Introduction
In our previous research, we showed that
susceptibility to 4-nitroquinoline 1-oxide (4NQO)-induced rat
tongue carcinogenesis is a polygenic trait involving a number of
susceptibility and resistance quantitative trait loci (QTLs)
(1,2). Dark Agouti (DA) rats are highly
susceptible to 4NQO-induced tongue cancer (TC), whereas
Wistar-Furth rats (WF) are very resistant (3,4). A
genome-wide association study in F2 progeny of these 2 strains
revealed 5 significant QTLs, namely Tscc1-5 (Tcas1-5
in the Rat Genome Database), which account for differences in
susceptibility to TC (2,5,6).
In the present study, we focused on the single-locus
effects of the tongue cancer susceptibility QTL 3 (Tcas3) on
rat chromosome 4 (RNO4) by constructing reciprocal speed congenic
strains; WF rats carrying a DA-derived Tcas3
(Tcas3DA) chromosomal segment and DA rats
carrying a WF-derived Tcas3
(Tcas3WF) chromosomal segment. The
modifier effects of Tcas3 on 4NQO-induced tongue
carcinogenesis were confirmed by allele type-dependent incidence of
TCs and frequent deletion of Tcas3WF in
TCs in (DA × WF) F1 rats. Subsequently, we identified genes
responsible for these modifying effects from the Tcas3
region by comparing microarray analyses of TCs and normal tongue
tissues, revealing a significant elevation in parathyroid
hormone-like hormone (Pthlh) and Kras2 expression.
Immunohistochemistry of TCs showed that the Pthlh signal was
more intense than the Kras2 signal. Subsequent sequencing of
DNA showed 3 unique single-nucleotide polymorphisms (SNP) in the WF
Pthlh gene. Rats carrying the
Tcas3DA and 4NQO-induced TCs had elevated
serum Pthlh and Ca2+ levels and their WF
Pthlh gene carried 3 single-nucleotide polymorphisms not
found in other rat strains. These findings suggest that
Pthlh is a promising candidate gene, located at Tcas3,
involved in the development and progression of rat TC.
Materials and methods
Animals
The following strains of rats and their F1 (DA × WF)
progeny were used in the present study. Dark Agouti/SIc rats were
purchased from Japan SLC Inc. (Hamamatsu, Japan); Fisher 344/DuCrj
(F344) rats were from Charles River Japan Inc. (Atsugi, Japan);
Sprague-Dawley/JcI (SD) rats were from Japan Clea Co. (Tokyo,
Japan); and DRH rats were from SEAC Yoshitomi (Fukuoka, Japan).
Long Evans/Stm (LE) rats were introduced from the Saitama Cancer
Center (Ina, Japan); ACI/Ms (ACI) rats were from the National
Institute of Genetics (Mishima, Japan); and Donryu rats were from
Osaka University (Osaka, Japan). The Wistar-Furth/Stm (WF) rats
were originally from Hiroshima University (Hiroshima, Japan). The
present study was carried out according to the Animal Care
Guidelines of Asahi University, Gifu, Japan (10-007).
Generation of speed congenic lines
Speed congenic strains for the Tcas3 locus
were developed using marker-assisted selection starting from an
intercross between DA and WF rats (7,8). The
chromosomal segment containing Tcas3 was defined by
microsatellite markers D4Rat206 and D4Rat72. A male
rat of F1 with DA-derived Tcas3
(Tcas3DA) from an F1 × WF cross was
selected and mated with WF females (8,9). After
serial selective backcrossing with WF rats over 6 generations,
heterozygous littermates were mated. Then a pair of littermates
that were homozygous for Tcas3DA were
selected and subjected to successive inbreeding. The resulting
strain was designated WF.DA-Tcas3. The reciprocal congenic
strain DA.WF-Tcas3 was generated using the same mating
protocol. All congenic rats used in the present study were of the
third or subsequent generation of Tcas3 homozygotes.
Genotype analysis for speed congenic
lines
All primers for PCR-based microsatellite analysis
were purchased from Research Genetics, Inc. (Huntsville, AL, USA).
We used 135 of the 360 polymorphic markers between DA and WF rats
to characterize congenic strains. Of these, 40 were found on RNO4
and the others were distributed 10–30 cM apart on each rat
chromosome (5,6). Theoretically, the percentage of DA/WF
segments was <1%. PCR and agarose electrophoresis of PCR
products were performed as described previously (1,2,10).
Tongue cancer induction
A stock solution of 4NQO (Nacalai Tesque Inc.,
Kyoto, Japan) at a concentration of 200 mg/l in 5% ethanol was
prepared and stored at 4°C until use. Starting at 6 weeks of age,
all rats were given drinking water containing 0.001% 4NQO ad
libitum from 5 p.m. to 9 a.m. Rats were inspected once a day
for TC development and were weighed once a week. All of the rats
given 4NQO (DA, WF, F1 and congenic rats) were sacrificed when they
became moribund or on day 180 of the experiment. Full necropsy and
histopathological examinations were performed. The diameter of the
largest TC tumor (DTCmax) and the number of TCs with a diameter ≥5
mm (TC#5) were recorded at necropsy. Paired samples of the largest
TC and kidney or tail from each rat were obtained and stored at
−80°C. High-molecular-weight DNA was extracted from the frozen
tissues as previously described (1,2).
Loss of heterozygosity (LOH)
analysis
LOH in the Tcas3 region, including that of
Kras2 (RGD ID: 5036392) and Pthlh (RGD ID: 11187)
genes of induced TCs in F1 rats was determined using PCR-based
microsatellite analysis (1,2). Fluorescently tagged primers were
purchased from Applied Biosystems (Foster City, CA, USA). Positions
of loci were mapped on the Rat Genome Database (6). LOH analysis was performed for tumor
samples from the F1 progeny of DA and WF rats as previously
described (11,12). Genomic DNA from TCs of 100
reciprocal F1 rats (50 females and 50 males) was used to survey
LOH. Samples were scanned using a PRISM 310 Genetic Analyzer, and
the data were collected automatically and analyzed using GeneScan
software (both from Applied Biosystems). A Genotyper 2.0 was used
to score alleles and assess LOH. A difference between the alleles
was expressed as the ratio of the tumor signal to the normal signal
(T1/T2 over N1/N2). Ratios <0.67 or >1.35 were considered
indicative of LOH for that locus (12).
Microarray analysis
TCs >5 mm in diameter and normal tissues from F1
rats were analyzed using microarrays. RNA was isolated from the
tissues and subjected to linear amplification by RiboAmp RNA
Amplification kit (Arcturus, Mountain View, CA, USA). RNA
amplification efficiency was compared with that of control RNA of
known quantity (0.1 μg) by running in parallel with the 18 samples.
Gene expression analysis was performed using the
GeneChip® Rat Gene 1.0 ST Array (Affymetrix Inc., Santa
Clara, CA, USA) technique according to the manufacturer’s protocol.
The arrays were scanned, and fluorescence intensity was measured
using Microarray Suite v5.0 software (Affymetrix Inc.). The arrays
were then imported into DNA-Chip analyzer software for
normalization and model-based analysis. All statistical analyses
were performed using S-Plus 6.0 (Insightful, Seattle, WA, USA)
software. Three criteria were applied to determine differentially
expressed gene transcripts as follows. First probe sets on the
array that were assigned as ‘absent’ call in all samples. Second, a
two-tailed Student’s t-test was used for comparison of the average
gene expression signal intensity between the F1 TCs (TC, 5–10 mm;
n=15) and normal samples (n=3), and differences were considered
significant at a critical α level of 0.05. Finally, fold ratios
were calculated for gene transcripts that showed a statistically
significant difference (P<0.05), and only gene transcripts that
exhibited >2-fold changes were included in further analyses.
Single-strand conformation polymorphism
(SSCP) analysis and sequencing in F1 TC rats
To detect mutations in genes of the Tcas3
region including Pthlh and Kras2, SSCP analysis was
performed in tumor samples from F1 progeny of DA and WF rats, as
previously described (12,13). Samples with altered electrophoretic
mobility were re-amplified, and direct sequencing of both strands
was performed to confirm and characterize mutations.
Quantitative real-time PCR
Total RNA was extracted from the tumors and normal
tongue mucosa from 4NQO-induced TCs from 50 F1 rats. Tissue
specimens were homogenized using an RNAqueous kit (Ambion, Grand
Island, NY, USA). cDNA was generated using a High Capacity cDNA
Transcription kit (Ambion) and was amplified by PCR using a TaqMan
Universal PCR Master Mix and TaqMan Gene Expression assays with 18S
(Rn03928990-g1), Pthlh (Rn00561818-g1) and Kras2
(Rn00580460-m1) (Applied Biosystems). PCR products were analyzed
using Applied Biosystems StepOne™ with Gene Amp software and
StepOne real-time PCR systems (13).
Direct sequence analysis of Pthlh in 8
rat strains
To detect SNPs in rat Pthlh (NC-005103.3,
ID24695, M34108), genomic DNA was obtained from the rat kidney, and
the entire sequence of the Pthlh locus was determined in 8
rat strains using an ABI 310 Genetic Analyzer (Applied Biosystems,
Foster City, CA, USA). Allele calling was performed using the
Genotyper 2.0 software (Applied Biosystems), and identified SNPs
were compared with those previously reported (12–15).
To determine whether SNPs in translational regions
led to structural or functional amino acid substitutions in the
Pthlh protein, we conducted in silico analysis using
freely available software, including MutPred (16), Panther (17), PhD-SNP (18), SNAP (19), PolyPhen2 (20) and SIFT (21). In addition, we evaluated the impact
of SNPs on transcription factor-binding sites by searching for
potential regulatory motifs at the 5′ UTR using Matlnspector
(22) and TESS (23). The software default values were used
in all bioinformatic analyses.
Plasma electrolytes, serum Pthlh-related
proteins and cytokines
Plasma electrolytes including Ca2+,
Na+, K+, IP and Cl− were
determined at Japan SLC, Inc. (24). The serum levels of Pthlh-N (normal
<3.9 pmol/l), Pthlh-intact (normal <1.1 pmol/l) and Pthlh-C
(normal <55.3 pmol/l), and interleukin (IL)-6, -8 and -11 (SRL
Inc., Tokyo, Japan) were also determined (25).
Immunohistochemistry of tongue
cancer
All tongue specimens from F1 rats were routinely
fixed in buffered 10% formalin and were embedded in paraffin.
Serial 4-μm sections were dewaxed in xylene and rehydrated in
graded ethanol. Sections were stained with hematoxylin and eosin
(H&E) to confirm histological diagnosis, and separate sections
were used for immunohistochemical analyses. Sections were incubated
with diluted rat polyclonal anti-PTHrP antibody (sc-9685; 1:100)
and mouse monoclonal anti-Kras2A antibody (sc-13794; 1:50) (both
from Santa Cruz Biotechnology, Inc., USA) as primary antibodies
overnight at 4°C.
Statistical analysis
Numerical data are presented as means ± standard
deviation (SD). Statistical analyses were performed using one-way
ANOVA or Fisher’s test with SPSS 17.0 (SPSS Inc., Chicago, IL, USA)
software.
Results
Generation of speed congenic rat strains
for Tcas3
To study the phenotypic effects of the single
Tcas3 locus on TC development, we established a set of speed
congenic strains for Tcas3 using marker-assisted
backcrossing of DA and WF rats and generated the WF.DA-Tcas3
and DA.WF-Tcas3 strains, respectively (8,9).
WF.DA-Tcas3 rats had an introgressed DA-derived RNO4 segment
spanning D4Rat140 to D4Rat70, whereas DA.WF-Tcas3 rats
carried a WF-derived RNO4 segment spanning D4Rat112 to D4Rat70
(Fig. 1). Both segments were ~6-cM
long, were partly overlapping and contained several cancer-related
genes including Pthlh and Kras2 (Fig. 1).
Effects of Tcas3 on tongue
carcinogenesis
We previously reported tumor incidence, number and
size in descending order in DA, F1 and WF rats (1,2,12). To
evaluate the effect of introgressed Tcas3 segments, rats of
both congenic strains, parental DA and WF strains and F1, were
administered 4NQO, and tongue and oral carcinogenesis was observed.
As shown in Fig. 2, DA rats
developed TCs with the shortest latency and highest incidence,
whereas WF rats developed TCs with the longest latency and lowest
incidence. In congenic strains, Tcas3DA
showed modest but significant accelerating effects on TCs in WF
rats, whereas Tcas3WF had TC-inhibitory
effects on DA rats. These differences in strains were reflected by
incidence, number and size of tumors (Table I). These observations clearly
indicate that Tcas3 alone exhibits significant phenotypic
effects on the development and growth of TCs and oral cancers. That
is Tcas3DA augmented grown whereas
Tcas3WF suppressed it.
 | Table I4NQO-induced rat tongue and other
cancers in the WF, F1, congenic and DA rats. |
Table I
4NQO-induced rat tongue and other
cancers in the WF, F1, congenic and DA rats.
| WF (n=55) | Congenic
WF.DA-Tcas3 (n=30) | F1 (n=100) | Congenic
DA.WF-Tcas3 (n=30) | DA (n=55) |
|---|
| Rats with TC#5
(incidence, %) | 5.5 (3/55) | 20 (6/30)b | 50 (50/100)c | 73.3
(22/30)c | 94.6
(52/55)d |
| No. of TC#5a | 0.23±0.22 | 0.33±0.12b | 0.61±0.71c | 0.68±1.20c | 1.28±1.56d |
| Total no. of
TCsa | 0.52±0.82 | 0.75±0.58b | 0.92±0.86c | 1.01±0.76c | 1.59±0.66d |
| DTCmax (mm) | 1.18±1.63 | 2.12±3.54b | 5.15±1.85c | 5.89±1.17c | 12.2±4.49d |
| Total no. of
cancersa | 1.26±1.45 | 1.39±5.46b | 1.56±1.67c | 2.89±1.64c | 3.45±1.58d |
Selective LOH of Tcas3WF in
larger TCs
To determine whether suppressive
Tcas3WF was selectively deleted in TCs,
genomic DNA of TCs induced in F1 rats was examined for LOH. Out of
100 F1 rats, 10 TCs of <5-mm in diameter, 29 TCs of 5–10 mm in
diameter and 40 non-TC tissues did not show any LOH. However, in 21
TCs of ≥10-mm diameter, LOH was observed between D4Mgh10 and
D4Mgh13 on chromosome 4q44. In the majority of cases, the WF allele
was selectively lost (Fig. 3).
Among the major cancer-related genes on the Tcas3 segment,
the incidence of LOH at Kras2 and Pthlh was 32.7 and
40.8%, respectively in the TC#5 (Table
II). Loss of the Tcas3WF allele only
in larger TCs indicates that it represents a late event in tumor
progression.
 | Table IIImmunostaining of the PTHrP protein
in tongue cancer (TC) samples from F1 rats. |
Table II
Immunostaining of the PTHrP protein
in tongue cancer (TC) samples from F1 rats.
| Rat case no. | DTCmax (mm) | TC#5 |
Microsatellite markersa on RNO4 |
|---|
|
|---|
| Mgh30 | Mgh10 | ENO2 | Mit27 | Rat140 | Kras2 | Got155 | Pthlh | Rat70 | Rat72 | Mgh13 |
|---|
| 30 | 10 | 2 | - | - | - | - | - | - | - | WF | - | - | - |
| 31 | 12 | 1 | - | - | - | - | - | - | - | WF | - | - | - |
| 32 | 12 | 2 | - | - | - | - | - | - | WF | WF | - | - | - |
| 33 | 13 | 1 | - | - | - | - | - | - | WF | WF | - | - | - |
| 34 | 13 | 1 | - | - | - | - | - | WF | WF | WF | - | - | - |
| 35 | 14 | 1 | - | - | - | - | - | WF | WF | WF | - | - | - |
| 36 | 14 | 2 | - | - | - | - | - | WF | WF | WF | - | - | - |
| 37 | 15 | 2 | - | - | - | - | - | WF | NI | WF | - | - | - |
| 38 | 15 | 2 | - | - | - | - | - | WF | WF | WF | WF | - | - |
| 39 | 15 | 2 | - | - | - | - | - | WF | WF | WF | WF | - | - |
| 40 | 16 | 1 | - | - | - | - | - | NI | WF | WF | WF | - | - |
| 41 | 17 | 2 | - | - | - | - | - | WF | WF | WF | WF | - | - |
| 42 | 18 | 1 | - | - | - | - | - | WF | WF | WF | WF | - | - |
| 43 | 19 | 2 | - | - | - | - | - | DA | DA | DA | WF | WF | - |
| 44 | 20 | 3 | - | - | - | - | WF | WF | WF | WF | WF | WF | - |
| 45 | 21 | 2 | - | - | - | - | WF | WF | WF | WF | WF | WF | - |
| 46 | 22 | 2 | - | - | - | - | WF | WF | WF | NI | WF | WF | - |
| 47 | 23 | 2 | - | - | - | - | WF | WF | WF | WF | WF | WF | - |
| 48 | 24 | 2 | - | - | - | - | WF | WF | WF | WF | WF | WF | - |
| 49 | 25 | 2 | - | - | - | WF | WF | WF | WF | WF | WF | WF | WF |
| 50 | 25 | 3 | - | WF | WF | WF | WF | WFb | WF | WF | WF | WF | WF |
| Incidence | | | 0/50 | 1/50 | 1/50 | 2/50 | 7/50 | 16/49 | 18/49 | 20/49 | 13/50 | 8/50 | 2/50 |
| Total (%) | | | 0 | 2.0 | 2.0 | 4.0 | 14.0 | 32.7 | 36.7 | 40.8 | 26.0 | 16.0 | 4.0 |
Microarray analysis
To identify genes that are responsible for the
effects of Tcas3, microarray analysis was performed in TCs
and normal tongue tissues from F1 rats. According to the Rat Genome
Database (6), 27 genes, 6 ESTs and
18 pseudogenes are present in the Tcas3 region. Table III shows the expression of the 27
genes as determined by microarray analysis. Significantly increased
expression was observed for Pthlh (7.33-fold, P<0.001)
and slightly less for Kras2 (5.21-fold, P<0.001).
 | Table IIIMicroarray analysis of the
Tcas3 region between tongue cancer and normal tissues in F1
rats. |
Table III
Microarray analysis of the
Tcas3 region between tongue cancer and normal tissues in F1
rats.
| Symbol | Accession no. | Gene name | Fold change | P-value |
|---|
| Abcc9 | NM-013040.2 | ATP-binding
cassette, subfamily C (CFTR/MRP), member 9 | −1.58 | 0.413 |
| Aebp2 | NM-001106626.1 | AE binding
protein 2 | 0.23 | 0.625 |
| Bcat1 | NM-017253.2 | Branched chain
amino acid transaminase 1, cytosolic | −2.11 | 0.073 |
| Bicd1 | NM-001108653.1 | Bicaudal D
homolog 1 | 2.31 | 0.071 |
| Capza3 | NM-017164.1 | Capping protein
(actin filament) muscle Z-line, α3 | −2.72 | 0.061 |
| Casc1 | BG377837.1 | Cancer
susceptibility candidate 1 | 2.29 | 0.085 |
| Cmas | NM-001009419.1 | Cytidine
monophosphate N-acetylneuraminic acid synthetase | 1.71 | 0.561 |
|
Fgfr1op2 | NM-201421 | FGFR1 oncogene
partner 2 | 2.89 | 0.051 |
| Gys2 | NM-013089.1 | Glycogen
synthase 2 | 1.12 | 0.761 |
| Iapp | NM-012586 | Islet amyloid
polypeptide | 0.56 | 0.982 |
| Itpr2 | NM-031046.3 | Inositol
1,4,5-trisphosphate receptor, type 2 | 1.26 | 0.781 |
| Kcnj8 | NM-017099.4 | Potassium
inwardly-rectifying channel, subfamily J, member 8 | 1.54 | 0.564 |
| Kras2 | NM-031515.1 | Kirsten rat
sarcoma viral oncogene homologue 2 | 5.21 | <0.001 |
| Ldhb | NM-012595.1 | Lactate
dehydrogenase B | 1.89 | 0.647 |
| Mgst1 | NM-134349.2 | Microsomal
glutathione S-transferase 1 | −2.54 | 0.062 |
| Pde3a | NM-017337.1 |
Phosphodiesterase 3A, cGMP
inhibited | 1.98 | 0.653 |
| Pik3c2g | NM-053923.1 |
Phosphatidylinositol-4-phosphate
3-kinase, catalytic subunit type 2γ | 2.01 | 0.076 |
| Plekha5 | AI009219.1 | Pleckstrin
homology domain-containing protein family A, member 5 | 1.51 | 0.711 |
| Pthlh | NM-012636.1 | Parathyroid
hormone-like peptide | 7.33 | <0.001 |
| Ptpro | NM-017336.1 | Protein tyrosine
phosphatase, receptor type, O | 1.09 | 0.881 |
| Rassf8 | NM-001191753 | Ras association
domain family (N-terminal) member 8 | 2.97 | 0.051 |
| Slc21a6 | NM-130736.1 | Solute carrier
organic anion transporter family, member 1a6 | 0.12 | 0.979 |
| Slco1a2 | NM-030838.1 | Solute carrier
organic anion transporter family, member 1A2 | 0.18 | 0.957 |
| Slco1c1 | NM-053441.1 | Solute carrier
organic anion transporter family, member 1c1 | 0.16 | 0.969 |
| Sox5 | NM-001014060 | SRY (gender
determining region Y)-box 5 | 1.11 | 0.652 |
| St8sia1 | NM-012813.2 | ST8
α-N-acetyl-neuraminide α-2,8-sialyltransferase 1 | 1.87 | 0.489 |
| Tm7sf3 | NM-001011970.1 | Transmembrane 7
superfamily member 3 | −2.32 | 0.065 |
The expression of genes in the Tcas3 region
was analyzed in 10 control and 60 TC samples using quantitative
real-time RT-PCR. Pthlh mRNA expression in large TCs (>10
mm) was consistently >30-fold higher than that in normal tongue
mucosa (Fig. 4), and expression
levels were higher in larger tumors.
Sequencing of cancer-related genes in the
Tcas3 segment
Pthlh and Kras2 were further examined
by sequencing germline and/or tumor DNA. Direct sequencing of the
germline Pthlh gene was performed for 8 laboratory rat
strains: DA, LE, SD, ACI, Fischer 344, Donryu, DRH and WF. The
Pthlh gene in resistant WF rats was found to carry 3 SNPs at
positions +2 bp (T→G), +17 bp (T→C) and +1485 (A→C) from the 5′ end
of exon 1, but these were not observed in the other strains
(Fig. 5). The base change at
position 2 and 17 does not cause amino acid substitutions, whereas
that at position +1485 [496 bp in the coding sequence (CDS)] bp
would substitute the polar residue threonine with the hydrophobic
residue proline at the 166th amino acid (ACC→CCC) of the precursor
protein (position 130th of the mature protein) in which the CDS
start is at 527 bp.
The potential impact of the 3 SNPs was determined
using computational analyses. Since 2 SNPs at +2 bp (T→G) and +17
bp (T→C) were found in the untranslated region of the Pthlh
gene, we searched for known transcription factor-binding sites
within the −950 to 526 bp region. TCF-4E (CTTTGCA) and ZFX
(GAGGCCTGGTG) motifs were found at −1 to +6 bp and +11 to +21 bp,
respectively, in sequences from DA, LE, SD, ACI, F344, Donryu and
DRH rats in which the +2 bp and 17 bp positions were occupied by T.
In the sequence from WF rats, PLU1-JARID1B.01 (TGGCTGTGC), SP1
(TGTGC) and ROAZ.01 (GTGCACCCAGAGGCCCG) motifs were found at −4 to
+5 bp, +1 to +5 bp and +2 to +8 bp, respectively, with G and C
residing at +2 bp and +17 bp, respectively. The agreement rates in
matrices of sequence motifs were 100, 98.8, 96.6, 100 and 73.1%,
respectively.
Using computational analyses, we evaluated the
impact of the 166th amino acid substitution on the structure and
function of the Pthlh protein. MedPred predicted a low impact, with
a probability of deleterious mutation score of 0.134 (>0.5
indicates impact). Panther also predicted that the substitution was
almost neutral, with a deleterious probability of 0.34315 (>0.5
indicates impact) and PhD-SNP predicted no impact. SNAP also
predicted that the impact was neutral with 85% expected accuracy.
Albeit with low confidence, SHIFT gave a score of 0 for affected
protein function. Polyphen2 did not yield a prediction. Thus, all
bioinformatics tools revealed almost no impact of these SNPs on
Pthlh protein structure and function.
Subsequently, Kras2 point mutations were
examined at codons 12, 13 and 61 in F1 TCs. One of the tumors had a
heterozygous CAA→CAT mutation in codon 61 of Kras2,
resulting in an amino acid substitution from glutamine to
histidine. No other activating mutations were found in the
Kras2 gene. Therefore, any involvement of Kras2 may
not be predominant in rat TCs.
Plasma electrolyte and antibody levels in
the WF, congenic and DA rats
Table IV shows the
titer of plasma electrolytes and immunoreactive PTHrP in control
and TC-bearing rats. Electrolyte levels in control rats did not
vary according to genetic background and Tcas3 genotype.
However, plasma Ca2+ levels were consistently higher in
TC-bearing rats than in control rats. We also evaluated levels of
PTHrP peptides derived from post-translational cleavage of the
whole protein. The PTHrP-C peptide levels were significantly
elevated in TC-bearing rats, whereas levels of PTHrP-N and
PTHrP-intact were unchanged.
 | Table IVPlasma electrolyte and antibody
levels in the WF, congenic and DA rats. |
Table IV
Plasma electrolyte and antibody
levels in the WF, congenic and DA rats.
| Rat | Cancer | No. of rats | Na+
(mEq/l) | K+
(mEq/l) | Ca2+
(mEq/l) | IP (mg/dl) | Cl−
(mEq/l) | PTHrP N-terminal
(pmol/l) | PTHrP intact
(pmol/l) | PTHrP
C-terminal |
|---|
| WF | No | 52 | 139.9±0.72 | 2.89±0.41 | 5.58±0.11 | 2.68±1.56 | 101.1±0.57 | <3.9 | <1.1 | 55.4±0.45 |
| TC | 3 | 140.1±0.22 | 2.92±0.26 | 6.71±0.16 | 2.89±0.66 | 102.3±0.98 | <3.9 | <1.1 | 68.4±1.76a |
|
WF.DA-Tcas3 | No | 24 | 140.6±0.53 | 2.91±0.35 | 5.59±0.27 | 2.91±0.67 | 101.3±0.39 | <3.9 | <1.1 | 57.4±0.55 |
| TC | 6 | 142.1±0.12 | 3.07±0.86 | 6.81±0.32a | 3.59±0.48 | 102.3±0.11 | <3.9 | <1.1 | 70.4±3.76b |
|
DA.WF-Tcas3 | No | 8 | 139.1±0.13 | 2.97±0.17 | 5.51±0.17 | 3.39±0.67 | 101.2±0.09 | <3.9 | <1.1 | 58.4±1.77 |
| TC | 22 | 142.2±0.63 | 3.05±0.85 | 6.91±0.17a | 4.27±4.49a | 102.8±0.19 | <3.9 | <1.1 | 131.4±0.47b |
| DA | No | 3 | 136.1±0.15 | 2.56±0.57 | 5.59±0.24 | 2.15±1.38 | 100.1±0.10 | >3.9 | >1.1 | 74.4±0.48 |
| TC | 52 | 137.8±0.46 | 3.06±0.68 | 7.89±0.65a | 5.45±1.59b | 103.4±0.61a | >3.9 | >1.1 | 149.4±0.88b |
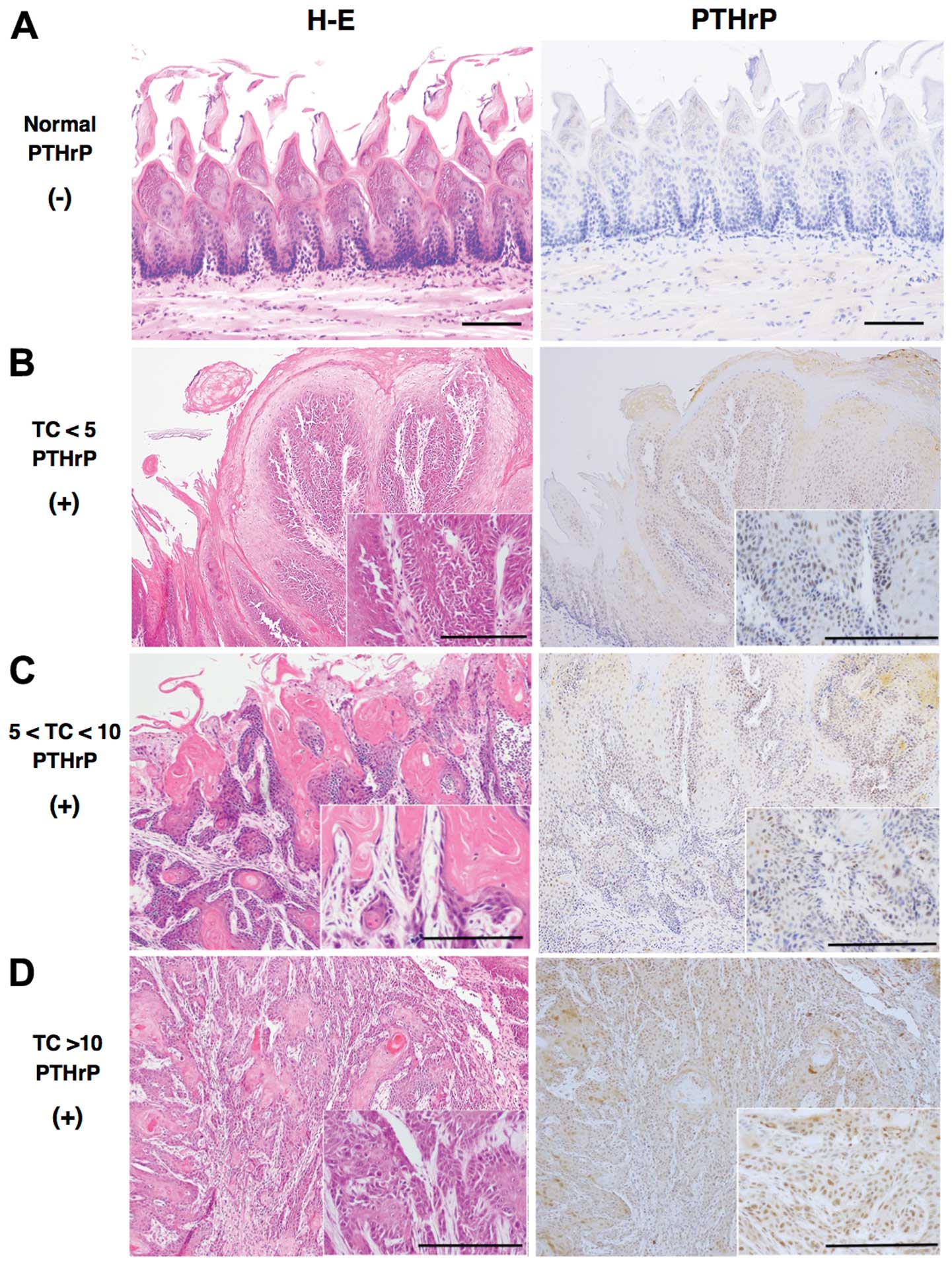
TC immunohistochemistry
Among 50 TC#5 and 10 rats with TCs of <5 mm in F1
rats, positive signals for PTHrP were detected in the nucleus and
cytoplasm of prickle-type cancer cells (Fig. 6). Staining intensity above or below
the cut-off score (10%) was classified as ‘positive’ or ‘negative’,
respectively, at a magnification of ×100 using computer-associated
image analyzer software (WinROOF 7.1; Mitani Co., Japan) (26). As shown in Table V, the fraction of positively stained
TCs increased with their size. There were significant differences
in TCs between the TC#5 and DTCmax groups (P<0.001), and among
the TCs <5 mm, 5–10 mm and TC >10 mm
(P<1×10−10, χ2=47.63). The expression of
Kras2 was consistently weaker than that of PTHrP (data not
shown).
 | Table VImmunostaining of the PTHrP protein
in tongue cancer (TC) samples from F1 rats. |
Table V
Immunostaining of the PTHrP protein
in tongue cancer (TC) samples from F1 rats.
| Positive | Negative |
|---|
| Rat normal tissue
(n=10) | 0 | 10 |
| TCs <5 mm
(n=10) | 1 | 9 |
| TCs 5–10 mm
(n=29) | 24 | 5 |
| TCs >10 mm
(n=21) | 21 | 0 |
Discussion
Out of 5 QTLs that affect susceptibility to TCs in
4NQO-treated rats, we focused on Tcas3 in the present study.
In genome-wide association studies in the F2 rat, the peak
logarithm of the odds score 6.88 was observed for Tcas3 at
4q44 (2). This chromosomal region
is homologous to human 12p12.1-q11.2 and harbors the cancer-related
genes Pthlh and Kras2(7,12). To
evaluate the effects of a single Tcas3, we generated speed
congenic strains for Tcas3 using marker-assisted
backcrossing.
As shown in Table I
and Fig. 2, the quantitative
parameters of carcinogenesis, namely tumor latency, incidence,
tumor number and size, were moderately but significantly modified
by Tcas3. Hence the Tcas3DA allele
increased susceptibility to tongue carcinogenesis and progression,
whereas the Tcas3WF allele bestowed
resistance. Selective loss of the resistance allele
Tcas3WF was frequent in larger TCs induced
in F1 rats, and the consensus stretch of LOH contained Pthlh
and Kras2. Microarray comparisons of TCs and normal tongue
mucosa also indicated that among genes in the Tcas3 region
these 2 genes were significantly highly expressed. We focused on
Pthlh following the observation that resistant WF
Pthlh rats carry 3 unique SNPs. The impact of these SNPs
will be discussed in greater detail below. Other observations that
suggest the relevance of Pthlh in tongue carcinogenesis
include (i) a 30-fold higher expression of Pthlh in TCs than
that in normal tongue epithelium, (ii) very strong PTHrP
immonostaining in larger TCs from F1 rats and (iii) increased
plasma levels of Ca2+ and PTHrP-C in TC-bearing DA rats
when compared with the control rats.
The anabolic effects of Pthlh have been
demonstrated in rodents and in humans. Pthlh is expressed in
many cell types and usually acts as an autocrine, paracrine, and/or
intracrine factor that plays numerous roles in embryonic
development and physiology (27,28).
PTHrP may also have important functions in tumor development. PTHrP
has been shown to stimulate proliferation and invasiveness of
cancer cells as well as protection from apoptosis. PTHrP has the
potential to cause malignant hypercalcemia and induce local
osteolysis, which facilitates the growth of tumor cells that have
metastasized to bone (28,29). Partial homology of PTHrP and
parathyroid hormone allows PTHrP to activate parathyroid hormone 1
receptor (PTH1R). PTH1R activation in bone and kidney leads to bone
absorption and renal calcium retention, respectively, inducing a
rise in blood calcium levels (30).
PTHrP consists of 3 molecular domains, the
parathyroid hormone-like domain in the N-terminal region, a
mid-region and a C-terminal domain. Post-translational cleavage of
the PTHrP protein allows these domains to function independently.
The parathyroid hormone-like domain stimulates protein kinase A,
protein kinase C, and/or the calcium-dependent pathways by
activating PTH1R (31,32). The mid-region domain can enter the
nucleus since it carries a bipartite nuclear localization sequence
at residues 88–91 and 102–106 and an importin β-binding site at
residues 66–94 (33). After
translocation into the nucleus, the PTHrP mid-region domain
modulates gene expression by an as yet undefined mechanism. Nuclear
transport of the mid-region domain is regulated by
CDK1(CDC2)/CDK2-dependent phosphorylation (34). The C-terminal domain of PTHrP
(osteostatin) physically interacts with β-arrestin (35), which regulates internalization and
desensitization of ligand-stimulated G-protein-coupled receptors
(36). Osteostatin bears a number
of phosphorylation sites that are important for the mitogenic
activity of PTHrP in vascular smooth muscle cells (37).
To understand the potential impact of SNPs in
untranslated regions of PTHrP genes, we performed in silico
analyses using computational tools with various prediction
algorithms and found several known regulatory motifs on the 5′-UTR
of the Pthlh sequence. These analyses identified only 2
transcription factors, TCF-4E and ZFX, in all strains apart from
WF, indicating that the SNPs at +2 bp (T→G) and +17 bp (T→C)
eliminate the transcriptional factor binding site. In contrast,
PLU1-JARID1B.01, SP1 and ROAZ.01 were found only in WF rats,
indicating that these 2 SNPs affect transcription factor binding to
these motifs. ZFX and ROAZ.01 are zinc finger proteins with a
DNA-binding motif, and SP1 is common to many eukaryotic
transcriptional regulatory pathways. The transcription factor
TCF-4E regulates expression of members of the Wnt pathway and its
splicing isoforms are related to the progression of renal cell
carcinoma (38). The motif
PLU1-JARID1B.01 was previously identified in the development and
progression of breast cancer (39).
However, no clear relationship between these transcription factors
and oral cancer development has been reported to date. Hence,
further research is necessary to understand the effect of these
motifs and SNPs at +2 and +17 bp. Using several bioinformatics
tools with varying predictive algorithms, we also analyzed
structural and functional effects of the SNP at +1485 bp, which
leads to an amino acid substitution at the 166th amino acid on the
Pthlh protein. Notably, none of these analyses suggested that this
SNP would have any effect. Potentially, these computational tools,
which are generated using characterized protein, may not predict
novel functional motifs, even though they yielded consistent
predictions. Benelli et al(15) reported an amino acid polymorphism in
Pthlh that elicited cancer-modifying effects in a mouse
squamous cell carcinoma cell line (15). Importantly, the SNP at position
+1485 [+496 bp in the coding sequence (CDS)] bp was common to both
mouse and rat models. This SNP can be expected to substitute the
polar residue threonine with the hydrophobic residue proline at the
166th amino acid (ACC→CCC) of the precursor protein (position 130th
of the mature protein), in which the CDS start is at 527 bp.
The present study revealed that Tcas3 alone
contributes moderate but significant phenotypic effects to
4NQO-induced rat tongue carcinogenesis. Several observations
suggest that Pthlh is a promising candidate gene in Tcas3,
although further studies using appropriate reporter assays and
in vivo transgenic analyses are required to provide direct
supporting evidence of this. In subsequent studies, we will focus
on structural and functional analyses of Pthlh and its
products.
Acknowledgements
The present study was supported by a grants-in-aid
for Scientific Research (C) from the Japan Society for Promotion of
Science (no. 24592850).
References
|
1
|
Tanuma J, Shisa H, Hiai H, et al:
Quantitative trait loci affecting 4-nitroquinoline 1-oxide-induced
tongue carcinogenesis in the rat. Cancer Res. 58:1660–1664.
1998.PubMed/NCBI
|
|
2
|
Tanuma J, Fujii K, Hirano M, et al: Five
quantitative trait loci affecting 4-nitroquinoline 1-oxide-induced
tongue cancer in the rat. Jpn J Cancer Res. 92:610–616. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kitano M, Hatano H and Shisa H: Strain
difference of susceptibility to 4-nitroquinoline 1-oxide-induced
tongue carcinoma in rats. Jpn J Cancer Res. 83:843–850. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kitano M, Hirayama Y, Tanuma J, et al:
Genetic controls of susceptibility and resistance to
4-nitroquinoline 1-oxide-induced tongue carcinomas in rats. Jpn J
Cancer Res. 87:1097–1101. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kwitek AE, Gullings-Handley J, Yu J, et
al: High-density rat radiation hybrid maps containing over 24,000
SSLPs, genes, and ESTs provide a direct link to the rat genome
sequence. Genome Res. 14:750–757. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
RGD-Rat Genome Database. http://rgd.mcw.edu/. May
1, 2011
|
|
7
|
Tanuma J, Hirano M, Hirayama Y, et al:
Genetic predisposition to 4NQO-induced tongue carcinogenesis in the
rat. Med Princ Pract. 14:297–305. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hirano M, Tanuma J, Hirayama Y, et al: A
speed congenic rat strain bearing the tongue cancer susceptibility
locus Tscc1 from Dark-Agouti rats. Cancer Lett. 231:185–191.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
NBRP. The National BioResource Project for
the Rat in Japan. NBRP Rat No. 0268http://www.nbrp.jp/.
Aug 10, 2010
|
|
10
|
Liu H, Higashi K and Hiai H: Role of
resistant Drh1 locus in chemical carcinogen-induced
hepatocarcinogenesis in rats: analysis with a speed congenic
strain. Cancer Sci. 96:164–169. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walentinsson A and Levan G: Ras
gene mutations in 7,12-dimethylbenz[a]anthracene (DMBA)-induced rat
sarcomas. Cancer Lett. 166:47–53. 2001. View Article : Google Scholar
|
|
12
|
Tanuma J, Hiai H, Shisa H, et al:
Carcinogenesis modifier loci in rat tongue are subject to frequent
loss of heterozygosity. Int J Cancer. 102:638–642. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogawa K, Tanuma J, Hirano M, et al:
Selective loss of resistant alleles at
p15INK4B and
p16INK4A genes in chemically-induced rat
tongue cancers. Oral Oncol. 42:710–717. 2006.PubMed/NCBI
|
|
14
|
Manenti G, Peissel B, Gariboldi M, et al:
A cancer modifier role for parathyroid hormone-related protein.
Oncogene. 19:5324–5328. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benelli R, Peissel B, Manenti G, et al:
Allele-specific patterns of the mouse parathyroid hormone-related
protein: influences on cell adhesion and migration. Oncogene.
22:7711–7715. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li B, Krishnan VG, Mort ME, et al:
Automated inference of molecular mechanisms of disease from amino
acid substitutions. Bioinformatics. 25:2744–2750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thomas PD, Campbell MJ, Kejariwal A, et
al: PANTHER: a library of protein families and subfamilies indexed
by function. Genome Res. 213:2129–2141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Capriotti E, Calabrese R and Casadio R:
Predicting the insurgence of human genetic diseases associated to
single point protein mutations with support vector machines and
evolutionary information. Bioinformatics. 22:2729–2734. 2006.
View Article : Google Scholar
|
|
19
|
Bromberg Y and Rost B: SNAP: predict
effect of non-synonymous polymorphisms on function. Nucleic Acids
Res. 35:3823–3835. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adzhubei IA, Schmidt S, Peshkin L, et al:
A method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ng PC and Henikoff S: Predicting
deleterious amino acid substitutions. Genome Res. 11:863–874. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cartharius K, Frech K, Grote K, et al:
Matlnspector and beyond: promoter analysis based on transcription
factor binding sites. Bioinformatics. 21:2933–2942. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schug J: Using TESS to transcription
factor binding sites in DNA sequence. Curr Protec Bioinfomatics.
21:2.6.1–2.6.15. 2008.
|
|
24
|
Laboratory animal data. Japan SLC Inc;
2007, http://jslc.co.jp. Feb 2,
2011
|
|
25
|
Fukunaga M, Eto S and Saito S:
Multicentric clinical studies on measurement of carboxyl-terminal
parathyroid hormone-related protein in normal subjects (the first
report). Clin Endocrinol. 40:977–986. 1992.
|
|
26
|
Takeda T, Sugihara K, Hirayama Y, et al:
Immunohistological evaluation of Ki-67, p63, CK19 and p53
expression in oral epithelial dysplasias. J Oral Pathol Med.
35:369–375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dittmer J: The importance of PTHrP for
cancer development. Gene Ther Mol Biol. 8:451–464. 2004.
|
|
28
|
Goltzman D, Karaplis AC, Kremer R and
Rabbani SA: Molecular basis of the spectrum of skeletal
complications of neoplasia. Cancer. 88(Suppl 12): 2903–2908. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Käkönen SM and Mundy GR: Mechanisms of
osteolytic bone metastases in breast carcinoma. Cancer. 97(Suppl
3): 834–839. 2003.PubMed/NCBI
|
|
30
|
Mannstadt M, Jüppner H and Gardella TJ:
Receptors for PTH and PTHrP: their biological importance and
functional properties. Am J Physiol. 277:F665–F675. 1999.PubMed/NCBI
|
|
31
|
Maioli E and Fortino V: The complexity of
parathyroid hormone-related protein signaling. Cell Mol Life Sci.
61:257–262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cataisson C, Lieberherr M, Cros M, et al:
Parathyroid hormone-related peptide stimulates proliferation of
highly tumorigenic human SV40-immortalized breast epithelial cells.
J Bone Miner Res. 15:2129–2139. 2000. View Article : Google Scholar
|
|
33
|
Henderson JE, Amizuka N, Warshawsky H, et
al: Nucleolar localization of parathyroid hormone-related peptide
enhances survival of chondrocytes under conditions that promote
apoptotic cell death. Mol Cell Biol. 15:4064–4075. 1995.
|
|
34
|
Lam MH, House CM, Tiganis T, et al:
Phosphorylation at the cyclin-dependent kinases site
(Thr85) of parathyroid hormone-related protein
negatively regulates its nuclear localization. J Biol Chem.
274:18559–18566. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Conlan LA, Martin TJ and Gillespie MT: The
COOH-terminus of parathyroid hormone-related protein (PTHrP)
interacts with β-arrestin 1B. FEBS Lett. 527:71–75. 2002.PubMed/NCBI
|
|
36
|
Ferrari SL, Behar V, Chorev M, et al:
Endocytosis of ligand-human parathyroid hormone receptor 1
complexes is protein kinase C-dependent and involves β-arrestin2.
Real-time monitoring by fluorescence microscopy. J Biol Chem.
274:29968–29975. 1999.PubMed/NCBI
|
|
37
|
Fiaschi-Taesch N, Takane KK, Masters S, et
al: Parathyroid-hormone-related protein as a regulator of pRb and
the cell cycle in arterial smooth muscle. Circulation. 110:177–185.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shiina H, Igawa M, Breault J, et al: The
human T-cell factor-4 gene splicing isoforms, Wnt signal pathway,
and apoptosis in renal cell carcinoma. Clin Cancer Res.
9:2121–2132. 2003.PubMed/NCBI
|
|
39
|
Scibetta AG, Santangelo S, Coleman J, et
al: Functional analysis of the transcription repressor
PLU-1/JARID1B. Mol Cell Biol. 27:7220–7235. 2007. View Article : Google Scholar : PubMed/NCBI
|