Introduction
The epidermal growth factor receptor (EGFR) is one
of the most important molecular targets for advanced colorectal
cancer. Activation of this transmembrane receptor tyrosine kinase
stimulates signaling pathways supporting cell proliferation,
adhesion, migration, evasion of apoptosis, angiogenesis and
survival (1–3). Oncogenic signaling pathways downstream
of EGFR, including RAS/Raf/MAPK and PI3K/PTEN/Akt pathways, are
important mechanisms of tumor progression.
Activating mutations in the RAS oncogene
family are present in ~30% of all human cancers. RAS genes
encode highly homologous proteins: KRAS, NRAS and
HRAS (4). Mutations in the
KRAS gene are frequency reported in various human neoplasms,
including pancreatic cancer, biliary tract cancer and lung
adenocarcinoma (4–6). Cancer types with a high rate of
NRAS mutations include myeloid leukemia and cutaneous
melanoma, whereas HRAS mutations are typical of bladder and
cervical cancers (4,7,8). In
colorectal cancer (CRC), rates of KRAS, NRAS and
HRAS mutation are 30–42%, 2.2–5% and 0–0.8%, respectively
(9–14). Relatively low rates of NRAS
and HRAS mutations in CRC remain unexplained.
Anti-EGFR antibody therapy exhibits antitumor
effects by inhibiting multiple EGFR signaling pathways, including
RAS/RAF/MAPK and PI3K/PTEN/AKT pathways. Clinical trials have
demonstrated that anti-EGFR monoclonal antibodies (i.e., cetuximab
or panitumumab) are largely ineffective for metastatic CRC patients
when tumors harbor mutations in the codon 12 or 13 of KRAS
exon 2 (15–20). These mutations cause constitutive
activation of the RAS/RAF/MAPK pathway, regardless of EGFR
inhibition. Therefore, KRAS exon 2 mutations are recognized
as predictive markers of anti-EGFR therapy resistance for
metastatic CRC patients. Accordingly, these clinical trials
routinely exclude CRC patients harboring KRAS exon 2
mutations.
Recent studies suggest that other activating
mutations in KRAS or NRAS, in addition to KRAS
exon 2, confer resistance to anti-EGFR therapy (9,11,13,21).
Since KRAS and NRAS mutations tend to be mutually
exclusive, they may be present in approximately half of metastatic
CRC patients (9–14). Therefore, personalized cancer
therapy should be tailored to the KRAS and NRAS
mutation profile of each patient to improve treatment outcomes.
Previous studies have evaluated the
clinicopathological features and prognostic influence of
KRAS or BRAF mutations in colorectal cancer. The
prognostic value of KRAS mutations in CRC remains
controversial (22–24). In contrast, BRAF mutations
are associated with proximal colon tumor location, poor
differentiation, mucinous component and microsatellite instability.
Patients with BRAF-mutated tumors revealed lower survival
rates compared with wild-type tumors, particularly those with
BRAF-mutated and microsatellite-stable CRC (22,25–27).
On the other hand, clinicopathological characteristics, molecular
features, and the prognostic value of the NRAS mutation
remain largely unknown (10,12).
To date, analyses of NRAS mutations in colorectal cancer
were performed as part of a subset analysis of clinical studies for
treatment of metastatic CRC with anti-EGFR antibodies, and few
studies have described NRAS mutations in the early stage of
CRC. Irahara et al (12)
associated NRAS mutations with left-sided cancers in
females, but the data did not reach statistical significance since
NRAS mutations were only detected in 5 (2.2%) of the 225
cases. Therefore, the prognostic role and clinical characteristics
of NRAS mutations should be clarified using large tissue
samples to guide future clinical studies on the predictive impact
of the NRAS gene.
The present study used 1,304 consecutive samples of
stage 0-IV CRC to investigate the impact of mutations in
NRAS exon 2 and 3, in addition to KRAS and
BRAF. We evaluated the relationship between NRAS
mutations and other clinicopathological or molecular features,
including KRAS and BRAF mutations, microsatellite
instability (MSI) status and patient survival.
Materials and methods
Patients and tissue samples
The present study was conducted on 1,304 consecutive
primary CRC patients at the Saitama Cancer Center from July 1999 to
July 2008. Information on clinical data, including age at
diagnosis, gender, tumor size, histological differentiation, tumor
location, International Union against Cancer (UICC) stage and
prognosis were collected from medical records. Tissue samples were
surgically excised after obtaining informed consent from each
patient. All tumor tissues were paired with normal colorectal
tissues and immediately stored at −80°C. The present study was
approved by the Ethics Committee of the Saitama Cancer Center.
Mutation analysis of KRAS, BRAF and
NRAS
Genomic DNA from each sample was extracted by
standard SDS-proteinase K procedure, followed by ethanol
precipitation. All tumor samples were tested for KRAS exon
2, 3 and 4; BRAF exon 15 (codon 600); NRAS exon 2 and
3; and MSI status.
KRAS mutations in exon 2 and 3 were detected
by denaturing gradient gel electrophoresis (DGGE), and BRAF
mutations in exon 15 by PCR-restriction fragment length
polymorphism (RFLP), as previously described (28,29).
High resolution melting (HRM) analysis was used to
identify mutations in NRAS exon 2 and 3 and in KRAS
exon 4 using a Rotor-Gene Q (Qiagen, Hilden, Germany). Primer sets
for NRAS were as follows: exon 2, 5′-GGTTTCCAACAGGT
TCTTGC-3′ (forward) and 5′-CACTGGGCCTCACCTCTA TG-3′ (reverse); exon
3, 5′-CACACCCCCAGGATTCTTAC-3′ (forward) and
5′-TGGCAAATACACAGAGGAAGC-3′ (reverse). The primer set for
KRAS exon 4 was as follows: 5′-GCCTTCTAGAACAGTAGACAC-3′
(forward) and 5′-GA CATAACAGTTATGATTTTGCAGA-3′ (reverse). The
reaction mixture contained 7 μl of 2× LightCycler 480 High
Resolution Melting Master Reaction Mix (Roche Diagnostics,
Mannheim, Germany) with 0.21 μM of each forward and reverse primer,
3.2 mM MgCl2, 20 ng purified genomic DNA, and water to a
total volume of 14 μl. PCR cycling and melting conditions were as
follows: initial denaturation at 95°C for 5 min, followed by 40
cycles of 10 sec at 95°C, 20 sec at 57°C, and 10 sec at 72°C. One
heteroduplex cycle was performed at 95°C for 1 min and 40°C for 1
min, followed by melting from 72°C to 95°C with 10 acquisitions per
°C. HRM data were analyzed using the Rotor-Gene Q software
ver.2.0.2.4.
The DNA sequence of NRAS exon 2 and 3
mutations was determined by HRM using primers particularly designed
for HRM. Amplified products were labeled with GenomeLab™DTCS Quick
Start kit (Beckman Coulter Inc., Fullerton, CA, USA) according to
the manufacturer’s instructions and sequenced using the GenomeLab™
GeXP Genetic Analysis System (Beckman Coulter). Sequencing was
performed in both directions, and sequence analysis was performed
using the GenomeLab Genetic Analysis System v10.2 (Beckman
Coulter).
Analysis of microsatellite status
The MSI status was determined using Bethesda
markers: BAT25, BAT26, D5S346, D2S123 and D17S250. PCR and
subsequent analyses were performed as previously described
(30). CRC samples showing
instability in two or more markers were defined as microsatellite
instability-high (MSI-H), and the ones with none or one marker as
microsatellite stable (MSS).
Statistical analysis
Possible associations between each mutation and
clinicopathological parameters of CRC were assessed by the
Chi-square or Fisher’s exact test for categorical variables and
Mann-Whitney U or Kruskal-Wallis test for continuous variables.
Overall survival (OS) time was calculated from the date of surgery
to the date of death by any cause or censored at the last follow-up
visit. Cox proportional hazards analysis was used to estimate
clinicopathological- and biomarker-specific survival hazard ratios
(HRs) and 95% confidence intervals (CIs). A multivariable model
stratification by UICC stage was performed. All P-values were
calculated from two-sided test, and P-values <0.05 were
considered statistically significant. All statistical analyses were
performed with the SPSS Statistics v.20 (SPSS, Inc., Chicago, IL,
USA).
Results
Patient characteristics
All 1,304 patients enrolled in the present study
were diagnosed with either CRC stage 0 (n=48), stage I (n=248),
stage II (n=407), stage III (n=384) or stage IV (n=217) (Table I). Three hundred and seventy-nine
cancers were from the proximal colon (cecum to transverse colon),
544 from the distal colon (descending colon to sigmoid colon) and
381 from the rectum. The median follow-up period was 5.6 years
(interquartile range, 4.1–7.8 years), during which there were 435
deaths (33%).
 | Table IClinicopathological and molecular
features of all of the CRC samples. |
Table I
Clinicopathological and molecular
features of all of the CRC samples.
| Features | Patients
(n=1,304)
n (%) |
|---|
| Gender |
| Male | 780 (59.8) |
| Female | 524 (40.2) |
| Age ± SD
(years) | 63.8±10.4 |
| Location |
| Proximal | 379 (29.1) |
| Distal | 544 (41.7) |
| Rectum | 381 (29.2) |
| Tumor size |
| Mean ± SD
(mm) | 45.4±24.2 |
| Histological
features |
|
Well-differentiated | 144 (11.0) |
| Moderately
differentiated | 1,078 (82.7) |
| Poorly
differentiated | 34 (2.6) |
| Others | 48 (3.7) |
| Stage |
| 0 | 48 (3.7) |
| 1 | 248 (19.0) |
| 2 | 407 (31.3) |
| 3 | 384 (29.4) |
| 4 | 217 (16.6) |
| KRAS
status |
| Mutated-type | 553 (42.4) |
| Wild-type | 751 (57.6) |
| NRAS
status |
| Mutated-type | 35 (2.7) |
| Wild-type | 1,269 (97.3) |
| BRAF
status |
| Mutated-type | 59 (4.5) |
| Wild-type | 1,245 (95.5) |
| MSI status |
| MSI-H | 72 (5.5) |
| MSS | 1,232 (94.5) |
Frequency of KRAS, NRAS and BRAF
mutations
All 1,304 CRC cases were examined for mutations in
KRAS (exon 2, 3 and 4), NRAS (exon 2 and 3) and
BRAF (exon 15), as well as MSI status and
clinicopathological factors (Table
I). KRAS mutations were detected in 42.4% (n=553),
NRAS in 2.7% (n=35) and BRAF in 4.5% (n=59) of
patients. MSI-H was detected in 5.5% (n=72) of cases. Table II presents changes in the
nucleotides and corresponding amino acids detected in NRAS,
with p.G12D (codon 12), p.G13R (codon 13) and p.Q61K (codon 61) as
the most frequently noted mutations.
 | Table IIFrequency and type of NRAS
mutation. |
Table II
Frequency and type of NRAS
mutation.
| Nucleotide
mutation | Amino acid
change | Mutation frequency
(n) | Coincidental
KRAS mutation |
|---|
| Codon12 |
| c.35G>A | p.G12D | 20.0% (7) | |
| c.34G>T | p.G12C | 5.7% (2) | |
| c.35G>T | p.G12V | 5.7% (2) | |
| Codon13 |
| c.37G>C | p.G13R | 11.0% (4) | |
| c.38G>A | p.G13D | 8.6% (3) | p.G57T |
| c.38G>T | p.G13V | 2.9% (1) | |
| Codon61 |
| c.181C>A | p.Q61K | 26% (9) | |
| c.182A>T | p.Q61L | 5.7% (2) | |
| c.183A>C | p.Q61H | 2.9% (1) | |
| c.183A>T | p.Q61H | 2.9% (1) | |
| c.182A>G | p.Q61R | 2.9% (1) | |
| Codon9 |
| c.26T>C | p.V9A | 2.9% (1) | p.G12D |
| Codon68 |
| c.204A>T | p.R68S | 2.9% (1) | p.G12V |
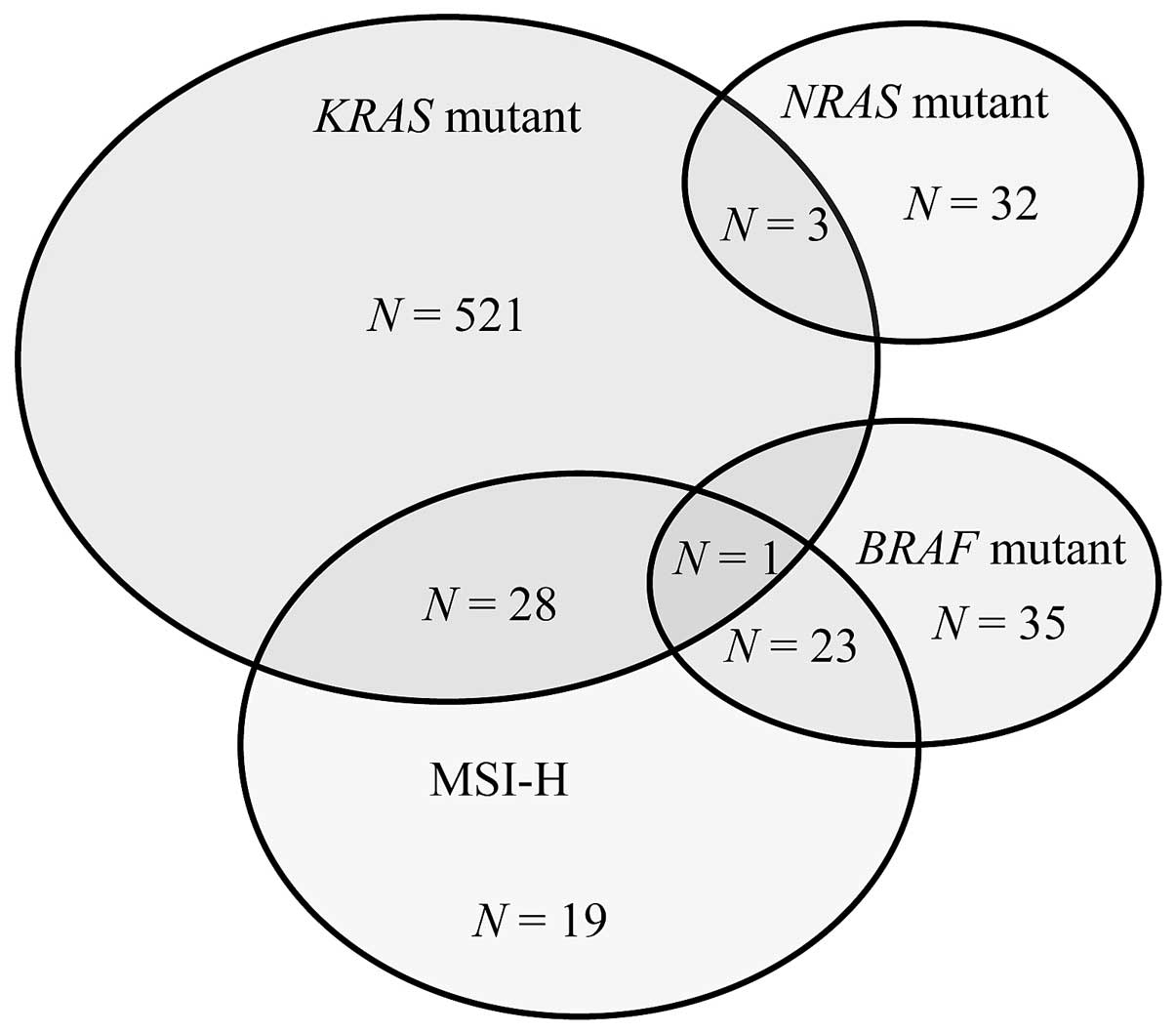
Mapping associations between molecular markers
revealed that 3 patients had both KRAS and NRAS
mutations, whereas 1 patient had both KRAS and BRAF
mutations (Fig. 1).
KRAS/NRAS mutation combinations were as follows:
p.G12D/p.V9A, pG12V/p.R68S and p.G57T/p.G13D. In contrast,
NRAS and BRAF mutations were mutually exclusive.
Regarding the MRI status, 28 patients with KRAS mutations
also had MSI-H tumors compared with 23 patients with BRAF
mutations. None of the patients with NRAS mutations had
MSI-H tumors.
BRAF mutations were significantly more
frequent in MSI-H than in MSS tumors (P<0.001), whereas no
significant association was observed between MSI status and
KRAS or NRAS mutations.
Frequency of KRAS and NRAS mutations in
each exon
In the KRAS gene, most mutations were located
in exon 2, with 495 of 1,304 cases (38.0%), whereas exon 3 or 4
mutations were detected in 26 (2.0%) and 32 (2.5%) cases,
respectively (Table III). In the
NRAS gene, 20 (1.5%) mutations were identified in exon 2 and
15 (1.2%) mutations in exon 3.
 | Table IIIMutation rates of KRAS and
NRAS genes for each exon. |
Table III
Mutation rates of KRAS and
NRAS genes for each exon.
| Gene | Patients with
mutations, n (%) |
|---|
| KRAS |
| Exon 2 | 495 (38.0) |
| Exon 3 | 26 (2.0) |
| Exon 4 | 32 (2.5) |
| NRAS |
| Exon 2 | 20 (1.5) |
| Exon 3 | 15 (1.2) |
Impact of KRAS, NRAS and BRAF mutation
status on clinicopathological and molecular characteristics of the
colorectal cancer patients
CRC patients were categorized into three groups on
the basis of KRAS, NRAS and BRAF mutations,
and they were compared in terms of gender, age, colorectal tumor
location, tumor maximum size, histological differentiation,
mucinous component, depth of tumor invasion, UICC stage, extramural
venous invasion and MSI status (Table
IV). BRAF-mutated tumors were more frequently associated
with mucinous component tumors (KRAS, P=0.003; NRAS,
P=0.002), poorly differentiated tumors (KRAS, P<0.001;
NRAS, P=0.013), female gender (KRAS, P=0.022) and
MSI-H (KRAS and NRAS, P<0.001).
NRAS-mutated tumors were more frequently located in the
distal colorectum compared with KRAS- or BRAF-mutated
tumors (P=0.015 and P<0.001, respectively). Compared with triple
wild-type tumors (KRAS, NRAS and BRAF
wild-type), KRAS- and BRAF-mutated tumors were more
commonly noted in the proximal colon (P<0.001 and P<0.001,
respectively), whereas no significant difference was observed
between NRAS-mutated tumors and triple wild-type tumors
(P=0.201).
 | Table IVClinicopathological characteristics
according to the KRAS, NRAS and BRAF mutation
status. |
Table IV
Clinicopathological characteristics
according to the KRAS, NRAS and BRAF mutation
status.
|
Characteristics | Triple wild-type n
(%) | KRAS mt n
(%) | NRAS mt n
(%) | BRAF mt n
(%) | P-value |
|---|
|
|---|
| KRAS vs.
NRAS | KRAS vs.
BRAF | NRAS vs.
BRAF |
|---|
| Patient | 661 | | | | 0.222 | 0.022 | 0.633 |
| Male | 431 (65.2) | 309 (55.9) | 16 (45.7) | 24 (40.7) | | | |
| Female | 230 (34.8) | 244 (44.1) | 19 (54.3) | 35 (59.3) | | | |
| Age ± SD
(years) | 63.3±10.3 | 64.2±10.4 | 65.5±9.5 | 64.2±11.5 | 0.716 | 0.64 | 0.997 |
| Location | | | | | 0.015 | <0.001 | <0.001 |
| Proximal | 142 (21.4) | 189 (34.2) | 4 (11.4) | 46 (77.9) | | | |
| Distal | 311 (47.1) | 209 (37.8) | 16 (45.7) | 9 (15.3) | | | |
| Rectum | 208 (31.5) | 155 (28.0) | 15 (42.9) | 4 (6.8) | | | |
| Tumor size | | | | | 0.456 | 0.417 | 0.928 |
| Mean ± SD
(mm) | 44.1±24.3 | 46.1±22.7 | 48.0±23.0 | 52.6±33.9 | | | |
| Histologic
feature | | | | | 0.916 | <0.001 | 0.013 |
|
Well-differentiated | 58 (8.8) | 78 (14.1) | 6 (17.1) | 3 (5.1) | | | |
| Moderately
differentiated | 573 (86.6) | 437 (79.0) | 29 (82.9) | 41 (69.4) | | | |
| Poorly
differentiated | 16 (2.4) | 11 (2.0) | 0 (0.0) | 8 (13.6) | | | |
| Mucinous | 11 (1.7) | 25 (4.5) | 0 (0.0) | 7 (11.9) | | | |
| Others | 3 (0.5) | 2 (0.4) | 0 (0.0) | 0 (0.0) | | | |
| Mucinous
component | | | | | 0.068 | 0.003 | 0.002 |
| + | 34 (5.1) | 98 (17.7) | 2 (5.7) | 20 (33.9) | | | |
| − | 627 (94.9) | 455 (82.3) | 33 (94.3) | 39 (66.1) | | | |
| Depth of tumor
invasion | | | | | 0.63 | 0.483 | 0.838 |
| Tis | 15 (2.3) | 33 (6.0) | 0 (0.0) | 0 (0.0) | | | |
| T1 | 62 (9.4) | 45 (8.1) | 4 (11.4) | 4 (6.8) | | | |
| T2 | 110 (16.6) | 68 (12.3) | 3 (8.6) | 7 (11.9) | | | |
| T3 | 404 (61.1) | 352 (63.7) | 24 (68.6) | 39 (66.0) | | | |
| T4 | 70 (10.6) | 55 (9.9) | 4 (11.4) | 9 (15.3) | | | |
| UICC stage | | | | | 0.353 | 0.151 | 0.75 |
| 0 | 15 (2.3) | 33 (6.0) | 0 (0.0) | 0 (0.0) | | | |
| 1 | 143 (21.6) | 89 (16.1) | 6 (17.1) | 10 (16.9) | | | |
| 2 | 210 (31.8) | 170 (30.7) | 10 (28.6) | 17 (28.8) | | | |
| 3 | 188 (28.4) | 173 (31.3) | 9 (25.7) | 17 (28.8) | | | |
| 4 | 105 (15.9) | 88 (15.9) | 10 (28.6) | 15 (25.5) | | | |
| Extramural venous
invasion | | | | | 0.231 | 0.105 | 0.689 |
| + | 471 (71.3) | 365 (66.0) | 24 (68.6) | 46 (78.0) | | | |
| − | 190 (28.7) | 188 (34.0) | 11 (31.4) | 13 (22.0) | | | |
| MSI-H | | | | | 0.171 | <0.001 | <0.001 |
| + | 20 (3.0) | 29 (5.2) | 0 (0.0) | 24 (40.7) | | | |
| − | 641 (97.0) | 524 (94.8) | 35 (100) | 35 (59.3) | | | |
Mutation rates of KRAS, NRAS and
BRAF for each UICC stage are presented in Fig. 2. KRAS mutations were detected
at similar frequencies in stage 0–I to IV. NRAS mutations
tended to occur more frequently in stage IV cancers than in stage
0–III cancers compared with KRAS mutations (P=0.061).
Impact of KRAS, NRAS and BRAF mutations
on CRC patient survival
Univariate analysis was conducted in regards to age,
gender, tumor location, stage, histological subtype, mucinous
component, extramural venous invasion, MSI status, KRAS,
NRAS and BRAF mutations (Table V). Patients with KRAS and
BRAF mutations had significantly worse survival compared
with wild-type cases [HR=1.25; 95% confidence interval (CI)
1.03–1.52; P=0.027 and HR=1.73; 95% CI, 1.15–2.60; P=0.009,
respectively]. Four other variables were significantly associated
with poor prognosis, namely age ≥65 years (HR=1.39; 95% CI,
1.15–1.69; P=0.001), UICC stage (stage II: HR=2.33; stage III:
HR=3.58; stage IV: HR=14.14; P<0.001 respectively), histological
subtype (HR=1.82; 95% CI, 1.31–2.52; P<0.001), and extramural
venous invasion (HR=3.28; 95% CI, 2.50–4.30; P<0.001). The only
predictor of good prognosis was female gender (HR=0.73; 95% CI,
0.60–0.89; P=0.002). In multivariable analysis, KRAS and
BRAF mutations were associated with significantly higher
mortality rates when stratified according to UICC staging (HR=1.44;
95% CI, 1.18–1.79; P<0.001 and HR=2.09; 95% CI, 1.36–3.28;
P=0.001, respectively). Notably, NRAS-mutated tumors
demonstrated a trend towards favorable prognosis (HR=0.53; 95% CI,
0.27–1.03; P=0.059).
 | Table VUnivariate and multivariate analyses
of the covariates associated with overall survival. |
Table V
Univariate and multivariate analyses
of the covariates associated with overall survival.
| Covariates | Univariate HR (95%
CI) | P-value | Multivariate HR(95%
CI) | P-value |
|---|
| Age ≥65 years | 1.39
(1.15–1.69) | 0.001 | 1.53
(1.27–1.86) | <0.001 |
| Female | 0.73
(0.60–0.89) | 0.002 | 0.69
(0.56–0.84) | <0.001 |
| Tumor location
(proximal vs. distal colorectum) | 0.88
(0.72–1.08) | 0.22 | 1.01
(0.81–1.25) | 0.96 |
|
KRAS-mutant | 1.25
(1.03–1.52) | 0.027 | 1.44
(1.18–1.76) | <0.001 |
|
NRAS-mutant | 0.83
(0.43–1.61) | 0.57 | 0.53
(0.27–1.03) | 0.059 |
|
BRAF-mutant | 1.73
(1.15–2.60) | 0.009 | 2.09
(1.33–3.28) | 0.001 |
| MSS (vs.
MSI-high) | 1.59
(0.98–2.58) | 0.062 | 1.56
(0.92–2.64) | 0.10 |
| UICC stage |
| II | 2.33
(1.58–3.44) | <0.001 | -a | |
| III | 3.58
(2.46–5.22) | <0.001 | -a | |
| IV (vs. Stage 0
and I) | 14.14
(9.68–20.7) | <0.001 | -a | |
| Histological
subtype (vs. well and mod ) | 1.82
(1.31–2.52) | <0.001 | 1.59
(1.09–2.32) | 0.016 |
| Mucinous
component | 1.23
(0.93–1.62) | 0.14 | 0.81
(0.59–1.11) | 0.2 |
| Extramural venous
invasion | 3.28
(2.50–4.30) | <0.001 | 1.78
(1.31–2.37) | <0.001 |
Discussion
The present study investigated clinicopathological
and prognostic features of KRAS, NRAS and BRAF
mutations in tumors from 1,304 consecutive CRC patients. An
important finding was that patients undergoing CRC tumor resection
at all stages were targeted by all these mutations. In addition, to
the best of our knowledge, this is the first large study to present
statistically significant comparisons between these three
categories of RAS/RAF mutations in CRC patients.
NRAS mutations were observed in 35 (2.7%) of
the 1,304 patients, of which 20 (1.5%) patients revealed a mutation
in exon 2 and others (1.2%) in exon 3. These data are consistent
with previous studies that reported NRAS mutations in
2.2–5.0% of CRC tumors, with approximately equal frequency in exon
2 and 3 (9,10,12,29).
Moreover, we showed that NRAS mutations are detected in
early stages of CRC and tend to occur more frequently in stage IV
cancers than in stage 0–III cancers. Therefore, NRAS
mutations appear to be acquired at early and advanced stages of CRC
(31). Nonetheless, the tendency of
higher NRAS mutation rates in stage IV CRC should be
ascertained in a larger scale study. The frequency of KRAS
mutations was also compatible with previous studies, but the
frequency of BRAF mutations (4.5%) declined below 7.4–10.1%
of the values previously reported (10,13,14).
On the other hand, Yokota et al (32) reported an incidence of 4.7% for
BRAF mutations (15/319 patients) in Japanese CRC patients.
Such agreement with our Japanese study suggests that racial or
environmental factors may affect the frequency of BRAF
mutations. The strong overlap between BRAF mutation and
MSI-H status that we detected suggests that the low frequency in
the BRAF mutation is affected by the low MSI-H frequency
reported among Asians compared with Westerners (33).
In the present study, 3 cases of 1,304 had mutations
in both KRAS and NRAS, which is inconsistent with
previous reports of mutual exclusivity. Notably, all three tumors
presented rare mutations (NRAS p.V9A, NRAS p.R68S and
KRAS p.G57T), whereas these tumors had common mutations
(KRAS p.G12D, KRAS p.G12V and NRAS p.G13D).
The oncogenic activity of these minor mutations is unknown, except
for KRAS p.G57T (28).
Therefore, KRAS and NRAS mutation detection only in
major mutation lesion may have missed these rare mutations.
Colorectal tumors with NRAS mutations were
found more frequently in the distal colon and rectum compared with
tumors with KRAS or BRAF mutations, while the
distribution of BRAF-mutated tumors was consistent with
previous studies reporting that BRAF-mutated tumors are
primarily located in the proximal colon (22,32,34).
It was proposed that cellular transformation and mutations occur
more frequently in the proximal colon due to close contact of
epithelial cells with stimulating bowel content (34). However, this theory does not explain
the higher frequency of NRAS-mutated tumors in the distal
colon and rectum compared with those with KRAS or
BRAF mutations. Elucidating factors responsible for distinct
locations of KRAS- and NRAS-mutated tumors may be
crucial to our understanding of NRAS mutations in CRC
patients.
The prognosis of advanced CRC patients carrying
NRAS mutations has been reported in the form of subset
analysis of clinical studies using anti-EGFR drugs such as
cetuximab and panitumumab (9,13,14).
Although there is currently no consistent view regarding the
efficacy of anti-EGFR drugs for CRC patients with NRAS
mutations, several reports consider this form of therapy
inappropriate for such CRC patients. However, the fundamental
malignant potential of the NRAS mutation should be
considered to determine the efficacy of anti-EGFR drugs. To the
best of our knowledge, this is the first study to compare patient
survival in NRAS-mutated and triple wild-type CRCs. Numerous
studies report an association between BRAF mutations and
poor clinical outcome (15,22,23,27,32,35).
Although the prognostic value of the KRAS mutation is still
controversial, several studies suggest a poor prognosis (24,35,36).
Since NRAS is a RAS family member, we expected
NRAS-mutated CRC patients to have a poor prognosis compared
with those without RAS mutations. However, multivariate
analysis showed a tendency toward a better prognosis for
NRAS-mutated CRC patients. Therefore, the present study
suggests that CRC cases with NRAS mutations exhibit
different characteristics than CRC cases with KRAS
mutations.
Recent studies report that common KRAS exon
2 mutations targeting the RAS/RAF/MAPK pathway are currently used
to determine patient eligibility for anti-EGFR monoclonal antibody
therapy (19,37,38).
Minor mutations (KRAS exon 3 and 4, NRAS,
BRAF) are expected to become the next predictive biomarkers.
In fact, a recent clinical trial on anti-EGFR monoclonal antibodies
excluded patients with the KRAS exon 2 mutation and those with
these less frequent mutations (39). In the present study, the incidence
of KRAS exon 2 mutations was 38%, whereas the combined
mutation rate for KRAS exon 3 and 4, NRAS and
BRAF was 49.3%. For stage IV CRC patients subjected to
anti-EGFR monoclonal antibody therapy, the total mutation rate
increased from 37.3 to 51.6%. If the validity of excluding CRC
patients with these minor RAS mutations from anti-EGFR
monoclonal antibody therapy is verified by future trials, more than
50% of CRC patients would greatly benefit from personalized
medicine for enhanced efficacy and a better prognosis.
In conclusion, the present study demonstrated that
KRAS- and NRAS-mutated CRC tumors exhibit distinct
characteristics and distributions along the colorectum. Future
molecular biology studies should address the significance of these
differences between NRAS- and KRAS-mutated CRC and
confirm possible positive prognoses associated with NRAS
mutations.
Acknowledgements
We would like to thank Akemi Takahashi, Mina Yamada
and Syuhei Takahashi for their excellent technical assistance. The
present study was supported by the Japanese Ministry of Health,
Labour and Welfare.
References
|
1
|
Scaltriti M and Baselga J: The epidermal
growth factor receptor pathway: a model for targeted therapy. Clin
Cancer Res. 12:5268–5272. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mendelsohn J and Baselga J: Epidermal
growth factor receptor targeting in cancer. Semin Oncol.
33:369–385. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schubbert S, Shannon K and Bollag G:
Hyperactive Ras in developmental disorders and cancer. Nat Rev
Cancer. 7:295–308. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ginesta MM, Mora J, Mayor R, et al:
Genetic and epigenetic markers in the evaluation of pancreatic
masses. J Clin Pathol. 66:192–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mascaux C, Iannino N, Martin B, et al: The
role of RAS oncogene in survival of patients with lung cancer: a
systematic review of the literature with meta-analysis. Br J
Cancer. 92:131–139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohlmann A, Grossmann V, Klein HU, et al:
Next-generation sequencing technology reveals a characteristic
pattern of molecular mutations in 72.8% of chronic myelomonocytic
leukemia by detecting frequent alterations in TET2, CBL, RAS, and
RUNX1. J Clin Oncol. 28:3858–3865. 2010.PubMed/NCBI
|
|
8
|
Uhara H, Ashida A, Koga H, et al:
NRAS mutations in primary and metastatic melanomas of
Japanese patients. Int J Clin Oncol. June 6–2013.(Epub ahead of
print).
|
|
9
|
De Roock W, Claes B, Bernasconi D, et al:
Effects of KRAS, BRAF, NRAS, and PIK3CA
mutations on the efficacy of cetuximab plus chemotherapy in
chemotherapy-refractory metastatic colorectal cancer: a
retrospective consortium analysis. Lancet Oncol. 11:753–762.
2010.
|
|
10
|
Vaughn CP, Zobell SD, Furtado LV, Baker CL
and Samowitz WS: Frequency of KRAS, BRAF, and
NRAS mutations in colorectal cancer. Genes Chromosomes
Cancer. 50:307–312. 2011.PubMed/NCBI
|
|
11
|
Loupakis F, Ruzzo A, Cremolini C, et al:
KRAS codon 61, 146 and BRAF mutations predict
resistance to cetuximab plus irinotecan in KRAS codon 12 and
13 wild-type metastatic colorectal cancer. Br J Cancer.
101:715–721. 2009. View Article : Google Scholar
|
|
12
|
Irahara N, Baba Y, Nosho K, et al:
NRAS mutations are rare in colorectal cancer. Diagn Mol
Pathol. 19:157–163. 2010. View Article : Google Scholar
|
|
13
|
Seymour MT, Brown SR, Middleton G, et al:
Panitumumab and irinotecan versus irinotecan alone for patients
with KRAS wild-type, fluorouracil-resistant advanced
colorectal cancer (PICCOLO): a prospectively stratified randomised
trial. Lancet Oncol. 14:749–759. 2013.PubMed/NCBI
|
|
14
|
Smith CG, Fisher D, Claes B, et al:
Somatic profiling of the epidermal growth factor receptor pathway
in tumors from patients with advanced colorectal cancer treated
with chemotherapy ± cetuximab. Clin Cancer Res. 19:4104–4113.
2013.PubMed/NCBI
|
|
15
|
Farina-Sarasqueta A, van Lijnschoten G,
Moerland E, et al: The BRAF V600E mutation is an independent
prognostic factor for survival in stage II and stage III colon
cancer patients. Ann Oncol. 21:2396–2402. 2010. View Article : Google Scholar
|
|
16
|
Van Cutsem E, Kohne CH, Hitre E, et al:
Cetuximab and chemotherapy as initial treatment for metastatic
colorectal cancer. N Engl J Med. 360:1408–1417. 2009.PubMed/NCBI
|
|
17
|
Bokemeyer C, Bondarenko I, Makhson A, et
al: Fluorouracil, leucovorin, and oxaliplatin with and without
cetuximab in the first-line treatment of metastatic colorectal
cancer. J Clin Oncol. 27:663–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar
|
|
19
|
Douillard JY, Siena S, Cassidy J, et al:
Randomized, phase III trial of panitumumab with infusional
fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4
alone as first-line treatment in patients with previously untreated
metastatic colorectal cancer: the PRIME study. J Clin Oncol.
28:4697–4705. 2010. View Article : Google Scholar
|
|
20
|
Amado RG, Wolf M, Peeters M, et al:
Wild-type KRAS is required for panitumumab efficacy in
patients with metastatic colorectal cancer. J Clin Oncol.
26:1626–1634. 2008.PubMed/NCBI
|
|
21
|
Douillard JY, Oliner KS, Siena S, et al:
Panitumumab-FOLFOX4 treatment and RAS mutations in
colorectal cancer. N Engl J Med. 369:1023–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Samowitz WS, Sweeney C, Herrick J, et al:
Poor survival associated with the BRAF V600E mutation in
microsatellite-stable colon cancers. Cancer Res. 65:6063–6069.
2005.PubMed/NCBI
|
|
23
|
Roth AD, Tejpar S, Delorenzi M, et al:
Prognostic role of KRAS and BRAF in stage II and III
resected colon cancer: results of the translational study on the
PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28:466–474.
2010.
|
|
24
|
Imamura Y, Morikawa T, Liao X, et al:
Specific mutations in KRAS codons 12 and 13, and patient
prognosis in 1075 BRAF wild-type colorectal cancers. Clin
Cancer Res. 18:4753–4763. 2012.PubMed/NCBI
|
|
25
|
Lochhead P, Kuchiba A, Imamura Y, et al:
Microsatellite instability and BRAF mutation testing in
colorectal cancer prognostication. J Natl Cancer Inst.
105:1151–1156. 2013.PubMed/NCBI
|
|
26
|
Gavin PG, Colangelo LH, Fumagalli D, et
al: Mutation profiling and microsatellite instability in stage II
and III colon cancer: an assessment of their prognostic and
oxaliplatin predictive value. Clin Cancer Res. 18:6531–6541. 2012.
View Article : Google Scholar
|
|
27
|
Ogino S, Shima K, Meyerhardt JA, et al:
Predictive and prognostic roles of BRAF mutation in stage
III colon cancer: results from intergroup trial CALGB 89803. Clin
Cancer Res. 18:890–900. 2012.PubMed/NCBI
|
|
28
|
Akagi K, Uchibori R, Yamaguchi K, Kurosawa
K, Tanaka Y and Kozu T: Characterization of a novel oncogenic
K-ras mutation in colon cancer. Biochem Biophys Res Commun.
352:728–732. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Asaka S, Arai Y, Nishimura Y, et al:
Microsatellite instability-low colorectal cancer acquires a
KRAS mutation during the progression from Dukes’ A to Dukes’
B. Carcinogenesis. 30:494–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ishikubo T, Nishimura Y, Yamaguchi K, et
al: The clinical features of rectal cancers with high-frequency
microsatellite instability (MSI-H) in Japanese males. Cancer Lett.
216:55–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Andreyev HJ, Norman AR, Cunningham D,
Oates JR and Clarke PA: Kirsten ras mutations in patients with
colorectal cancer: the multicenter ‘RASCAL’ study. J Natl Cancer
Inst. 90:675–684. 1998.
|
|
32
|
Yokota T, Ura T, Shibata N, et al:
BRAF mutation is a powerful prognostic factor in advanced
and recurrent colorectal cancer. Br J Cancer. 104:856–862. 2011.
View Article : Google Scholar
|
|
33
|
Oh JR, Kim DW, Lee HS, et al:
Microsatellite instability testing in Korean patients with
colorectal cancer. Fam Cancer. 11:459–466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamauchi M, Morikawa T, Kuchiba A, et al:
Assessment of colorectal cancer molecular features along bowel
subsites challenges the conception of distinct dichotomy of
proximal versus distal colorectum. Gut. 61:847–854. 2012.
View Article : Google Scholar
|
|
35
|
Richman SD, Seymour MT, Chambers P, et al:
KRAS and BRAF mutations in advanced colorectal cancer
are associated with poor prognosis but do not preclude benefit from
oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin
Oncol. 27:5931–5937. 2009. View Article : Google Scholar
|
|
36
|
Phipps AI, Buchanan DD, Makar KW, et al:
KRAS-mutation status in relation to colorectal cancer survival: the
joint impact of correlated tumour markers. Br J Cancer.
108:1757–1764. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tol J, Koopman M, Cats A, et al:
Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal
cancer. N Engl J Med. 360:563–572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bokemeyer C, Bondarenko I, Hartmann JT, et
al: Efficacy according to biomarker status of cetuximab plus
FOLFOX-4 as first-line treatment for metastatic colorectal cancer:
the OPUS study. Ann Oncol. 22:1535–1546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fornaro L, Lonardi S, Masi G, et al:
FOLFOXIRI in combination with panitumumab as first-line treatment
in quadruple wild-type (KRAS, NRAS, HRAS,
BRAF) metastatic colorectal cancer patients: a phase II
trial by the Gruppo Oncologico Nord Ovest (GONO). Ann Oncol.
24:2062–2067. 2013. View Article : Google Scholar : PubMed/NCBI
|