Glioma, the most common brain tumor, accounts for
~30% of all brain and central nervous system tumors and 80% of all
malignant brain tumors (1). Over
the past 10 years, the 5-year survival rate of glioblastoma
patients was only 2%, and the median survival was 1 year (2,3). In
fact, the prognosis for glioblastoma has not improved for an
extensive period of time, due to its resistant to radiotherapy,
chemotherapy and adjuvant therapies (4). As a result, development of a promising
therapeutic target is urgently needed.
Mitochondria are fundamental to metabolic
homeostasis (5). Mitochondrial
transcription factor A (TFAM), a high-mobility group (HMG) protein,
plays a central role in mitochondrial DNA (mtDNA) replication,
transcription and inheritance (6).
Aberrant function of mitochondria has been reported to be
associated with various metabolic disorders, such as cancer
(7). Thus, investigating the role
of TFAM in malignant tumors may provide critical knowledge for
better understanding the regulatory mechanism of cancer as well as
a promising therapeutic strategy.
microRNAs (miRNAs), endogenous non-coding RNAs,
regulate gene expression by binding to the 3′ untranslational
region (UTR) of target mRNAs and act as endogenous agents of RNA
interference, resulting in either mRNA degradation or translational
repression (8,9). The role of miRNAs in brain physiology
and tumorigenesis has confirmed their use as promising therapy
targets for glioma (10). It has
been shown that miR-23b is highly expressed in tumors and is
associated with tumorigenesis (11,12);
however, the role of miR-23b in glioma remains unclear. The aim of
the present study was to elucidate the role of TFAM and miR-23b as
well as the related molecular mechanism in malignant glioma.
Materials and methods
Materials and reagents
Human glioma U251 cells were purchased from the
American Type Culture Collection (ATCC, Manassas, VA, USA). Fetal
bovine serum (FBS) and TRIzol reagent were obtained from Invitrogen
(Carlsbad, CA, USA). Dulbecco’s modified Eagle’s medium (DMEM) and
MTT were purchased from Sigma (St. Louis, MO, USA). The cell cycle
detection kit was purchased from Nanjing Jikai (Nanjing, China).
All antibodies were purchased from Santa Cruz Biotechnology (Santa
Cruz, CA, USA). SYBR-Green qRCR Mix was purchased from Toyobo
(Osaka, Japan). The reverse transcription kit was obtained from
Fermentas (Hanover, MD, USA). TaqMan qRT-PCR miRNA assay kit was
purchased from Applied Biosystems (Foster City, CA, USA).
Cell culture
Human glioma U251 cells were cultured in DMEM
supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C
with 5% CO2.
Tissue specimen collection
All protocols were approved by the Ethics Committee
of the Central South University. Informed consent was obtained from
each patient. All normal brain tissues and glioma tissues were
collected from patients at the Department of Neurosurgery, Third
Xiangya Hospital of Central South University, Changsha, Hunan,
China. Patients with no history of other tumors were diagnosed with
gliomas and were untreated. Following surgical removal, all tissues
were immediately snap-frozen in liquid nitrogen and stored until
use.
RNA extraction and real-time RT-PCR
analysis
Total RNA was extracted using TRIzol reagent
according to the manufacturer’s protocol. RNA was then reverse
transcribed into a cDNA template using the reverse transcription
kit. miR-23b expression was examined using the TaqMan qRT-PCR miRNA
assay kit. The relative expression of miR-23b was normalized to U6.
For TFAM assay, SYBR-Green qRCR Mix was then used to perform
qRT-PCR. The TFAM primer is as follows: forward
5′-CCATCTACCGACCGGATGTTA-3′ and reverse
5′-CAGACCTTCCCAGGGCACTCA-3′. β-actin primer is as follows: forward
5′-AGGGGCCGGACTCGTCATACT-3′ and reverse
5′-GGCGGCACCACCATGTACCCT-3′.
Western blotting
Glioma tissues or U251 cells were solubilized in
cold RIPA lysis buffer. Subsequently, protein was separated with 5%
SDS-PAGE. After SDS-PAGE, proteins were transferred from the gel to
PVDF membranes. Membranes were blocked in 5% non-fat dried milk in
PBST for 3 h and then incubated overnight with specific primary
antibodies (Santa Cruz Biotechnology) with β-actin as a control.
After incubation with the appropriate secondary antibody (Santa
Cruz Biotechnology), immune complexes were detected using an ECL
kit (Huyu Co., Shanghai, China).
Dual luciferase reporter assays
A normal and a mutated 3′-UTR of TFAM were
constructed by PCR, and then inserted into the multiple cloning
sites in the psiCHECK™-2 luciferase miRNA expression reporter
vector.
For the luciferase assay, 20,000 cells were cultured
to ~50–60% confluence in a 24-well plate. For each well, medium was
replaced by 300 μl OPTI-MEM medium (Invitrogen). Cells were
cotransfected with psiCHECK™-2-TFAM-3′-UTR or psiCHECK™-2-mut
TFAM-3′-UTR vector plus 50 nM miR-23b or 100 mM miR-23b inhibitor
using Cellfectin II reagent (Invitrogen), according to the
manufacturer’s instructions. Cells were incubated with transfection
reagent/DNA complex for 5 h and then refreshed with fresh complete
medium. A dual-luciferase reporter assay system (Promega, Madison,
WI, USA) was used to determine the luciferase activities 48 h after
cotransfection. Renilla luciferase activity was normalized
to firefly luciferase activity.
Plasmid construction and
transfection
The plasmids for TFAM, miR-23b, miR-SCR as well as
the retroviral supernatants were obtained from Aijia Bio (Changsha,
China). U251 cells were transduced using 20 mg/ml polybrene for
over 48 h.
MTT assays
For all groups, 5,000 cells/well were plated in a
96-well plate. MTT (0.5%) was added to each well and incubated for
4 h at 37°C in 5% CO2. The reaction liquid was
discarded. The formazan product was dissolved by adding 150 μl DMSO
to each well. The absorbance was detected at 570 nm with a
microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Cell cycle analysis
For all groups, 106 cells were collected
in 1X PBS and resuspended in 70% ethanol to fix overnight at −20°C.
Cells were pelleted at 1,000 rpm for 5 min, washed in 1X PBS, and
then pelleted at 1,000 rpm for 5 min. Cells were resuspended in 300
μl propidium iodide (PI) staining buffer and incubated for 30 min
at room temperature. DNA content analyses were performed using flow
cytometry (FACSCalibur, Beckman Coulter).
Colony formation assay
For all groups, 3 ml complete medium containing 150
cells was added to each well of a 6-well plate. Plates were
incubated at 37°C in 5% CO2 for 14–21 days. After that,
cells were gently washed and stained with Giemsa. Colonies
containing at least 50 cells were counted.
Transwell assay
For all groups, the invasive ability of cells was
determined by using 24-well Transwell chambers (Chemicon, Temecula,
CA, USA). After a 24-h incubation at 37°C, the migrated cells were
stained and counted.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
statistical software. All data are expressed as the mean value ± SD
of triplicate experiments, and all experiments were repeated at
least three times. The data were analyzed by one-way analysis of
variance (ANOVA) and the Student’s t-test. A P-value of <0.05
was considered to indicate a statistically significant result.
Results
TFAM protein levels are upregulated with
the malignancy of glioma
The protein levels of TFAM in normal brain tissues
and glioma tissues of different grades were examined by western
blot analysis. Data showed that the protein expression levels of
TFAM in glioma were significantly increased, when compared with
those in normal brain tissues. Moreover, the protein levels of TFAM
were positively correlated to the malignancy of glioma (Fig. 1).
miR-23b levels are decreased with the
malignancy of glioma
The miR-23b expression levels in normal brain
tissues, adjacent tissues and gliomas were determined by real-time
RT-PCR. As shown in Fig. 2, the
expression levels of miR-23b in glioma were significantly lower
than those in the normal and adjacent tissues (P<0.05).
Additionally, the miR-23b expression levels were negatively
correlated with the malignancy of glioma.
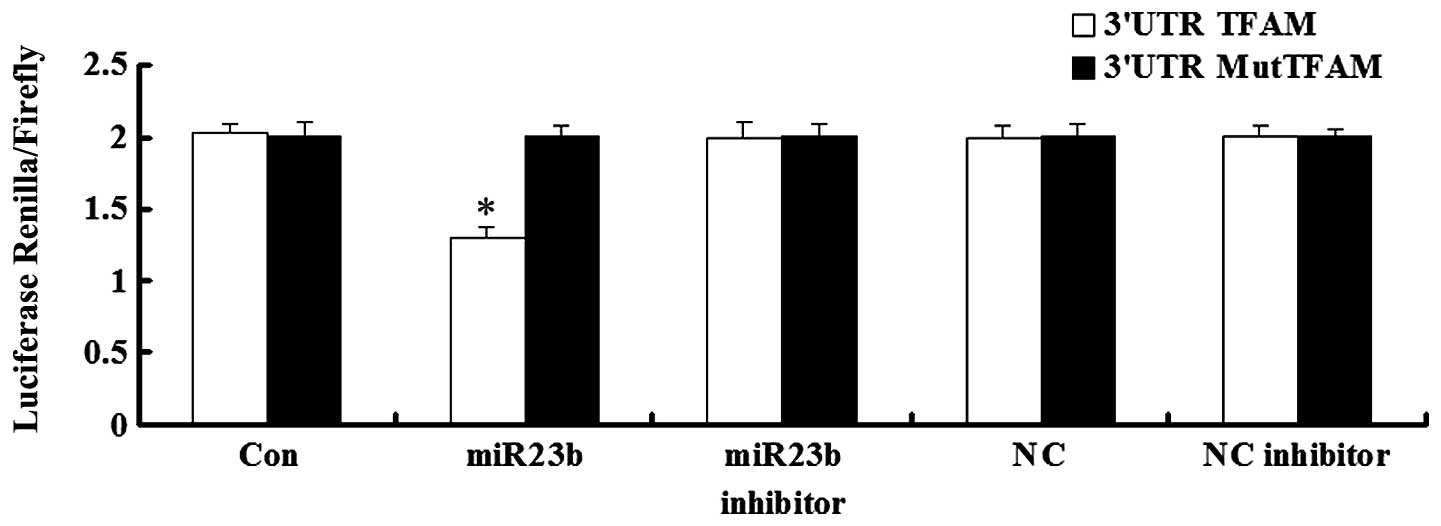
TFAM is the direct target of miR-23b
Luciferase assay was applied to test whether TFAM is
a direct target of miR-23b. As demonstrated in Fig. 3, the Renilla/firefly value of
luciferase was significantly lower in miR-23b and 3′UTR of TFAM
cotransfected cells, when compared with each control. However, the
Renilla/firefly value of luciferase in cells cotransfected
with miR-23b and mutated 3′UTR of TFAM showed no difference with
that in each control. These findings demonstrated that 3′UTR of
TFAM is the direct target of miR-23b.
Expression levels of miR-23b or TFAM
after transfection
After transfection of miR-23b or TFAM lentiviral
vectors into U251 cells, we determined the expression levels of
miR-23b or TFAM, respectively. As shown in Fig. 4A, the miR-23b expression level in
U251 cells was markedly increased after transfection when compared
with the level in the controls (P<0.05). In addition, the TFAM
expression in U251 cells was significantly upregulated after
transfection when compared with that in the controls (P<0.05)
(Fig. 4B). These data demonstrated
that the transient transfection was successful.
Effects of TFAM and miR-23b
overexpression on cell proliferation in U251 cells
To further study the roles of TFAM and miR-23b in
glioma in vitro, an MTT assay was applied to determine the
effects of miR-23b and TFAM overexpression on U251 cell
proliferation. MTT results showed that in TFAM-overexpressing U251
cells, the cell proliferation was upregulated when compared with
that in the controls. In contrast, however, in the
miR-23b-overexpressing U251 cells, the cell proliferation was
decreased when compared with that in the controls (Fig. 5). These findings revealed that
miR-23b inhibited cell proliferation of U251 cells while TFAM
promoted it.
Effects of TFAM and miR-23b
overexpression on cell cycle progression in U251 cells
We further examined the cell cycle distribution in
the different groups. As shown in Fig.
6, the miR-23b-overexpressing U251 cells showed the highest
percentage of cells in the G2 stage when compared with other three
groups, indicating that mitosis was blocked in the G2 stage. The
TFAM-overexpressing U251 cells exhibited the highest percentage of
cells in the G1 and S stages, and only a few cells were in the G2
stage, suggesting that TFAM-overexpressing U251 cells were in a
rapidly dividing state. These findings demonstrated that miR-23b
blocked the cell cycle progression of U251 cells, entirely contrary
to the effect of TFAM on human glioma U251 cells.
Effects of TFAM and miR-23b
overexpression on colony-formation efficiency of U251 cells
The effects of TFAM and miR-23b overexpression on
colony-formation efficiency in U251 cells were studied. As
demonstrated in Fig. 7, the
miR-23b-overexpressing U251 cells exhibited the lowest
colony-formation efficiency, while the TFAM-overexpressing U251
cells showed the highest colony-formation efficiency, when compared
with controls (P<0.05).
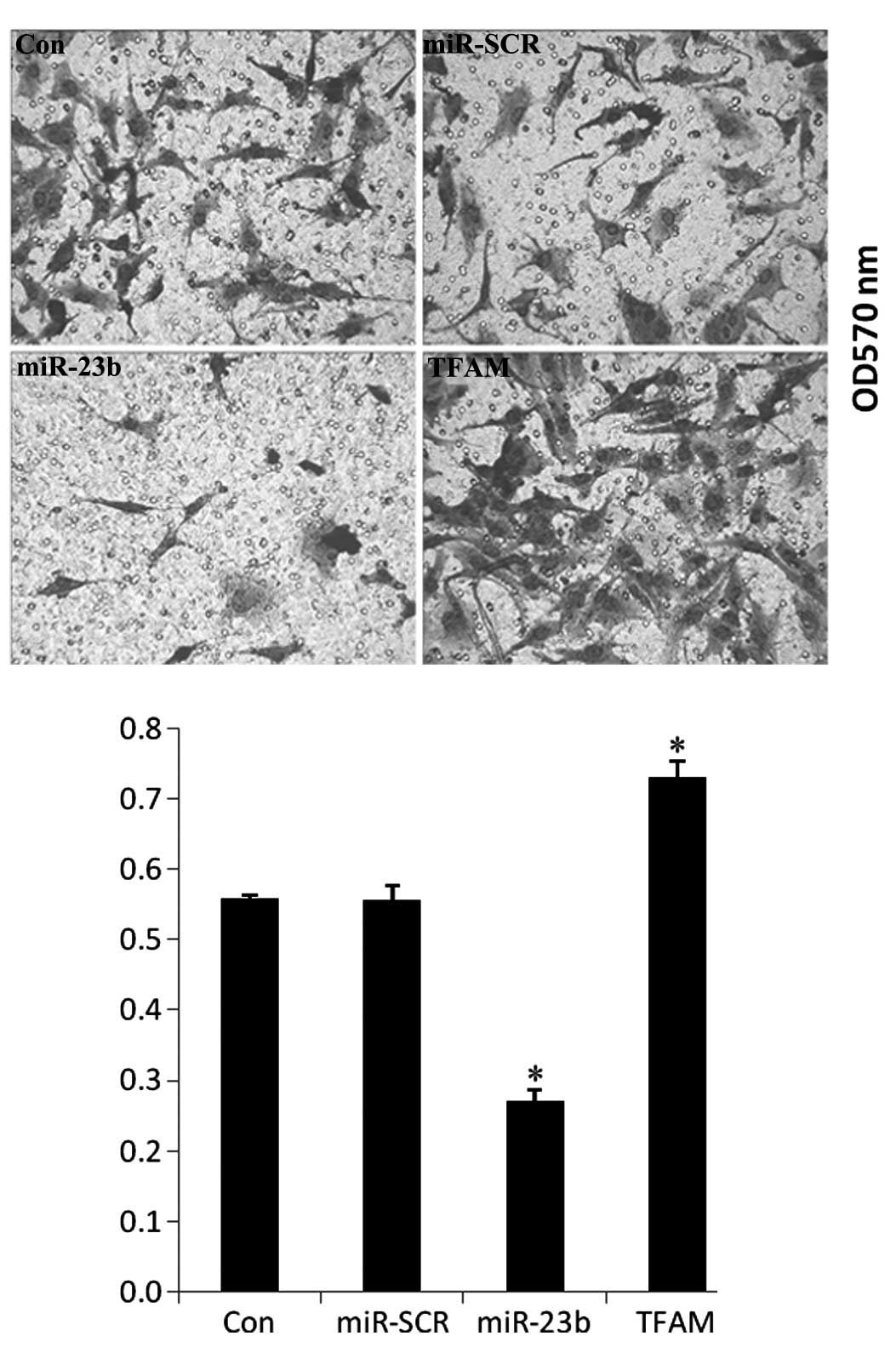
Effects of TFAM and miR-23b
overexpression on the invasion of U251 cells
The changes in the cell invasive ability of U251
cells were examined after transfection with TFAM or miR-23b. As
shown in Fig. 8, TFAM
overexpression significantly promoted cell invasion, while miR-23b
overexpression notably inhibited the invasion of U251 cells, when
compared with the controls (P<0.05).
Effects of TFAM and miR-23b
overexpression on the PI3K/Akt signaling pathway in U251 cells
Changes in the PI3K/Akt signaling pathway and
invasion-related proteins in U251 cells were studied after
transfection with TFAM or miR-23b. The protein levels of PI3K,
p-PI3K, AKT, p-Akt, MMP2 and MMP9 were examined by western blot
analysis. As shown in Fig. 9, the
expression levels of PI3K, p-PI3K, AKT, p-Akt, MMP2 and MMP9 were
all decreased in the miR-23b-overexpressing U251 cells, but
upregulated in the TFAM-overexpressing U251 cells, when compared
with the controls (P<0.05), suggesting that miR-23b suppresses
PI3K/Akt signaling activity while TFAM enhances it in U251
cells.
Discussion
TFAM, a transcription factor for mitochondrial DNA,
is required for mtDNA replication and transcription. TFAM has been
reported to participate in the regulation of cell survival,
proliferation and migration. In injured rat carotid artery,
vascular smooth muscle cell proliferation is dependent on the
upregulation of TFAM expression (13). It has been demonstrated that TFAM
regulates p21 (WAF1/CIP1), a critical regulator of cell cycle
progression, as knockdown of TFAM expression was found to induce
p21-dependent G1 cell cycle arrest (14,15).
Moreover, TFAM is involved in the development and progression of
malignant tumors. Frequent truncating mutation of TFAM has been
shown to induce mtDNA depletion and apoptotic resistance in
microsatellite-unstable colorectal cancer (16). However, the role of TFAM in glioma
remains uncovered. In the present study, the protein levels of TFAM
were significantly increased with the malignancy of glioma,
suggesting that TFAM may act as an important regulator in the
growth and progression of glioma.
miRNAs regulate gene expression through binding to
the 3′-untranslational region (UTR) of target mRNAs and act as
endogenous agents of RNA interference, and thus play a critical
role in gene silencing and function (8,9).
Recently, the association between miRNAs and malignant tumors has
become the focus of scientific research. miR-23b has been
implicated to function as a tumor suppressor in several cancer
types, including breast, prostate and renal cancer (12,17,18).
In fact, methylation-mediated silencing of miR-23b expression
exists in glioma stem cells, and miR-23b was found to regulate cell
migration and invasion via targeting of Pyk2 in migrating
glioblastoma cells, indicating that miR-23b may play a regulatory
role in glioma (19,20). Additionally, it has been recently
demonstrated that VHL regulates the effects of miR-23b on glioma
survival and invasion via suppression of HIF-1α/VEGF and
β-catenin/Tcf-4 signaling (21).
In the present study, we found that the expression
of miR-23b was significantly decreased in glioma tissues, when
compared with normal brain and adjacent tissues. Moreover, its
expression was positively correlated with the malignancy of
glioma.
Furthermore, expression of TFAM was inversely
correlated with miR-23b in glioma samples, suggesting that TFAM may
be a direct target of miR-23b. Luciferase assay data showed that
miR-23b directly deregulated the expression of TFAM. Based on these
findings, we speculated that TFAM may affect the pathological
process of glioma under the negative regulation of miR-23b. Further
investigation was performed to test our hypothesis. Data showed
that forced overexpression of TFAM enhanced cell proliferation,
cell cycle progression and invasion in vitro, contrary to
the results of forced overexpression of miR-23b in U251 cells.
These findings indicate that as a direct target of miR-23b, TFAM
plays a positive regulatory role in the growth and progression of
malignant glioma.
The molecular regulatory mechanisms of miR-23b and
TFAM in the glioma cell line U251 were further studied. The
activity of the PI3K/Akt signaling pathway and the protein levels
of invasion-related genes MMP2 and MMP9 were examined after
upregulation of miR-23b or TFAM. Western blot data showed that the
increased expression of TFAM notably promoted the protein levels of
PI3K, p-PI3k, AKT, p-AKT, suggesting that PI3K/Akt signaling was
activated. It has been well established that the PI3K/Akt signaling
pathway plays a central role in the growth, progression and
invasion of various types of cancers (22–24).
Moreover, the protein expression levels of MMP2 and MMP9 were also
upregulated after TFAM overexpression. MMP2 and MMP9 are two
typical enzymes secreted by cancer cells and are associated with
the invasion of malignant tumors, and are primarily regulated
through the PI3K/Akt signaling pathway (25,26).
Both the PI3K/Akt signaling pathway and MMPs were upregulated in
the TFAM-overexpressing U251 cells, which contributed to the
promotion of cell proliferation, cell cycle progression, clone
formation and invasion of human glioma cells. In contrast, as we
expected, miR-23b overexpression in glioma U251 cells significantly
deregulated the protein levels of PI3K, p-PI3k, AKT, p-AKT, MMP2
and MMP9. Accordingly, we showed that the PI3K/Akt signaling
pathway as well as its downstream effectors MMP2 and MMP9
participated in the regulatory network of miR-23b and TFAM in
glioma U251 cells.
In conclusion, the present study elucidated the
expression pattern of TFAM and miR-23b in gliomas of different WHO
grades, and demonstrated for the first time that TFAM is the direct
target of miR-23b. Furthermore, we revealed that the PI3K/Akt
signaling pathway is involved in the TFAM regulatory network, which
may contribute to the development of a promising therapeutic
strategy for malignant glioma.
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Surawicz TS, Davis F, Freels S, Laws ER Jr
and Menck HR: Brain tumor survival: results from the National
Cancer Data Base. J Neurooncol. 40:151–160. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bondy ML, Scheurer ME, Malmer B, et al:
Brain tumor epidemiology: consensus from the Brain Tumor
Epidemiology Consortium. Cancer. 113:1953–1968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chargari C, Moncharmont C, Levy A, et al:
Cancer stem cells, cornerstone of radioresistance and perspectives
for radiosensitization: glioblastoma as an example. Bull Cancer.
99:1153–1160. 2012.(In French).
|
|
5
|
Warda M, Kim HK, Kim N, Ko KS, Rhee BD and
Han J: A matter of life, death and diseases: mitochondria from a
proteomic perspective. Expert Rev Proteomics. 10:97–111. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uchiumi T and Kang D: The role of
TFAM-associated proteins in mitochondrial RNA metabolism. Biochim
Biophys Acta. 1820:565–570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng Z and Ristow M: Mitochondria and
metabolic homeostasis. Antioxid Redox Signal. Mar 28–2013.(Epub
ahead of print).
|
|
8
|
Li X, Zhang G, Luo F, et al:
Identification of aberrantly expressed miRNAs in rectal cancer.
Oncol Rep. 28:77–84. 2012.PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nikaki A, Piperi C and Papavassiliou AG:
Role of microRNAs in gliomagenesis: targeting miRNAs in
glioblastoma multiforme therapy. Expert Opin Investig Drugs.
21:1475–1488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Phang JM and Liu W: Proline metabolism and
cancer. Front Biosci. 17:1835–1845. 2012. View Article : Google Scholar
|
|
12
|
Jin L, Wessely O, Marcusson E, Ivan C,
Calin G and Alahari SK: Pro-oncogenic factors miR-23b and miR-27b
are regulated by Her2/Neu, EGF, and TNFα in breast cancer. Cancer
Res. 73:2884–2889. 2013.PubMed/NCBI
|
|
13
|
Yoshida T, Azuma H, Aihara K, et al:
Vascular smooth muscle cell proliferation is dependent upon
upregulation of mitochondrial transcription factor A (mtTFA)
expression in injured rat carotid artery. Atherosclerosis.
178:39–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim AJ, Jee HJ, Song N, Kim M, Jeong SY
and Yun J: p21WAF1/C1P1 deficiency induces mitochondrial
dysfunction in HCT116 colon cancer cells. Biochem Biophys Res
Commun. 430:653–658. 2013.PubMed/NCBI
|
|
15
|
Han B, Izumi H, Yasuniwa Y, et al: Human
mitochondrial transcription factor A functions in both nuclei and
mitochondria and regulates cancer cell growth. Biochem Biophys Res
Commun. 408:45–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo J, Zheng L, Liu W, et al: Frequent
truncating mutation of TFAM induces mitochondrial DNA depletion and
apoptotic resistance in microsatellite-unstable colorectal cancer.
Cancer Res. 71:2978–2987. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ishteiwy RA, Ward TM, Dykxhoorn DM and
Burnstein KL: The microRNA -23b/-27b cluster suppresses the
metastatic phenotype of castration-resistant prostate cancer cells.
PLoS One. 7:e521062012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zaman MS, Thamminana S, Shahryari V, et
al: Inhibition of PTEN gene expression by oncogenic miR-23b-3p in
renal cancer. PLoS One. 7:e502032012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geng J, Luo H, Pu Y, et al: Methylation
mediated silencing of miR-23b expression and its role in glioma
stem cells. Neurosci Lett. 528:185–189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Loftus JC, Ross JT, Paquette KM, et al:
miRNA expression profiling in migrating glioblastoma cells:
regulation of cell migration and invasion by miR-23b via targeting
of Pyk2. PLoS One. 7:e398182012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen L, Han L, Zhang K, et al: VHL
regulates the effects of miR-23b on glioma survival and invasion
via suppression of HIF-1α/VEGF and β-catenin/Tcf-4 signaling. Neuro
Oncol. 14:1026–1036. 2012.PubMed/NCBI
|
|
22
|
Berg M and Soreide K: EGFR and downstream
genetic alterations in KRAS/BRAF and PI3K/AKT pathways in
colorectal cancer: implications for targeted therapy. Discov Med.
14:207–214. 2012.PubMed/NCBI
|
|
23
|
Papadimitrakopoulou V: Development of
PI3K/AKT/mTOR pathway inhibitors and their application in
personalized therapy for non-small-cell lung cancer. J Thorac
Oncol. 7:1315–1326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Luca A, Maiello MR, D’Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16(Suppl 2):
S17–S27. 2012.PubMed/NCBI
|
|
25
|
Rietz A and Spiers J: The relationship
between the MMP system, adrenoceptors and phosphoprotein
phosphatases. Br J Pharmacol. 166:1225–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.PubMed/NCBI
|