Introduction
Hepatocellular carcinoma (HCC) remains one of the
most prevalent malignant diseases and has the fourth highest
mortality rate worldwide (1).
Chronic HBV infection remains the major etiological factor of HCC
worldwide with >50% of HCC patients being chronic carriers
(2). The activities of HBx are
known to play major roles in the onset and progression of
HBV-associated HCC. HBx, encoded by the smallest open reading frame
of mammalian hepadnaviruses, is 154 amino acids in size, with a
molecular mass of ~17.5 kDa (3).
HBx protein interacts with different host factors to modulate cell
signal transduction pathways, transcriptional regulations, cell
cycle progression, DNA repair, apoptosis and genetic stability
(1).
HBx is predominately localized in the cytoplasm with
a low level of nuclear distribution (4). Mounting evidence from
immunofluorescence microscopy and subcellular fractionation
techniques have demonstrated that a fraction of cytosolic HBx
co-localizes with mitochondria in primary rat hepatocytes (5), in the human WRL68 liver cell line and
human HepG2 hepatocarcinoma cell line (6), in Huh7 (7) and in HBx-expressing COS cells, a
monkey kidney cell line (8), and in
HBx-transgenic mouse hepatocytes (9), suggesting a key role of HBx in
affecting mitochondrial physiology, metabolism and other relevant
functions. The abovementioned studies explain using different cell
lines that HBx localizes to the outer mitochondrial membrane
(10). However, our recent studies
have shown that the inner mitochondrial membrane protein, COXIII,
also interacts with hepatitis B virus X protein in vivo by
the yeast two-hybrid system, further verified by mating experiment,
coimmunoprecipitation and confocal laser scanning microscopy
(11–13).
Mitochondria are key organelles that regulate
apoptosis, cellular energetics and signal transduction pathways
(14), and are the source of
HBx-induced ROS (15). Accumulating
evidence has suggested that inflammation contributes to HCC
development due to the adverse effects of inflammatory mediators
such as proinflammatory cytokine and reactive oxygen species (ROS).
They play a key role on DNA repair, DNA methylation, DNA oxidation
and lipid peroxidation (16,17).
There are five enzyme complexes (complexes I–V) involved in
oxidative phosphorylation in mitochondria. Cytochrome c
oxidase (COX; or complex IV), composed of the COXI, COXII and
COXIII mtDNA encoded subunits and 10 nuclear DNA encoded subunits,
is an essential component of the respiratory chain that catalyzes
the reduction of molecular oxygen by reduced cytochrome c
(18). COX is the terminal enzyme
of the ETC and plays a pivotal role in the generation of ATP and
maintenance of the mitochondrial transmembrane potential (ΔΨm).
Increased expression of COX subunits is associated with restoration
of COX activity, increased cellular ATP levels, and a delayed
restoration of ΔΨm (19). In human
colorectal carcinoma cell lines metastasis is closely associated
with COXIII, which functions in the process of growth and
differentiation, transcription, apoptosis and signal transduction
(20). Furthermore, upregulation of
mitochondrial energy production-associated genes, COXIII and
COXVb, are capable of enhancing cell growth by supporting
higher energy requirements for cells (21,22).
The abovementioned findings indicate that changes of mitochondrial
function may be important in the HBX-associated HCC.
In the present study, we used HL-7702 cells which
expressed HBx stably as a biologically relevant system to explore
the physiological effect of HBx. We found that HBx located to the
inner mitochondrial membrane protein COXIII, leading to alterations
of mitochondrial function and the subsequent upregulation of
inflammatory mediator, ROS.
Materials and methods
Antibodies and reagents
Anti-DYKDDDDK-Tag (anti-flag) antibody was purchased
from Abmart (Shanghai, China). Anti-cytochrome c oxidase
subunit III (anti-COXIII) (N-20) antibody was purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Anti-β-actin
antibody was purchased from ZSGB-BIO (Beijing, China).
CF™488-conjugated donkey anti-mouse IgG (H+L), CF™350-conjugated
donkey anti-goat IgG (H+L), and puromycin were obtained from Sigma
(St. Louis, MO, USA). MitoTracker Red was obtained from Molecular
Probes (Eugene, OR, USA). Primers were produced by Biosune
(Shanghai, China).
Plasmids
The HBx expression plasmid PcDNA3.1-x (HBV subtype
ayw) was a gift from Professor Michael J. Bouchard (Drexel
University College of Medicine, Philadelphia, PA, USA). The
lentivirus packaging system: pLOV.CMV.eGFP.2A. EF1a.PuroR (pLOV),
psPAX2 and pMD2.G were provided by Neuron Biotech Co., Ltd.
(Shanghai, China). The lentivirus vector pLOV contains the gene
coding for green fluorescent protein (GFP), puromycin and the
peptide DYKDDDDK (flag).
Cells cultures
The human 293T embryonic kidney cell line was
provided by ATCC (UK). The human HL-7702 hepatocytes cell line was
purchased from the Shanghai Cell Bank (Shanghai, China). The 293T
and HL-7702 cells were grown in Dulbecco’s modified Eagle’s medium
(DMEM) containing 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin solution and maintained at 37°C in a
humidified atmosphere composed of 95% air and 5%
CO2.
Generation of the recombinant lentivirus
and establishment of the stably transfected HL-7702 cell line
Full-length HBx (465 bp) was amplified from
PcDNA3.1-x by polymerase chain reaction (PCR) and then infused with
flag epitope at the N-terminus and then cloned into pLOV. The
plasmid was digested with XbaI to construct the recombinant
lentiviral vector pLOV.flag-HBx. The recombinant lentiviral
particles were generated by transient cotransfection with 293T
cells by the three-plasmid expression system, and harvested by
filtration through a 0.45 μm filter and ultracentrifugation
at 100,000 × g for 2 h at 4°C. The recombinant lentiviral particles
titer was calculated by qPCR and used to transduce HL-7702 cells.
The cell clones were then treated with 0.2 μg/ml puromycin
for 10 days. Expression of GFP in the objective clones was examined
directly by fluorescence microscopy. Positive clones expressing GFP
and resistant to puromycin were screened and designated as
HL-7702-HBx and HL-7702-mock, respectively. HBx, mock and control
were considered to represent HL-7702-HBx, HL-7702-mock and HL-7702
cells, respectively. The expression of flag-HBx was detected by
qPCR and western blot analysis.
Indirect immunofluorescence
To eliminate the impact of green fluorescence caused
by GFP in HL-7702-HBx cells, we used 100% methanol as a fixative
(23). As a result, green
fluorescence disappeared (data not shown). The distributions of HBX
and COXIII proteins were measured by confocal microscopy. The cells
were seeded directly onto coverslips and incubated for 48 h. When
the density of cells achieved 60–70%, the culture medium was
removed by repeated washes with phosphate-buffered saline (PBS),
and the cells were stained for 30 min with 150 nM MitoTracker Red.
The cells were first fixed with a 100% ice methanol solution,
preincubated in blocking solution (5% donkey serum albumin in PBS)
and incubated with the appropriate primary antibodies (anti-flag
and anti-COXIII) at 4°C overnight. After overnight incubation, the
cells were washed. The fluorescence-labeled secondary antibodies
(CF™488-conjugated donkey anti-mouse, CF™350-conjugated donkey
anti-goat) were added and incubated for 60 min at 37°C in the dark,
and the sections were mounted using glycerol. Fluorescence images
were captured by confocal laser scanning microscopy (Leica,
Germany).
Isolation of mitochondria and measurement
of COX activity
Mitochondria were isolated using the mitochondrial
isolation kit for mammalian cells (Thermo Scientific, Waltham, MA,
USA) according to the manufacturer’s instructions. Briefly,
following lysis of ~2×107 cells, cell debris and nuclei
were pelleted at 700 × g for 10 min at 4°C, followed by
centrifugation at 3,000 × g for 15 min at 4°C to pellet a
mitochondrially enriched fraction, and then 12,000 × g for 5 min at
4°C to pellet the isolated mitochondria. The isolated mitochondria
were vortexed with 30 μl of 1X enzyme dilution buffer (10 mM
Tris-HCl, pH 7.0, containing 250 mM sucrose). Protein
concentrations were measured using the bicinchoninic acid (BCA)
assay and then adjusted to 1 μg/μl. COX activity was
determined using the Cytochrome c Oxidase Assay kit (GenMed,
Shanghai, China) according to the manufacturer’s instructions. COX
activity was based on a colorimetric assay that quantifies the
oxidation of ferrocytochrome c to ferricytochrome c
via cytochrome c oxidase, a reaction that results in a
decrease in absorbance at 550 nm. This decrease was monitored by an
enzyme mark instrument (Bio-Tek, Winooski, VT, USA) calibrated to
zero. Isolated mitochondria (10 μl) were combined with 10
μl of lysis buffer, then mixed with 205 μl of buffer
solution in 96-well plates. The reaction was initiated by the
addition of 25 μl of reaction liquid, and the decrease in
absorbance at 550 nm was measured for 1 min. Activity was
calculated based on the equation: U/ml = [(ΔAbs550/min for the
sample - ΔAbs550/min for the blank) x dilution factor x total
reaction volume]/[mitochondria isolate volume x the difference in
extinction coefficients between ferro- and ferri-cytochrome
c at 550 nm (21.84)]. One unit was considered the amount
that oxidizes 1 μmol reduced cytochrome c/min at pH
7.0 and 25°C.
Flow cytometric analysis
Cells in the three groups were harvested with
trypsin and resuspended in PBS (1×106 cells/ml) to
analyze ΔΨm, ROS and cell cycles by flow cytometry. The ΔΨm was
assessed using the cationic fluorescent dye, TMRM. When cells
reached 1×106 cells/ml, they were incubated in 100 nM
TMRM solution at 37°C in the dark for 20 min. The cells were washed
twice with PBS and resuspended in PBS for analysis by a flow
cytometer. Intracellular ROS levels were subsequently detected by
staining cells with 50 μM dihydroethidium (DHE) fluorescence
probe (Vigorous Biotechnology, Beijing, China) for 30 min in the
dark. The cells were collected by trypsinization and the mean
fluorescence intensity was quantified by flow cytometry.
Transmission electron microscopy
(TEM)
The morphological changes of cell and mitochondria
were observed by TEM. Cells (1×106) were collected and
subjected to fixation with 3% fresh glutaraldehyde and 1.5%
paraformaldehyde solution at 4°C for 1 h, followed by post-fixation
with 1% osmium tetroxide and 1.5% potassium ferrocyanide solution
at 4°C for 1.5 h. The cells were dehydrated with a graded series of
ethanol solution and embedded in Epon-618. Ultrathin sections were
cut to stain with uranyl acetate and lead citrate, and observed
under TEM (Phillips, Madison, WI, USA).
qPCR detects gene expression
Total RNA was extracted using TRIzol reagent (Life
Technologies, Rockville, MD, USA). The primer sequences of each
gene are listed in Table I. The
first-strand cDNA was generated using MMLV transcriptase [New
England Biolabs (NEB), Ipswich, MA, USA], and qPCR was performed
with FastStar Universal SYBR-Green Master (Roche, Foster City, CA,
USA) in triplicate in an Applied Biosystems StepOnePlus Real-Time
PCR system (Life Technologies). Endogenous mRNA values were
normalized to the level of β-actin mRNA.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene name | Primer sequence (5′
to 3′) |
|---|
| HBV X | F:
ACTCTCTCGTCCCCTTCTCC |
| R:
GGTCGTTGACATTGCTGAGA |
| COXIII | F:
CCCGCTAAATCCCCTA′AAG |
| R:
GGAAGCCTGTGGCT′CAAAA |
| β-actin | F:
CTCCATCCTGGCCTCGCTGT |
| R:
GCTGTCACCTTCACCGTTCC |
Western blot analysis
The cells were prepared by washing with ice-cold PBS
and lysed. Protein concentrations were measured using the
bicinchoninic acid (BCA) assay. Equal amounts of protein were
loaded and separated by SDS-PAGE, and transferred to a
nitrocellulose membrane. The membrane was blocked in 5% milk in
TBST [0.1% Tween-20, 20 mM Tris (pH 7.4) and 150 mM NaCl] for 2 h
at room temperature, followed by overnight incubation with the
primary antibody at 4°C. The membrane was washed and incubated with
a horseradish peroxidase-conjugated secondary antibody for 1 h at
room temperature. Proteins were visualized using an enhanced
chemiluminescence (ECL) kit (ZSGB-BIO Co. Ltd., Beijing, China).
Images of the blots were captured using the Image Scanner (Epson,
Nagano, Japan). The band intensity was quantified using ImageJ
software.
Statistical analysis
Each set of experiments was repeated at least three
times with similar results. Results were presented as means ±
standard deviations (SD). Statistical evaluation was carried out by
one-way analysis of variance (ANOVA). P<0.05 was considered to
indicate a statistically significant result.
Results
Construction and identification of an
HL-7702 cell line stably transduced with a lentivirus expressing
the HBx gene
Lentiviral vectors lead to prolonged transgene
expression (24). In the present
study, we used the vectors to establish long-term HBx expressing
HL-7702 cells in vitro. The sequences of the recombinant
vector PCMV.flag-HBx examined by the sequencing process showed that
the full-length HBx gene had been successfully subcloned
into the lentiviral vector (data not shown). The titre of the
recombinant lentivirus was 8.61×107 TU/ml. GFP
expression of HL-7702-HBx and HL-7702-mock were observed by
fluorescence microscopy. The GFP-positive cells were 95-98%
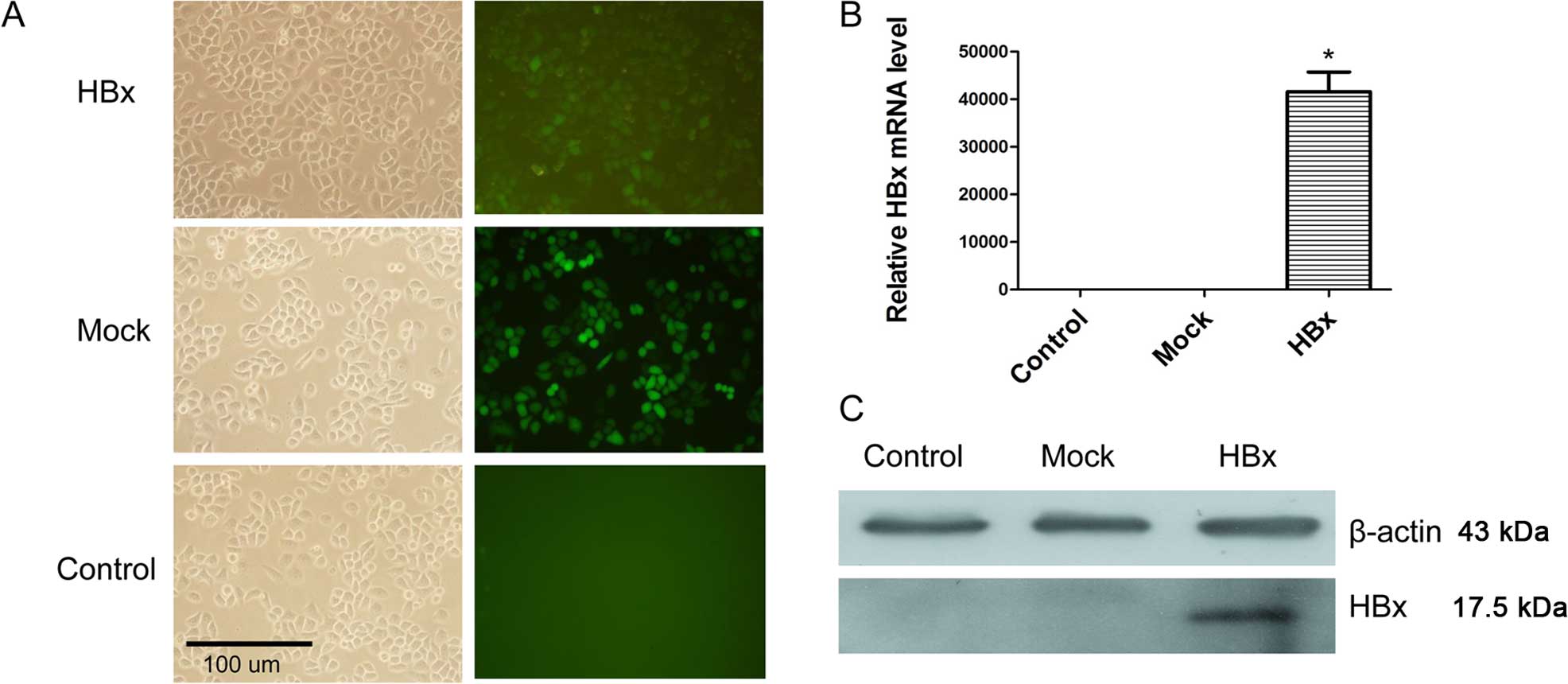
(Fig. 1A). Expression of HBx mRNA
and protein were detected in HL-7702-HBx cells (Fig. 1B and C). This showed that the
HL-7702 cell line stably expressing HBx was established.
HBx co-localizes with inner mitochondrial
protein COXIII in HL-7702 cells
Our recent studies have verified that COXIII
interacts with HBx (11–13). To confirm this finding, we subjected
HL-7702-HBx and control cells to fluorescent staining and
examination by confocal microscopy. The co-localization of HBx
protein (green color) with COXIII protein (blue color) in
mitochondria (red color) was in white in HL-7702-HBx cells, while
in HL-7702 cells the co-localization of COXIII protein (blue color)
and mitochondria (red color) was in purple (Fig. 2). In addition, HBx localized in the
nucleus and cytoplasm, with a fraction of cytosolic HBx
co-localized with mitochondria as identified in previous studies
(4–9). Thus, our study demonstrated that HBx
co-localized with inner mitochondrial protein COXIII in HL-7702
cells.
Upregulation of the mitochondrial
function in HL-7702-HBx cells
COXIII is an essential component of the
mitochondrial respiratory chain. Thus, the co-localization of HBx
and COXIII was expected to induce an alteration in the normal
mitochondrial electron flow. To determine mitochondrial function
alteration in HL-7702-HBx cells, we compared the expression of
mitochondrial protein subunits of complexes IV (COXIII subunit).
The expression of COXIII protein was increased in HL-7702-HBx
cells, while the RNA level did not change (Fig. 3A and B). The result indicated that
HBx regulated COXIII gene function at the post-transcription
level. Then we measured the activities of COX (Fig. 3C). As expected, COX activity of
HL-7702-HBx cells was significantly increased, correlating with
their upregulated protein activities. However, there was no
difference in the ΔΨm of any of the cells (Fig. 3D).
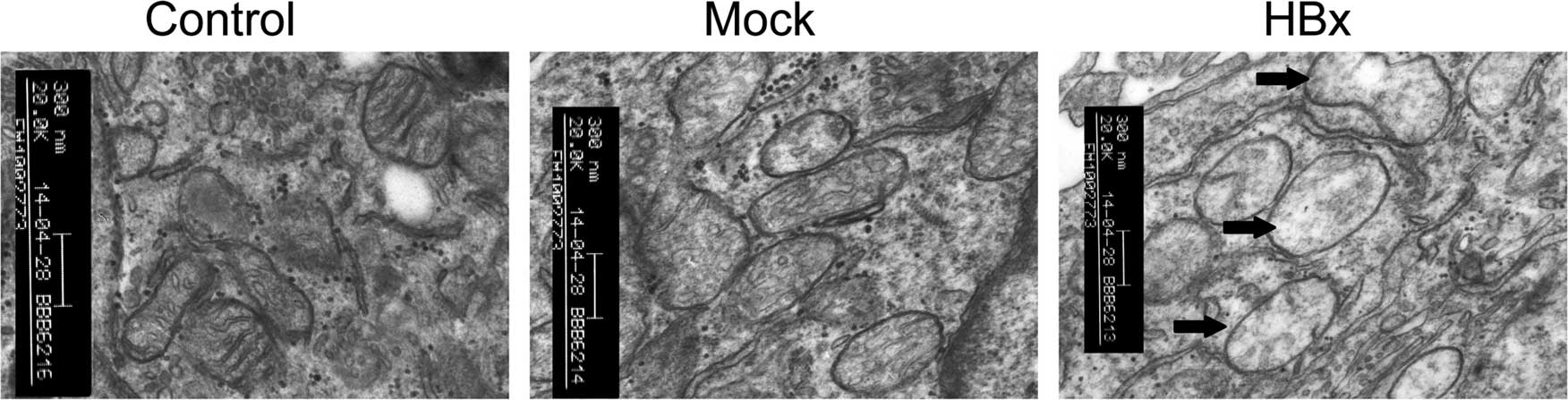
Changes of mitochondrial ultrastructure
in HBx steadily expressed cells
Abnormal aggregation of mitochondria has been found
in HBx-expressing cells (9,25). To assess the mitochondrial
morphological consequences in HL-7702-HBx cells, we observed the
mitochondria, nuclei and other intracellular membrane structures by
TEM. As shown in Fig. 4,
mitochondria had only slight swelling in HL-7702-HBx cells.
Mitochondria in HL-7702-mock and HL-7702 cells were round or oval,
and their membrane was integral, crest was dense and arranged in
order. Our results indicated that cell and nuclear morphology were
not significantly changed and there was no mitochondrial
aggregation in any of the three groups.
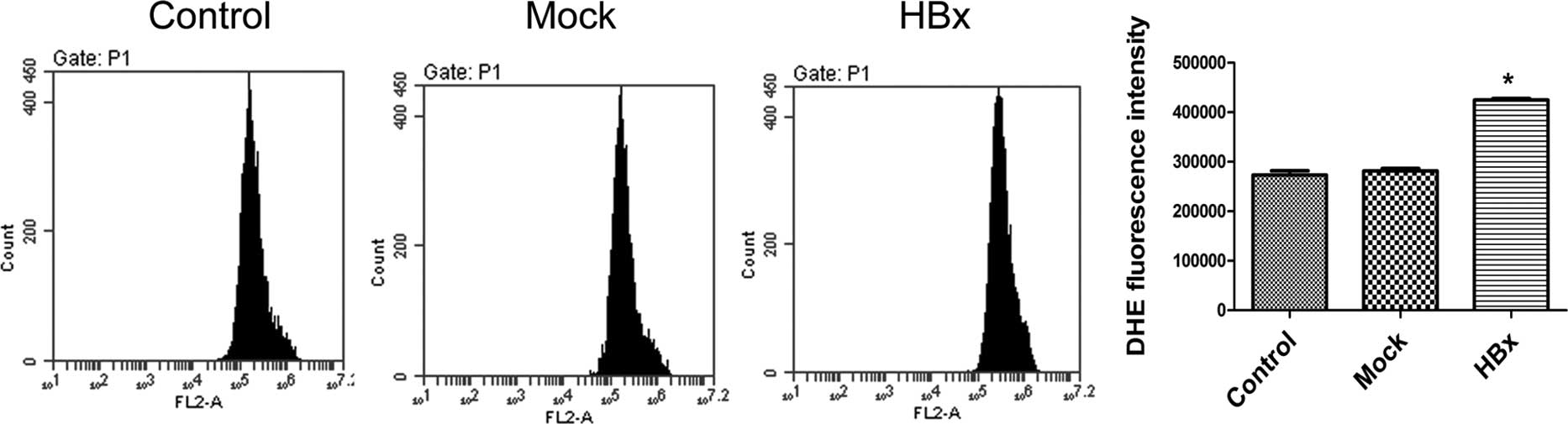
Upregulation of ROS production in
HL-7702-HBx cells
Since we demonstrated that HBx upregulates genes are
involved in oxidative phosphorylation at mitochondria, a major ROS
generator, we examined whether HBx induces ROS production in
HBx-transfected cells. Therefore, we examined ROS levels by flow
cytometry using the DHE probe. Fig.
5 shows that DHE fluorescence revealed an increase in the
HL-7702-HBx cells.
Discussion
HBx plays a key role in the development of
HBV-associated HCC (3). Using a
variety of methods such as liposome preparation, electrical
transduction and adenovirus can deliver HBx. However, lentiviral
vectors are more suitable for long-term expression in non-dividing
or infrequently dividing cells due to advantages of low toxicity
and lack of immunogenicity (26,27).
In the present study, we successfully established HL-7702 cells
that stably expressed HBx in vitro using lentiviral vectors.
They served as a biologically relevant system for exploring the
effect of HBx in human normal hepatocytes in physiology.
HBx protein is located in the cytoplasm and partly
in the nucleus (4). Accumulating
evidence has demonstrated that a fraction of cytosolic HBx
co-localizes with the mitochondria in different cell lines
(5–10) in addition to the outer mitochondrial
membrane protein, VDAC3 (8,10). HBx has been found to localize to
mitochondria, leading to mtDNA damage, which encodes COXIII subunit
(18,28). Recent findings have shown that
COXIII also interacts with HBx (11–13).
These findings strongly predict the interaction between HBx and
COXIII. However, the localization of HBx within the inner
mitochondrial membrane and the effect of HBx on mitochondrial
physiology in HL-7702 cells have not been previously addressed. To
the best of our knowledge, our results were the first to suggest
the inner mitochondrial membrane localization of HBx in HL-7702
cells (Fig. 2). These results
provide insight into the mitochondrial localization of HBx.
COX is the terminal complex of the electron
transport chain and its biogenesis is a critical part of
mitochondrial biogenesis (29,30).
Mounting evidence suggests that the upregulation of COX has been
found in human prostate (31) and
breast carcinoma (32), and human
colon cancers (22). In the present
study, we characterized the upregulation of COX biogenesis in
HL-7702-HBx cells, including the increasing protein expression of
COXIII and upregulation of the activities of COX, which may provide
more energy for cells to grow. These results were consistent with
recent studies showing HBx (HBV serotype ayw) upregulated the
protein expression of COXIII and activities of COX in HepG2 cells
which stably expressed HBx using lentiviral vectors (13). By contrast, HBx (HBV genotype B)
downregulated COXIII expression and inhibited COX activity in
HL-7702 cells that expressed HBx using eukaryotic vector (33). The reasons for this discrepancy
include that HBx plays pro-apoptotic and anti-apoptotic roles in
different situations (34), and
different genotypes of HBV may play different roles. On the other
hand, HBx in long-term transfected cells is different from that in
transiently transfected cells. COX accepts electrons from
cytochrome c and transfers them to molecular oxygen. At the
same time protons are pumped across the inner mitochondrial
membrane leading to the generation of ΔΨm (35). It has been reported that HBx located
in mitochondria may cause loss of ΔΨm in hepatoma cells (8,36). By
contrast, in our system, the co-localization of HBx with COXIII
protein did not induce changes in ΔΨm. We consider that long-term
HBx-transfected cells in our system were more stable than the
abovementioned HBx transiently transfected cells. Thus, the
upregulation of COX biogenesis did not result in ΔΨm alteration.
Additionally, the role of HBx in human normal hepatocytes (HL-7702
cells) may be different from that in hepatoma cells. We then showed
the slight swelling of mitochondria in HL-7702-HBx cells
accompanied by increased respiratory enzyme activities and protein
levels in mitochondria, as indicated by electron microscopy. This
is an adaptive state for mitochondria, which adapt to the change of
microenvironment. These results suggest that the co-localization of
HBx with COXIII led to changes of mitochondrial function and
morphology.
Mitochondria are the source of ROS (15). COX encoded gene expression changes
are associated with the development of tumors mainly through an
increase in ROS in mitochondria oxidative phosphorylation (37–40).
In the present study, we observed that HBx expression elevated the
generation of ROS (Fig. 5). HBx
increases the level of mitochondrial ROS, which is associated with
hepatocellular carcinogenesis (15,41),
although the mechanism involved remains controversial.
In summary, we have demonstrated that the HBx
protein co-localizes with the inner mitochondrial membrane protein
COXIII in HL-7702 cells, leading to changes of mitochondrial
function, morphology and increasing the generation of mitochondrial
ROS. These results provide insight into the molecular mechanism of
HBV-associated HCC.
Acknowledgments
The present study was supported by the National
Natural Science (grant no. 81300321) from the Foundation of China,
the Key Clinical Specialty Discipline Construction Program of
Fujian Province (2012-149), and Young and Middle-Aged Personnel
Training Project of Fujian Province Health Department
(2014-ZQN-ZD-9). We thank Professor Bouchard for the PcDNA3.1-X
plasmid.
References
|
1
|
Motavaf M, Safari S, Saffari Jourshari M
and Alavian SM: Hepatitis B virus-induced hepatocellular carcinoma:
The role of the virus x protein. Acta Virol. 57:389–396. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Neuveut C, Wei Y and Buendia MA:
Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol.
52:594–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bouchard MJ and Schneider RJ: The
enigmatic X gene of hepatitis B virus. J Virol. 78:12725–12734.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Henkler F, Hoare J, Waseem N, Goldin RD,
McGarvey MJ, Koshy R and King IA: Intracellular localization of the
hepatitis B virus HBx protein. J Gen Virol. 82:871–882.
2001.PubMed/NCBI
|
|
5
|
Clippinger AJ and Bouchard MJ: Hepatitis B
virus HBx protein localizes to mitochondria in primary rat
hepatocytes and modulates mitochondrial membrane potential. J
Virol. 82:6798–6811. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li SK, Ho SF, Tsui KW, Fung KP and Waye
MY: Identification of functionally important amino acid residues in
the mitochondria targeting sequence of hepatitis B virus X protein.
Virology. 381:81–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J and Siddiqui A: Hepatitis B virus X
protein stimulates the mitochondrial translocation of Raf-1 via
oxidative stress. J Virol. 81:6757–6760. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rahmani Z, Huh KW, Lasher R and Siddiqui
A: Hepatitis B virus X protein colocalizes to mitochondria with a
human voltage-dependent anion channel, HVDAC3, and alters its
trans-membrane potential. J Virol. 74:2840–2846. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takada S, Shirakata Y, Kaneniwa N and
Koike K: Association of hepatitis B virus X protein with
mitochondria causes mitochondrial aggregation at the nuclear
periphery, leading to cell death. Oncogene. 18:6965–6973. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huh KW and Siddiqui A: Characterization of
the mitochondrial association of hepatitis B virus X protein, HBx.
Mitochondrion. 1:349–359. 2002. View Article : Google Scholar
|
|
11
|
Li D, Wang XZ, Yu JP, Chen ZX, Huang YH
and Tao QM: Cytochrome c oxidase III interacts with hepatitis B
virus X protein in vivo by yeast two-hybrid system. World J
Gastroenterol. 10:2805–2808. 2004.PubMed/NCBI
|
|
12
|
Wang XZ, Li D, Tao QM, Lin N and Chen ZX:
A novel hepatitis B virus X-interactive protein: Cytochrome C
oxidase III. J Gastroenterol Hepatol. 21:711–715. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng BY, Fang XF, Zou LY, Huang YH, Chen
ZX, Li D, Zhou LY, Chen H and Wang XZ: The co-localization of HBx
and COXIII upregulates COX-2 promoting HepG2 cell growth. Int J
Oncol. 45:1143–1150. 2014.PubMed/NCBI
|
|
14
|
Salvioli S, Bonafè M, Capri M, Monti D and
Franceschi C: Mitochondria, aging and longevity - a new
perspective. FEBS Lett. 492:9–13. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee YI, Hwang JM, Im JH, Lee YI, Kim NS,
Kim DG, Yu DY, Moon HB and Park SK: Human hepatitis B virus-X
protein alters mitochondrial function and physiology in human liver
cells. J Biol Chem. 279:15460–15471. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nishida N, Arizumi T, Takita M, et al:
Reactive oxygen species induce epigenetic instability through the
formation of 8-hydroxy-deoxyguanosine in human
hepatocarcinogenesis. Dig Dis. 31:459–466. 2013. View Article : Google Scholar
|
|
17
|
Castello G, Costantini S and Scala S:
Targeting the inflammation in HCV-associated hepatocellular
carcinoma: A role in the prevention and treatment. J Transl Med.
8:1092010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mkaouar-Rebai E, Ellouze E, Chamkha I,
Kammoun F, Triki C and Fakhfakh F: Molecular-clinical correlation
in a family with a novel heteroplasmic Leigh syndrome missense
mutation in the mitochondrial cytochrome c oxidase III gene. J
Child Neurol. 26:12–20. 2011. View Article : Google Scholar
|
|
19
|
Bauerfeld CP, Rastogi R, Pirockinaite G,
Lee I, Hüttemann M, Monks B, Birnbaum MJ, Franchi L, Nuñez G and
Samavati L: TLR4-mediated AKT activation is MyD88/TRIF dependent
and critical for induction of oxidative phosphorylation and
mitochondrial transcription factor A in murine macrophages. J
Immunol. 188:2847–2857. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang L, Qu L and Ding Y: Protein and mRNA
characterization in human colorectal carcinoma cell lines with
different metastatic potentials. Cancer Invest. 25:427–434. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bafna S, Singh AP, Moniaux N, Eudy JD,
Meza JL and Batra SK: MUC4, a multifunctional transmembrane
glycoprotein, induces oncogenic transformation of NIH3T3 mouse
fibroblast cells. Cancer Res. 68:9231–9238. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H, Rao GN, Dai B and Singh P: Autocrine
gastrins in colon cancer cells up-regulate cytochrome c oxidase Vb
and down-regulate efflux of cytochrome c and activation of
caspase-3. J Biol Chem. 275:32491–32498. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nybo K: GFP imaging in fixed cells.
Biotechniques. 52:359–360. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu B, Tai A and Wang P: Immunization
delivered by lentiviral vectors for cancer and infectious diseases.
Immunol Rev. 239:45–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim
K and Cho H: Hepatitis B virus x protein induces perinuclear
mitochondrial clustering in microtubule- and Dynein-dependent
manners. J Virol. 81:1714–1726. 2007. View Article : Google Scholar :
|
|
26
|
Naldini L, Blömer U, Gallay P, Ory D,
Mulligan R, Gage FH, Verma IM and Trono D: In vivo gene delivery
and stable transduction of nondividing cells by a lentiviral
vector. Science. 272:263–267. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ohashi K, Park F, Schwall R and Kay M:
Efficient gene transduction to cultured hepatocytes by HIV-1
derived lentiviral vector. Transplant Proc. 34:1431–1433. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jung SY and Kim YJ: C-terminal region of
HBx is crucial for mitochondrial DNA damage. Cancer Lett.
331:76–83. 2013. View Article : Google Scholar
|
|
29
|
Zaslavsky D and Gennis RB: Proton pumping
by cytochrome oxidase: Progress, problems and postulates. Biochim
Biophys Acta. 1458:164–179. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qi Z, He J, Su Y, et al: Physical exercise
regulates p53 activity targeting SCO2 and increases mitochondrial
COX biogenesis in cardiac muscle with age. PLoS One. 6:e211402011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang FL, Wang Y, Wong WK, Liu Y,
Addivinola FJ, Liang P, Chen LB, Kantoff PW and Pardee AB: Two
differentially expressed genes in normal human prostate tissue and
in carcinoma. Cancer Res. 56:3634–3637. 1996.PubMed/NCBI
|
|
32
|
Bini L, Magi B, Marzocchi B, et al:
Protein expression profiles in human breast ductal carcinoma and
histologically normal tissue. Electrophoresis. 18:2832–2841. 1997.
View Article : Google Scholar
|
|
33
|
Lin N, Li D, Chen HY, Chen ZX and Wang XZ:
Effect of HBV X gene on mitochondria in HL-7702 cells. Xi Bao Yu
Fen Zi Mian Yi Xue Za Zhi. 24:972–974. 2008.In Chinese. PubMed/NCBI
|
|
34
|
Lee WP, Lan KH, Li CP, Chao Y, Lin HC and
Lee SD: Pro-apoptotic or anti-apoptotic property of X protein of
hepatitis B virus is determined by phosphorylation at Ser31 by Akt.
Arch Biochem Biophys. 528:156–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Adrie C, Bachelet M, Vayssier-Taussat M,
Russo-Marie F, Bouchaert I, Adib-Conquy M, Cavaillon JM, Pinsky MR,
Dhainaut JF and Polla BS: Mitochondrial membrane potential and
apoptosis peripheral blood monocytes in severe human sepsis. Am J
Respir Crit Care Med. 164:389–395. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shirakata Y and Koike K: Hepatitis B virus
X protein induces cell death by causing loss of mitochondrial
membrane potential. J Biol Chem. 278:22071–22078. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ray AM, Zuhlke KA, Levin AM, Douglas JA,
Cooney KA and Petros JA: Sequence variation in the mitochondrial
gene cytochrome c oxidase subunit I and prostate cancer in African
American men. Prostate. 69:956–960. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Athar M, Chaudhury NK, Hussain ME and
Varshney R: Hoechst 33342 induced reactive oxygen species and
impaired expression of cytochrome c oxidase subunit 1 leading to
cell death in irradiated human cancer cells. Mol Cell Biochem.
352:281–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bernstein C, Facista A, Nguyen H, et al:
Cancer and age related colonic crypt deficiencies in cytochrome c
oxidase I. World J Gastrointest Oncol. 2:429–442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gutierrez-Gonzalez L, Graham TA,
Rodriguez-Justo M, et al: The clonal origins of dysplasia from
intestinal metaplasia in the human stomach. Gastroenterology.
140:1251–1260.e1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ha HL and Yu DY: HBx-induced reactive
oxygen species activates hepatocellular carcinogenesis via
dysregulation of PTEN/Akt pathway. World J Gastroenterol.
16:4932–4937. 2010. View Article : Google Scholar : PubMed/NCBI
|