Introduction
Anthracyclines such as doxorubicin and epirubicin
are chemotherapeutic drugs used to effectively treat various types
of cancer, including leukemia and breast, ovarian, uterine, and
lung cancers (1). Despite being
introduced over 30 years ago, anthracyclines remain part of the
gold standard chemotherapy for breast cancer (2). However, a significant number of breast
cancer patients acquire resistance to these drugs during
chemotherapy (3,4). Drug resistance can be classified into
two main categories: intrinsic drug resistance, in which previously
untreated cancer cells are inherently insensitive to the
chemotherapeutic drugs; and acquired drug resistance (ADR), in
which the cancer cells become insensitive after drug exposure
(5,6). The mechanisms of drug resistance have
been intensively studied with the aim of overcoming this major
obstacle in chemotherapy, and, although the exact mechanisms of ADR
remain unclear, numerous theories have been proposed. One mechanism
that may underlie acquired resistance to anthracycline is the
active cellular extrusion of drugs by the overexpression of
multidrug resistance protein 1 (MDR1) (7). MDR1, also known as
P-glycoprotein 1 (permeability glycoprotein, abbreviated as P-gp or
Pgp) or ATP-binding cassette (ABC) sub-family B member 1
(ABCB1), acts as an efflux pump for various xenobiotics such
as toxins or drugs. While in vitro studies have demonstrated
the efficacy of certain MDR1 inhibitors, in clinical
studies, these compounds have yet to show a consistent advantage
(8,9). Other than drug efflux pumps,
mechanisms that may contribute to anthracycline resistance include:
changes in intracellular drug distribution, apoptotic and DNA
repair responses and alteration of topoisomerase II, which is the
major cellular target of anthracyclines (10,11).
However, the validity of these particular mechanisms of action
remains to be ascertained.
ADR is multifactorial because it involves host
factors, various molecular events, and numerous genetic changes
(12). In developing targeted
therapeutic strategies to overcome drug resistance, it is essential
to understand the basic genetic changes associated with acquisition
of drug resistance. High-throughput gene sequencing technologies,
such as microarrays, are widely used to comprehensively analyze
gene expression, and to detect mutations and single-nucleotide
polymorphisms (13). By applying
these technologies, investigators have improved their understanding
of the cellular and molecular changes that occur during the
development of ADR in breast cancer (14,15).
Previous studies have provided lists of differentially expressed
genes (DEGs); however, findings tend to be inconsistent across
studies due to small sample sizes, and differences in sample
quality, laboratory protocol, platform, and analytical technique
(16). In order to overcome the
limitations presented by these inconsistencies, it is possible to
take a systematic approach and perform integrated analyses of
multiple microarray datasets.
Interest in using integrated analysis to investigate
multiple independent microarray datasets has been on the increase
(17). Accumulating evidence has
shown that meta-analysis increases the statistical power of
expression profiling and enables an assessment of between-study
heterogeneity, which may lead to more robust and reliable gene
signatures (17,18). To the best of our knowledge, a
meta-analysis focusing on data for acquired resistance to
anthracycline in breast cancer has yet to be performed. Therefore,
in the present study, we performed the first cross-platform
meta-analysis of multiple gene expression profiles taken from
various independent studies with the aim of identifying novel
candidate genes and biological processes that are involved in
acquired anthracycline resistance, and overcoming the limitations
presented by inconsistencies in individual studies.
Materials and methods
Extraction of eligible microarray
datasets containing data on anthracycline-resistant breast cancer
cell lines
Gene expression studies related to acquired
anthracycline resistance in breast cancer were collected in July
2014 by searching the PubMed database, NCBI Gene Expression Omnibus
(GEO) (available at: http://www.ncbi.nlm.nih.gov/geo/), and ArrayExpress
(AE) (available at: http://www.ebi.ac.uk/arrayexpress/). When searching
these resources, the following keywords and their combinations were
used: ‘anthracycline’, ‘drug resistance’, ‘breast cancer’, and
‘gene expression’. Two independent reviewers extracted data from
the original studies. Any discrepancies between the reviewer’s data
were resolved either by consensus or a third reviewer. Inclusion
criteria for the study were: i) gene expression profiling of
stepwise-selected, anthracycline-resistant, derivative breast
cancer cell lines; and ii) sufficient data and the correct platform
to facilitate the meta-analysis. We retained only those original
experimental articles in which gene expression profiles of
stepwise-selected, anthracycline-resistant, derivative breast
cancer cell lines were analyzed relative to parental control cells.
Non-human data, review articles, and integrated analysis of
expression profiles were also excluded from the meta-analysis. The
following information was extracted from each selected study: GEO
accession number, platform and sample type, and gene expression
data.
Meta-analysis of gene expression in
multiple microarray data-sets
We used the meta-analysis of the gene expression
profiles in the selected microarray datasets to identify DEGs.
Prior to processing of data, all the gene and probe IDs were
annotated as Entrez IDs for consistency, and intensity values were
log2-transformed and normalized in order that their mean and unit
variance was zero. A meta-analysis was performed using rank product
methods (RankProd package in R) implemented in the web-based INMEX
program. RankProd (developed from the non-parametric rank product
method) was used to apply a statistically rigorous algorithm, which
included biological intuition of fold-change (FC) criteria and
determined the ranks of the DEGs based on FC scores in all possible
pairwise comparisons, to the integrated datasets. With the RankProd
algorithm, genes that were consistently identified as up- or
downregulated DEGs in whole datasets were assigned a higher rank
depending on their P-value and FC level in a given number of
replicates multiplied across the given datasets, and these were
considered the most significantly regulated DEGs. The expression
profiles of DEGs across different data-sets/conditions were
visualized as heat maps by implementing the ‘Pattern Extractor’
tool.
Functional and pathway enrichment
analyses of DEGs
To investigate the cellular function of DEGs, we
performed a gene ontology (GO) enrichment analysis based on the GO
database (http://www.geneontology.org/), and a pathway analysis
based on the Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (http://www.genome.ad.jp/) contained
in the functional analysis module of the INMEX program.
Analysis of protein-protein interaction
(PPI) network
To determine the function of the proteins that they
encoded, DEGs were imported into the PPI network constructed by
using the Biological General Repository for Interaction Datasets
(BioGRID) (http://thebiogrid.org/) in Cytoscape
software (http://www.cytoscape.org/). The PPI
network identified for the DEGs was screened at a genome-wide
scale, with both end nodes having DEGs.
Results
Microarray datasets used in the
meta-analysis
Three micro-array datasets were found to meet our
study criteria and these were extracted from the GEO database as a
GEO series (GSE, an original record in GEO that summarizes an
experiment). The datasets, GSE24460 and GSE3926, were microarray
expression profiles of breast cancer cell lines that acquire drug
resistance by stepwise treatment with doxorubicin. The other
dataset, GSE54326, was a microarray expression profile of breast
cancer cell lines that acquire drug resistance by stepwise
treatment with epirubicin. As shown in Table I, from the 3 GSEs, we used 31 GEO
samples (GSM, an identifier of specific experimental conditions)
from 2 different GEO platforms (GPL, an identifier of specific
microarray designs) in the meta-analysis.
 | Table ICharacteristics of individual studies
analyzed in the meta-analysis. |
Table I
Characteristics of individual studies
analyzed in the meta-analysis.
| No. of samples | | | |
|---|
|
| | | |
|---|
| GEO dataset | PC | AR | Drug | Cell line | Platform |
|---|
| GSE24460 | 2 | 2 | Doxorubicin | MCF-7 | Affymetrix human
genome U133A array |
| GSE3926 | 1 | 2 | Doxorubicin | MCF-7 | Affymetrix human
genome U133A array |
| GSE54326 | 12 | 12 | Epirubicin | MDA-MB-231, MCF-7,
SKBR-3, ZR-75-1 | Illumina human HT-12
V4.0 expression beadchip |
Identification of DEGs commonly regulated
in multiple data-sets
We selected DEGs with P<0.05 based on the
estimated percentage of false-positives and P-values produced by
the algorithm in RankProd. We identified 413 DEGs from GSMs in
which acquired anthracycline-resistant breast cancer cell lines
were compared with a parental control, including 255 up- and 158
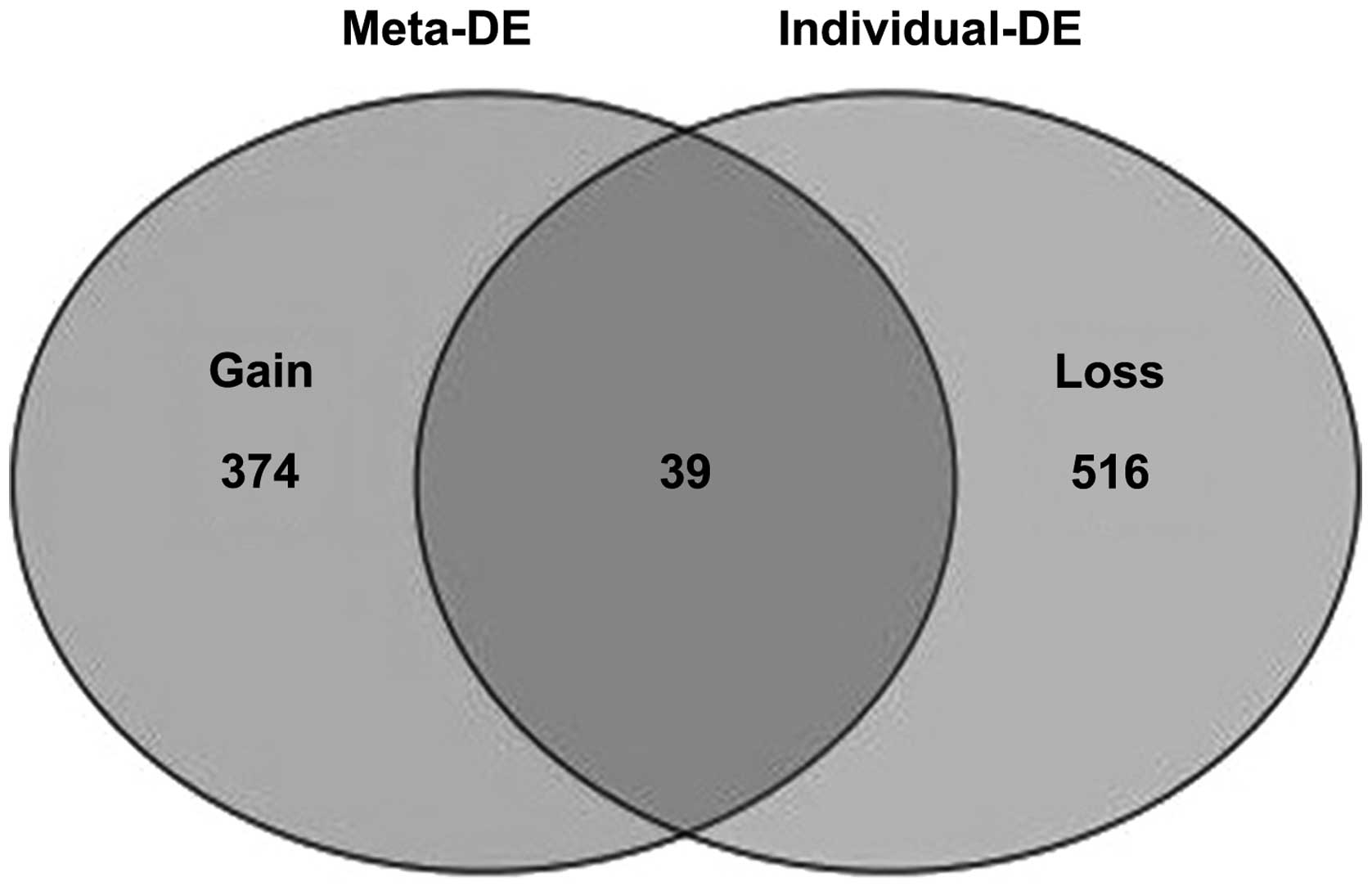
downregulated genes. Additionally, 374 ‘gain’ genes were identified
in the meta-analysis, but not in any individual analysis (Fig. 1). The 20 most significantly up- or
downregulated DEGs, with P<1.0E-5, are shown in Table II. The upregulated genes were
TRIM29, PTPRM, EPB41L3, VTN,
ABCB1, LY6D, ADAMTS9, OAS2,
CXCR7, and AKR1B10. The downregulated genes were
carbonic anhydrase VIII (CA8), ARMC4, SNTB1,
CCNA1, karyopherin α 5 (KPNA5), POPDC3,
ZNF711, FOLH1, SLC16A10, DPYSL3,
TFF3, AZGP1, VTCN1, and CPVL. Among
these, the up- and downregulated DEGs with the largest mean logFC
were TRIM29 and CA8, respectively (Table II). Heat maps, based on the
meta-analysis of individual data sets, were used to visualize the
correlation in expression patterns for a subset of genes from the
three studies (Fig. 2).
 | Table IIThe top 20 most strongly up- or
downregulated DEGs by meta-analysis. |
Table II
The top 20 most strongly up- or
downregulated DEGs by meta-analysis.
| Enterz ID | Gene symbol | Average log FC | P-value | Gene name |
|---|
| Upregulated
genes | | | | |
| 23650 | TRIM29 | −11.5579 | <1.0E-5 | Tripartite motif
containing 29 |
| 5797 | PTPRM | −11.2551 | | Protein tyrosine
phosphatase, receptor type, M |
| 23136 | EPB41L3 | −10.6225 | | Erythrocyte
membrane protein band 4.1-like 3 |
| 7448 | VTN | −9.83827 | | Vitronectin |
| 5243 | ABCB1 | −9.73049 | | ATP-binding
cassette, sub-family B (MDR/TAP), member 1 |
| 8581 | LY6D | −6.59411 | | Lymphocyte antigen
6 complex, locus D |
| 56999 | ADAMTS9 | −6.5461 | | ADAM
metallopeptidase with thrombospondin type 1 motif, 9 |
| 4939 | OAS2 | −5.92265 | |
2′-5′-oligoadenylate synthetase 2, 69/71
kDa |
| 57007 | CXCR7 | −5.88834 | | Atypical chemokine
receptor 3 |
| 57016 | AKR1B10 | −5.12808 | | Aldo-keto reductase
family 1, member B10 |
| 972 | CD74 | −8.34455 | 0.00050 | CD74 molecule,
major histocompatibility complex, class II invariant chain |
| 54894 | RNF43 | −7.23015 | 0.00053 | Ring finger protein
43 |
| 6590 | SLPI | −6.86764 | 0.00056 | Secretory leukocyte
peptidase inhibitor |
| 7124 | TNF | −7.69007 | 0.00059 | Tumor necrosis
factor |
| 10964 | IFI44L | −6.05839 | 0.00063 | Interferon-induced
protein 44-like |
| 6707 | SPRR3 | −5.51409 | 0.00067 | Small proline-rich
protein 3 |
| 79132 | DHX58 | −7.722 | 0.00071 | DEXH
(Asp-Glu-X-His) box polypeptide 58 |
| 7078 | TIMP3 | −6.28697 | 0.00077 | TIMP
metallopeptidase inhibitor 3 |
| 9429 | ABCG2 | −6.67666 | 0.00083 | ATP-binding
cassette, sub-family G (white), member 2 |
| 4973 | OLR1 | −7.30092 | 0.00091 | Oxidized
low-density lipoprotein (lectin-like) receptor 1 |
| Downregulated
genes | | | | |
| 767 | CA8 | 10.97236 | <1.0E-5 | Carbonic anhydrase
VIII |
| 55130 | ARMC4 | 10.22814 | | Armadillo repeat
containing 4 |
| 6641 | SNTB1 | 9.843391 | | Syntrophin, β 1
(dystrophin-associated protein A1, basic component 1) |
| 8900 | CCNA1 | 9.58363 | | Cyclin A1 |
| 3841 | KPNA5 | 9.107488 | | Karyopherin α 5
(importin α 6) |
| 64208 | POPDC3 | 8.982291 | | Popeye domain
containing 3 |
| 7552 | ZNF711 | 8.752859 | | Zinc finger protein
711 |
| 2346 | FOLH1 | 8.277195 | | Folate hydrolase
(prostate-specific membrane antigen) 1 |
| 117247 | SLC16A10 | 8.21847 | | Solute carrier
family 16 (aromatic amino acid transporter), member 10 |
| 1809 | DPYSL3 | 8.109953 | |
Dihydropyrimidinase-like 3 |
| 7033 | TFF3 | 6.606418 | | Trefoil factor 3
(intestinal) |
| 563 | AZGP1 | 5.309637 | | α-2-glycoprotein 1,
zinc-binding |
| 79679 | VTCN1 | 3.976014 | | V-set domain
containing T-cell activation inhibitor 1 |
| 54504 | CPVL | 2.726046 | | Carboxypeptidase,
vitellogenic-like |
| 26154 | ABCA12 | 7.400289 | 0.00067 | ATP-binding
cassette, sub-family A (ABC1), member 12 |
| 26047 | CNTNAP2 | 2.945366 | 0.00125 |
Contactin-associated protein-like 2 |
| 23493 | HEY2 | 3.766967 | 0.00167 | Hes-related family
bHLH transcription factor with YRPW motif 2 |
| 6578 | SLCO2A1 | 5.874261 | 0.00176 | Solute carrier
organic anion transporter family, member 2A1 |
| 241 | ALOX5AP | 3.967401 | 0.00200 | Arachidonate
5-lipoxygenase-activating protein |
| 89874 | SLC25A21 | 6.886683 | 0.00211 | Solute carrier
family 25 (mitochondrial oxoadipate carrier), member 21 |
Functional analysis of DEGs
The 413 DEGs were classified by GO biological
processes. The most enriched terms of biological process were
‘steroid metabolic process’ (Table
III). The KEGG pathway enrichment analysis was performed to
select significantly overrepresented biochemical pathways (Table IV). Among the significantly
enriched pathways (determined by a hypergeometric test, where
P<0.05), ‘steroid hormone biosynthesis’ was the most
significant. Additionally, ‘cytokine-cytokine receptor interaction’
and ‘drug metabolism-cytochrome P450’ were highly enriched. The
network of proteins encoded by the top 10 up- and downregulated
DEGs were identified using the BioGRID PPI network (Fig. 3). The size of nodes representing
proteins indicates the degree of interaction in the PPI, where
larger nodes have more interactions. The proteins with
significantly more interactions were encoded by the upregulated
DEGs TRIM29, VTN, and ABCB1, and the
downregulated DEGs CCNA1 and KPNA5.
 | Table IIIThe top 5 enriched terms in
biological process of GO analysis. |
Table III
The top 5 enriched terms in
biological process of GO analysis.
| GO ID | Term | P-value | Genes |
|---|
| GO:0008202 | Steroid metabolic
process | 2.15E-11 | AKR1B10; TNF;
UGT2B15; LRP2; NPC1L1; CYP3A5; CELA3A; ESR1; AKR1C3; ACADL; LCAT;
HSD3B1; SOAT2; LMF1; UGT2B17; HSD17B1; EPHX2; HSD17B14; SLCO1B3;
HSD17B2; STS; NR5A2; MT3; SULT1B1; TFCP2L1; APOA1; HSD3B2; DKK3;
LIPC; CACNA1H |
| GO:0042221 | Response to
chemical stimulus | 1.28E-10 | VTN; ABCB1; PTPRM;
CXCR7; LY6D; OAS2; DPYSL3; OLR1; ABCG2; TIMP3; TNF; CD74; KPNA5;
UGT2B15; ACP5; ALOX5AP; CCL20; POU3F2; TFF1; MYL9; BRCA2; LRP2;
DLG4;ALDH3A1; LY96; IL1R1; CYP4F8; CYP3A5; CYP3A7; NTF3; AFF3;
CCL16; TH; SLC6A14; ESR1;AKR1C3; GCKR; LCAT; TESC; SPARC;S100A12;
HSPB7; GHR; NGF; PTGS2; COLEC12; FOXA1; BMP7; VN1R1;KRT13; MGMT;
SLC1A3; CIITA; RARRES2; GATM; KYNU; PDE1C; PTGER2; PLK3; CA2;
PDE3B;PSMB8; NRAS; CPB2; LHX2; GNB3; FGFBP1; CALCR; NPPB; EPHX2;
CX3CL1; GIP; LMO2; NNMT; MAP1B; GH2; GSTM3; CUX2; EBI3; PGR;
SERPINA1; FMO3; IFIT3; HTR2B; NRP2; PLA2G7; HERC5; HSD17B2; FADS1;
LUM; STS; NRCAM; HTR1B; MT3; SULT1B1; IRAK3; MICB; ABCB4; FES;
PDGFRB; MAT1A; GNAI1; ARTN; APOA1; S100A7; IL6; FZD5; IL15RA; RAC2;
CACNA1H; REN; CD14; ACSL5; SEMA3A; TRPC6; MPP1; TRPM6; GPR77 |
| GO:0050896 | Response to
stimulus | 7.84E-09 | CA8; VTN; ABCB1;
PTPRM; CXCR7;LY6D; OAS2; DPYSL3; OLR1; ABCG2; TIMP3; DHX58;
SPRR3;IFI44L; TFF3; TNF; RNF43; CD74; KPNA5; AZGP1; FSTL1; IL32;
CEACAM6; UGT2B15;CNTNAP2; ACP5; HEY2; ALOX5AP; CCL20; NPM1; POU3F2;
TFF1; CLEC1A; ABCA4; MYL9; BRCA2; LRP2; MAGEA1; DLG4; ALDH3A1;
GUCY1B3; PRRX2; GP2; CDH2; LY96; CPQ; IL1R1; CYP4F8; STRA6; NEDD9;
LGALS9; CYP3A5; CYP3A7; NTF3; AFF3; GPX2; CCL16; TH; NT5E; SLC6A14;
ITGA6; NR2F1;ESR1; CD3D; HRK; SRGN; CD19; AKR1C3; GCKR; LCAT;
TAAR5; TESC; CD33; IQGAP2; CSPG4; SPARC; S100A12; CUL3; TLE4;
INSL4; HSPB7; GHR; NGF; CEACAM1; PTGS2; COLEC12; FOXA1; DTX3; BMP7;
SIRPA; VN1R1; KRT13; MGMT; BFSP2; SLC1A3; CDC42EP3; FGD1;CIITA;
RARRES2; GATM; KYNU; PDE1C; PTGER2; PLK3; ENDOU; PDIA3; NINJ2; CA2;
ARHGDIG; PDE3B; CLEC4M; SLC7A10; PSMB8; NRAS; FGF21; CPB2; LHX2;
EHD3; NREP; EIF2C4; GPR15; ZNF175;IL2; GNB3; FGFBP1; AMHR2; CALCR;
CSF2; NFE2; NPPB; EPHX2; NOX3;CX3CL1; GIP; SAG; GAP43; ARL14; LMO2;
C8B; NNMT; MAP1B; KLK8; GH2; GSTM3; CUX2; EBI3; NPBWR2; RSAD2; PGR;
AVIL; SERPINA1; FMO3; CD300C; ORM1; IFIT3; RAMP3; RAB3B; KSR1;
HTR2B; RAB25; PDPN; STAB1; NRP2; PLA2G7; RPGRIP1; HERC5; HSD17B2;
RRH; FADS1; EMR1; LUM; STS; NODAL; NRCAM; HTR1B; NR5A2; NRG1; MT3;
SULT1B1; IRAK3; GIMAP5; MICB; CNGA3; ABCB4; FES; PDGFRB; MAT1A;
ACTN2; GNAI1; ARTN; APOA1; S100A7; CD8A; FZD9; IL6; DKK3;
FZD5;P2RX6; IL15RA; RAC2; A2M; CACNA1H; REN; GULP1; IGFBP6;
CD14;MIP; PRAME; ACSL5; SEMA3A; ZIC1; ARHGAP29; BIK; TRPC6; MPP1;
CAMP; TRPM6; GPR77; GNA15 |
| GO:0009605 | Response to
external stimulus | 1.59E-08 | VTN; PTPRM; DPYSL3;
TIMP3; TNF; CD74; ACP5; CCL20; ABCA4; MYL9; BRCA2; LRP2; DLG4;
ALDH3A1; STRA6; NTF3; CCL16; TH; NT5E; HRK; AKR1C3; TESC; SPARC;
GHR; NGF; PTGS2; BMP7; KRT13; MGMT; SLC1A3; RARRES2; GATM; KYNU;
NRAS; CPB2; LHX2; IL2; NOX3; CX3CL1; GIP; SAG; MAP1B; KLK8; PDPN;
NRP2;PLA2G7; HSD17B2; RRH; FADS1; NRCAM; MT3; MICB; FES; PDGFRB;
ARTN; APOA1; S100A7; IL6; RAC2;A2M; CACNA1H; ACSL5; SEMA3A; TRPC6;
MPP1; GPR77 |
| GO:0032501 | Multicellular
organismal process | 4.81E-08 | VTN; PTPRM; CXCR7;
SNTB1; CCNA1; AKR1B10; LY6D; ADAMTS9; DPYSL3; OLR1;TIMP3; DHX58;
SPRR3; TFF3; TNF; CD74; CNTNAP2; ODAM; ACP5; HEY2; ALOX5AP; PAQR5;
POU3F2;TFF1; IGF2BP3; ABCA4; MYL9; PTPRB; BRCA2; LRP2; DLG4; NME5;
KCND2; GUCY1B3; PRRX2; TBX2;CDH2; MAL; IL1R1; STRA6; NPC1L1;
PPP1R9A; NTF3; AFF3; SLC1A4; CDH11; TH; CELA3A; CELF3; TNNC1;ITGA6;
NR2F1; ESR1; CD3D; SRGN; AKR1C3; ACADL; LCAT; TAAR5; TESC; OLFM1;
CSPG4; SPARC; CUL3; INSL4; GPM6B; HSPB7; GHR; NGF; CEACAM1; PTGS2;
COLEC12; FOXA1; LEPREL1; BMP7; HOXC10; SIRPA; KRT13; MGMT; BFSP2;
SLC1A3; PCP4; FGD1; CIITA; SOAT2; RARRES2; GATM; LMF1; NINJ2;
PCDHA5; CA2; PDE3B; CLEC4M; SLC7A10; NRAS; FGF21; CPB2; LHX2; EHD3;
PCDHB12; NREP; IL2; ZNF287; GNB3; AMHR2; CSF2; NFE2; NPPB; EPHX2;
NOX3; CX3CL1; GIP; CRYAB; SAG; GAP43;LMO2; KCNQ4; MAP1B; KLK8;
CKMT2; GSTM3; EBI3; RSAD2; BARX2; AMELY; PGR; AVIL; SERPINA1; LBX1;
HOXA10; TNFAIP2; HTR2B; PDPN; PCDHB11; STAB1; NRP2; PLA2G7;RPGRIP1;
HERC5; CHODL; HSD17B2; RRH; LUM; STS; NODAL; NRCAM; HTR1B; NR5A2;
NRG1; MT3; CLGN; IRAK3; GIMAP5; CNGA3; FES; PDGFRB; TFCP2L1; ACTN2;
GNAI1; ARTN; APOA1; S100A7; CD8A; FZD9; IL6; NEB; DKK3; NEUROD6;
FZD5; LIPC; P2RX6; IL15RA; RAC2; A2M; CACNA1H; REN; CAPN9;CD14;
MIP; SEMA3A; ZIC1; LAMC2; BIK; CSGALNACT1; TRPC6; GNA15; HAND1 |
 | Table IVThe top 15 KEGG pathway enrichment of
the identified DEGs. |
Table IV
The top 15 KEGG pathway enrichment of
the identified DEGs.
| KEGG ID | Pathway | No. of genes | P-value |
|---|
| hsa00140 | Steroid hormone
biosynthesis | 9 | 1.91E-05 |
| hsa04060 | Cytokine-cytokine
receptor interaction | 15 | 0.00707 |
| hsa00982 | Drug metabolism -
cytochrome P450 | 5 | 0.00811 |
| hsa00980 | Metabolism of
xenobiotics by cytochrome P450 | 6 | 0.01756 |
| hsa00590 | Arachidonic acid
metabolism | 5 | 0.03716 |
| hsa04726 | Serotonergic
synapse | 6 | 0.04440 |
| hsa04540 | Gap junction | 6 | 0.04651 |
| hsa05145 | Toxoplasmosis | 6 | 0.05552 |
| hsa00040 | Pentose and
glucuronate interconversions | 3 | 0.05704 |
| hsa05152 | Tuberculosis | 9 | 0.07015 |
| hsa00350 | Tyrosine
metabolism | 3 | 0.07198 |
| hsa00260 | Glycine, serine and
threonine metabolism | 3 | 0.07198 |
| hsa04810 | Regulation of actin
cytoskeleton | 9 | 0.08736 |
| hsa04010 | MAPK signaling
pathway | 12 | 0.08955 |
| hsa04612 | Antigen processing
and presentation | 4 | 0.10430 |
Discussion
A major obstacle in breast cancer chemotherapy is
treatment failure due to anticancer drug resistance. Anthracyclines
are one of the most commonly used chemotherapy agents in breast
cancer; however, development of anthracycline resistance is a
common limitation. In order to manage
chemotherapy-resistant/refractory breast cancer, a comprehensive
analysis of the mechanisms underlying the development of
anthracycline resistance is essential. In the present study, we
used publically available data sets and a meta-analysis approach,
in which DEGs from various microarray datasets were combined and
analyzed to identify genes that were consistently and significantly
differentially expressed, to investigate the common biological
signatures in the development of anthracycline resistance in breast
cancer. Additionally, we investigated the DEGs by analysis of GO
enrichment, KEGG pathways, and a constructed PPI network.
We identified 413 DEGs potentially involved in the
development of anthracycline resistance. The upregulated gene with
the most statistical significance was TRIM29 (tripartite
motif containing 29), which is a member of the TRIM family, which
is involved in hematologic and solid tumor cancers. It may also
function in the suppression of radiosensitivity because it is
associated with the ataxia telangiectasia phenotype (19). Upregulation of TRIM29
reportedly promotes cancer cell proliferation and predicts poor
survival in gastric, prostate and pancreatic cancer (19–21),
however, its role with the development of anthracycline resistance
has yet to be associated. By contrast, some of our DEGs had already
linked to chemotherapeutic drug resistance. For example, consistent
with previous studies, we found upregulation of the genes that
encode proteins belonging to the multidrug resistance-associated
protein (MRPs) family. MDR1/ABCB1 (or Pgp), a member of the ABC
transporter superfamily, is a major contributor to resistance. A
variety of Pgp inhibitors have been identified, but they show no
consistent advantage in clinical studies (8,9). The
ABCG2 gene encodes a unique member of the ABC
half-transporter group that hydrolyzes ATP to efflux a large number
of chemotherapeutic agents. The substrates of the ABCG2 protein
include anticancer drugs primarily targeting topoisomerases, which
include the anthracyclines (22).
ABCG2-positive cells show increased tumorigenicity, and
overexpression of ABCG2 enhances the capacity for proliferation and
resistance to doxorubicin (23).
The most statistically significant downregulated DEG
was CA8. The protein encoded by this gene was initially
termed ‘CA-related protein’ because of sequence homologues with
other known carbonic anhydrase genes. However, CA8 lacks
carbonic anhydrase activity. Little is known with regard to how
CA8 functions in physiological processes, and its role has
yet to be reported in relation to the development of drug
resistance; therefore, it is a gene that remains to be
investigated. KPNA5, also known as importin α 6, was also
identified as a significantly downregulated DEG. The KPNA5 protein
belongs to the importin α protein family and is thought to be
involved in nuclear localization signal (NLS)-dependent protein
import into the nucleus. The mechanism underlying the acquisition
of drug-resistance is probably linked to nuclear trafficking
machinery. For example, the nuclear sparing phenomenon has been
reported in drug resistant cells treated with various
anthracyclines (24). Additionally,
drug-sensitive cancer cells transport anthracyclines in their
nuclei bound to a protein carrier (25). Therefore, the downregulation of
nuclear trafficking-associated genes may contribute to the
mechanism of anthracycline resistance.
In the GO term enrichment analysis of the 413 DEGs,
enriched terms included the biological processes ‘steroid metabolic
process’ and ‘response to chemical and external stimuli’, the
molecular functions ‘xenobiotic-transporting ATPase activity’ and
‘steroid dehydrogenase activity’, and the cellular components
‘plasma membrane’ and ‘extracellular region’. Of the 98
statistically significant pathways in our KEGG analysis, steroid
hormone biosynthesis, cytokine-cytokine receptor interaction, drug
metabolism-cytochrome P450, and metabolism of xenobiotics by
cytochrome P450 were the pathways most differentially regulated in
relation to acquired anthracycline-resistant breast cancer. A
number of drug efflux pumps are involved in the production and
secretion of steroid hormone, and the expression is usually
upregulated in tissues that partticipate in steroid hormone
biosynthesis (26). Cytochrome
P450s are enzymes that play a vital role in activating and
inactivating many anticancer drugs, including anthracyclines
(27). Therefore, cytochrome P450
pathways may be central to anthracycline resistance.
In our analysis, we identified a PPI network
comprising the encoded proteins from the top 10 up- and
downregulated DEGS. We found that TRIM29, ABCB1 and
VTN were significant hub proteins in the upregulation of the
PPI network, while CCNA1 and KPNA5 were hubs in the
downregulation network. Taken together, our meta-analysis and PPI
network strongly suggests that TRIM29 and KPNA5 are
involved in the development of acquired anthracycline resistance in
breast cancer. However, we acknowledge that further validation of
the DEGs is required, and suggest that additional investigation
could lead to the identification of new targets for anthracycline
resistance and possibly the development of better cancer
chemotherapy strategies.
In the present study, we followed a rigorous
protocol for the systematic review, in which we comprehensively
identified and analyzed data from three different databases.
However, the results of our meta-analysis should be interpreted
with caution in light of some unavoidable limitations. First,
potential heterogeneity and confounding factors may have affected
the analysis. For example, samples may be heterogeneous with
respect to culture conditions, drug exposure time, drug
concentrations, and microarray platforms. Second, ADR is a complex
and multifactorial phenomenon, and thus potential gene-gene and
gene-environment interactions must be considered. Despite these
limitations, our meta-analysis, which is the most up-to-date review
of the current evidence, provides a comprehensive view of gene
expression patterns and new regulatory insight for acquired
anthracycline-resistant breast cancer.
Acknowledgements
This study was supported by the National Research
Foundation of Korea Grant funded by the Korean Government
(NRF-2013R1A1A1075999).
References
|
1
|
Langer SW, Sehested M and Jensen PB:
Anthracycline extravasation: a comprehensive review of experimental
and clinical treatments. Tumori. 95:273–282. 2009.PubMed/NCBI
|
|
2
|
Gluck S: Adjuvant chemotherapy for early
breast cancer: optimal use of epirubicin. Oncologist. 10:780–791.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreno-Aspitia A and Perez EA:
Anthracycline- and/or taxane-resistant breast cancer: results of a
literature review to determine the clinical challenges and current
treatment trends. Clin Ther. 31:1619–1640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu BH: Strategy in the treatment of
anthracycline-resistant breast cancer. Zhonghua Zhong Liu Za Zhi.
29:241–244. 2007.(In Chinese). PubMed/NCBI
|
|
5
|
Stavrovskaya AA: Cellular mechanisms of
multidrug resistance of tumor cells. Biochemistry (Mosc).
65:95–106. 2000.
|
|
6
|
Banerjee D, Mayer-Kuckuk P, Capiaux G,
Budak-Alpdogan T, Gorlick R and Bertino JR: Novel aspects of
resistance to drugs targeted to dihydrofolate reductase and
thymidylate synthase. Biochim Biophys Acta. 1587:164–173. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Faneyte IF, Kristel PM and van de Vijver
MJ: Multidrug resistance associated genes MRP1, MRP2 and MRP3 in
primary and anthracycline exposed breast cancer. Anticancer Res.
24:2931–2939. 2004.PubMed/NCBI
|
|
8
|
Liscovitch M and Lavie Y: Cancer multidrug
resistance: a review of recent drug discovery research. IDrugs.
5:349–355. 2002.
|
|
9
|
van Zuylen L, Nooter K, Sparreboom A and
Verweij J: Development of multidrug-resistance convertors: sense or
nonsense? Invest New Drugs. 18:205–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ravdin PM: Anthracycline resistance in
breast cancer: clinical applications of current knowledge. Eur J
Cancer. 31A(Suppl 7): S11–S14. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chien AJ and Moasser MM: Cellular
mechanisms of resistance to anthracyclines and taxanes in cancer:
intrinsic and acquired. Semin Oncol. 35(Suppl 2): S1–S14; quiz S39.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang HC, Kim IJ, Park JH, et al:
Identification of genes with differential expression in acquired
drug-resistant gastric cancer cells using high-density
oligonucleotide microarrays. Clin Cancer Res. 10:272–284. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calcagno AM, Salcido CD, Gillet JP, et al:
Prolonged drug selection of breast cancer cells and enrichment of
cancer stem cell characteristics. J Natl Cancer Inst.
102:1637–1652. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Munkacsy G, Abdul-Ghani R, Mihaly Z, et
al: PSMB7 is associated with anthracycline resistance and is a
prognostic biomarker in breast cancer. Br J Cancer. 102:361–368.
2010. View Article : Google Scholar :
|
|
16
|
Siddiqui AS, Delaney AD, Schnerch A,
Griffith OL, Jones SJ and Marra MA: Sequence biases in large scale
gene expression profiling data. Nucleic Acids Res. 34:e832006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moreau Y, Aerts S, De Moor B, De Strooper
B and Dabrowski M: Comparison and meta-analysis of microarray data:
from the bench to the computer desk. Trends Genet. 19:570–577.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cohn LD and Becker BJ: How meta-analysis
increases statistical power. Psychol Methods. 8:243–253. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosaka Y, Inoue H, Ohmachi T, et al:
Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph
node metastasis in gastric cancer. Ann Surg Oncol. 14:2543–2549.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Yang H, Palmbos PL, et al:
ATDC/TRIM29 phosphorylation by ATM/MAPKAP kinase 2 mediates
radioresistance in pancreatic cancer cells. Cancer Res.
74:1778–1788. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanno Y, Watanabe M, Kimura T, Nonomura K,
Tanaka S and Hatakeyama S: TRIM29 as a novel prostate basal cell
marker for diagnosis of prostate cancer. Acta Histochem.
116:708–712. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dezi M, Fribourg PF, Di Cicco A, et al:
The multidrug resistance half-transporter ABCG2 is purified as a
tetramer upon selective extraction from membranes. Biochim Biophys
Acta. 1798:2094–2101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang G, Wang Z, Luo W, Jiao H, Wu J and
Jiang C: Expression of potential cancer stem cell marker ABCG2 is
associated with malignant behaviors of hepatocellular carcinoma.
Gastroenterol Res Pract. 2013:7825812013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Featherstone JM, Speers AG, Lwaleed BA,
Hayes MC, Cooper AJ and Birch BR: The nuclear membrane in multidrug
resistance: microinjection of epirubicin into bladder cancer cell
lines. BJU Int. 95:1091–1098. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kiyomiya K, Matsuo S and Kurebe M:
Mechanism of specific nuclear transport of adriamycin: the mode of
nuclear translocation of adriamycin-proteasome complex. Cancer Res.
61:2467–2471. 2001.PubMed/NCBI
|
|
26
|
Fujise H, Annoura T, Sasawatari S, Ikeda T
and Ueda K: Trans-epithelial transport and cellular accumulation of
steroid hormones and polychlorobiphenyl in porcine kidney cells
expressed with human P-glycoprotein. Chemosphere. 46:1505–1511.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Doehmer J, Goeptar AR and Vermeulen NP:
Cytochromes P450 and drug resistance. Cytotechnology. 12:357–366.
1993. View Article : Google Scholar : PubMed/NCBI
|