Introduction
Regulator of G protein signaling (RGS) proteins is a
family of GTPase activating proteins, which interact directly with
α subunit of heterotrimeric G proteins to accelerate the GTP
hydrolysis activity of the α subunit and negatively regulate the G
protein signaling (1,2). All the RGS family members contain an
essential 120-amino acid RGS domain. Regulator of G protein
signaling 5 (RGS5) belongs to the R4 subfamily of RGS proteins,
which consists mostly of small RGS proteins containing only short
sequences beyond the RGS domain (3). RGS5 expression is abundant in
pericytes and vascular smooth muscle cells (4,5) and
high levels of RGS5 have been detected in the heart, lung, skeletal
muscle and small intestine, while lower levels have been found in
the brain, placenta, liver, colon and leukocytes (6). RGS5 has been implicated in many
pathophysiological processes including cardiac hypertrophy
(7), blood pressure regulation
(8), atherosclerosis (9,10),
lipid metabolism (11), bronchial
smooth muscle cell contraction and asthma (12). RGS5 was markedly downregulated
during the endothelial cell morphogenesis in an in vitro
human capillary tube formation model (13). RGS5 levels were elevated in
pericytes during wound healing and ovulation, suggesting a strong
correlation between the RGS5 expression and the active vessel
remodeling (14,15).
Substantial evidence indicates an active role of
RGS5 in tumor angiogenesis. RGS5 is found highly expressed in the
vasculature of several tumors. RGS5 mRNA is expressed in the
endothelial cells of tumor vessels, but not in the endothelial
cells of the normal tissues in renal cell carcinoma (RCC) (16), hepatic cell carcinoma (HCC)
(17) and ovarian carcinoma
(18). RGS5 mRNA is specifically
upregulated in the vasculature of premalignant lesions during the
‘angiogenic switch’ and further elevated in tumor vessels in a
mouse model of pancreatic islet cell carcinogenesis (14). Elevated expression of RGS5 has been
reported in tumor cells in gastric tumor (19), parathyroid (20) and non-small cell lung cancer (NSCLC)
(21). Yet, the fundamental role
RGS5 played in the neoplasm and its mechanisms are still to be
clarified.
In the present study, we investigated the potential
role of the high levels of RGS5 in human lung cancer cells and
further explored the underlying molecular mechanisms.
Materials and methods
Ethics statement
The present study conforms to the principles
outlined in the Declaration of Helsinki, and was approved by the
Medical Ethics Committee of Sun Yat-Sen University. Written
informed consents were obtained from the donors.
Cells and cultures
Human lung cancer cell lines A549, Calu-3, H1299 and
SK-MES-1 were obtained from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China), and cultured in RPMI-1640 medium
supplemented with 10% (v/v) fetal bovine serum (FBS) (both from
Life Technologies, Grand Island, NY, USA) at 37°C, 5%
CO2 in a humidified incubator. Human umbilical vein
endothelial cells (HUVECs) were freshly isolated from the human
umbilical cord veins, and incubated in the human endothelial-SFM
basal growth medium supplemented with 20% fetal calf serum, 100
μg/ml streptomycin, 100 U/ml penicillin, 5 μg/ml
amphotericin B (Life Technologies), 2 mmol/l l-glutamine, 15 mg/l
ECGS (Upstate Biotechnology, Lake Placid, NY, USA) (22).
mRNA extraction and reverse transcription
PCR
mRNA extractions were performed with an ultrapure
RNA purification kit (Kangweishiji Biotech Co., Ltd., China)
according to the manufacturer instructions. Approximately 500 ng of
RNA was reverse transcribed into complementary DNA using a
PrimeScript RT Reagent kit (Takara, Shiga, Japan). Relative
quantity of RGS5 mRNA was determined using GAPDH as an internal
control. PCR was performed with the primers for RGS5,
5′-atgtgcaaaggacttgcagc-3′/5′-ctacttgattaactcctgat-3′; and GAPDH,
5′-tccaccaccctgttgctgta-3′/5′-accacagtccatgc atcac-3′.
Cloning of RGS5 coding region and cell
transfection
The complete RGS5 coding region was cloned into the
pTRiEX Neo expression vector (Novagen, USA) for transient
transfection of cancer cell lines. The RGS5 coding region was
amplified from cDNA of HUVECs using primers 5′-cgcggatccgggatcc
gatgtgcaaaggacttgcagc-3′ and 5′-ccgctcgagcttgattaactcctgataaaac-3′
which contain restriction site tags for BamHI and
XhoI, respectively. The RGS5 coding region was then cloned
into the pTRiEX Neo vector (Clontech, USA). Human lung cancer cell
lines A549 and Calu-3 were transiently transfected with pTRiEX
vectors or pTRiEX-RGS5 plasmids using Attractene Transfection
Reagent (Qiagen, Germany) according to the protocol of the
manufacturer.
Cell viability
Cell viability was measured by MTT assays as
previously described (23). At
various time-points following transfection, MTT solution was added
at a final concentration of 0.5 mg/ml and the cells were incubated
for 4 h at 37°C. Formazan crystals in the viable cells were
solubilized with dimethyl sulfoxide and the absorbance was measured
at 570 nm.
Colony formation assay
The cells were seeded into 6-well plates at a
density of 500 cells/well. For radiation groups, 6-well plates were
irradiated with 2 or 6 Gy X-ray 12 h after seeding. Irradiation was
performed at room temperature using RS 2000 X-ray Biological
Irradiator (Rad Source Technologies, USA) at a dose rate of 1
Gy/min through a 0.2-mm copper filter. The plates were incubated
for 10 days at 37°C with the growth medium being replaced every 3
days. The colonies were fixed with methanol and stained with 0.5%
crystal violet (in methanol:water, 1:1). The number of the colonies
containing at least 50 cells was then counted. Data are presented
as means ± SE (SEM) of at least three independent experiments.
Apoptosis assay
The cells were collected at 36 h after transfection
and the apoptotic cells were analyzed using flow cytometry (FACS)
with an Annexin V/propidium iodide (PI)-staining assay according to
the instructions of the manufacturer (Kaiji Biotechnology Co.,
Nanjing, China).
Cell adhesion assay
Ninety-six-well plates were coated with human
fibronectin (BD Biosciences) at a final concentration of 2
μg/ml overnight at 4°C. The plates were washed with 1%
bovine serum albumin in phosphate-buffered saline (PBS) to block
non-specific cell adhesion. Twelve hours after being transfected
with either pTRiEX or pTRiEX-RGS5 plasmids, the cells were
trypsinized and seeded at 5×104cells/well, and then
incubated for 1 h. The plates were washed with PBS to remove
non-adherent cells, fixed with formalin and followed by staining
with crystal violet for 15 min and solubilized with 1% SDS. The
absorbance was measured at 570 nm by a microplate reader (Tecan
Sunrise, Germany). All the experiments were performed in
triplicates and repeated at least three times.
Cell migration assay
The in vitro migration ability of the cells
was measured using an 8-μm Boyden chamber assay (Corning,
USA). Twelve hours after transfection with pTRiEX or pTRiEX-RGS5
plasmids, the cells were trypsinized and seeded into the upper
chamber. A total of 2x104 cells in 0.2 ml of 2% FBS
medium was placed in the upper chamber, whereas the lower chamber
was loaded with 0.8 ml medium containing 20% FBS. After incubation
at 37°C for 12 h, non-migratory cells were wiped off with
PBS-rinsed cotton swabs. The filters were then fixed in ethanol,
stained with crystal violet. The stained cells were counted under a
microscope (Olympus, Tokyo, Japan). Values for migration were
expressed as the average number of migrated cells/microscopic field
over six fields/assay from three independent experiments.
Western blot analysis
The cells were lysed in lysis buffer [20 mM Tris (pH
7.4), 250 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 0.4 mM PMSF] and
heated at 100°C for 10 min. The protein concentration was
determined using the Bio-Rad Protein Assay kit (Bio-Rad, Hercules,
CA, USA). The samples were separated by SDS-PAGE and transferred to
the PVDF membranes (Bio-Rad). The membranes were blocked in 5% milk
for 1 h. Protein expression was detected using primary antibodies
incubated overnight at 4°C. The membranes were washed and incubated
for 1 h with HRP-conjugated secondary antibodies at room
temperature. The following primary antibodies were used: anti-RGS5
(1:1,000), anti-PARP (1:1,000), anti-caspase 3 (1:1,000),
anti-caspase 9 (1:1,000), anti-bax (1:2,000), anti-bcl (1:1,000),
anti-Rock1 (1:1,000), anti-Rock2 (1:1,000), anti-CDC42 (1:1,000),
anti-Phosphop53 (Serine 15) (1:1,000) and anti-p53 (1:1,000).
Horseradish peroxidase-coupled anti-rabbit or anti-mouse antibodies
(Vector Laboratories) were used at a dilution of 1:1,000. The blots
were probed with an anti-β-actin antibody (Sigma) for equal
loading.
Statistical analysis
Data are presented as mean ± SEM. Differences of the
variables between the groups were analyzed by one-way ANOVA test.
Differences were considered significant at P<0.05.
Results
Expression of RGS5 in human lung cancer
cells
Four human non-small cell lung cancer cell lines
were tested for the expression levels of RGS5 using RT-PCR and
western blot analysis with HUVECs as positive controls. RGS5 was
differentially expressed in human lung cancer cells, with high
levels in H1299 cells and undetectable levels in A549, Calu-3 and
SK-MES-1 cells (Fig. 1).
Overexpressing of RGS5 in A549 and Calu-3
cells inhibits growth and clongenic survival
As shown in Fig. 2,
A549 and Calu-3 cells that were transfected with RGS5-expressing
plasmid had markedly increased levels of RGS5 mRNA and protein,
compared to the cells transfected with the vector control.
Upon transfection with RGS5-expressing plasmid, both
A549 and Calu-3 cells showed reduced cell density at 36 h
post-transfection, compared to the cells receiving vector control
or no plasmid (Fig. 3A). An MTT
assay was employed to study the effects of overexpression of RGS5
on the cell growth. As shown in Fig.
3B, expression of RGS5 was able to significantly inhibit the
growth of both A549 and Calu-3 cells. Compared to the cells
receiving vector control, growth inhibition of A549 and Calu-3
cells was 44.4 and 39.27% at 48 h, and 54.3, 44.7% at 72 h,
respectively (Fig. 3C). The in
vitro clonogenic assay further confirmed that overexpression of
RGS5 asserted an inhibitory effect on the cell proliferation.
Compared to the non-transfected control or the vector transfected
cells, overexpression of RGS5 inhibited colony formation of both
A549 and Calu-3 cells by 46.8 and 52.5%, respectively (Fig. 3D).
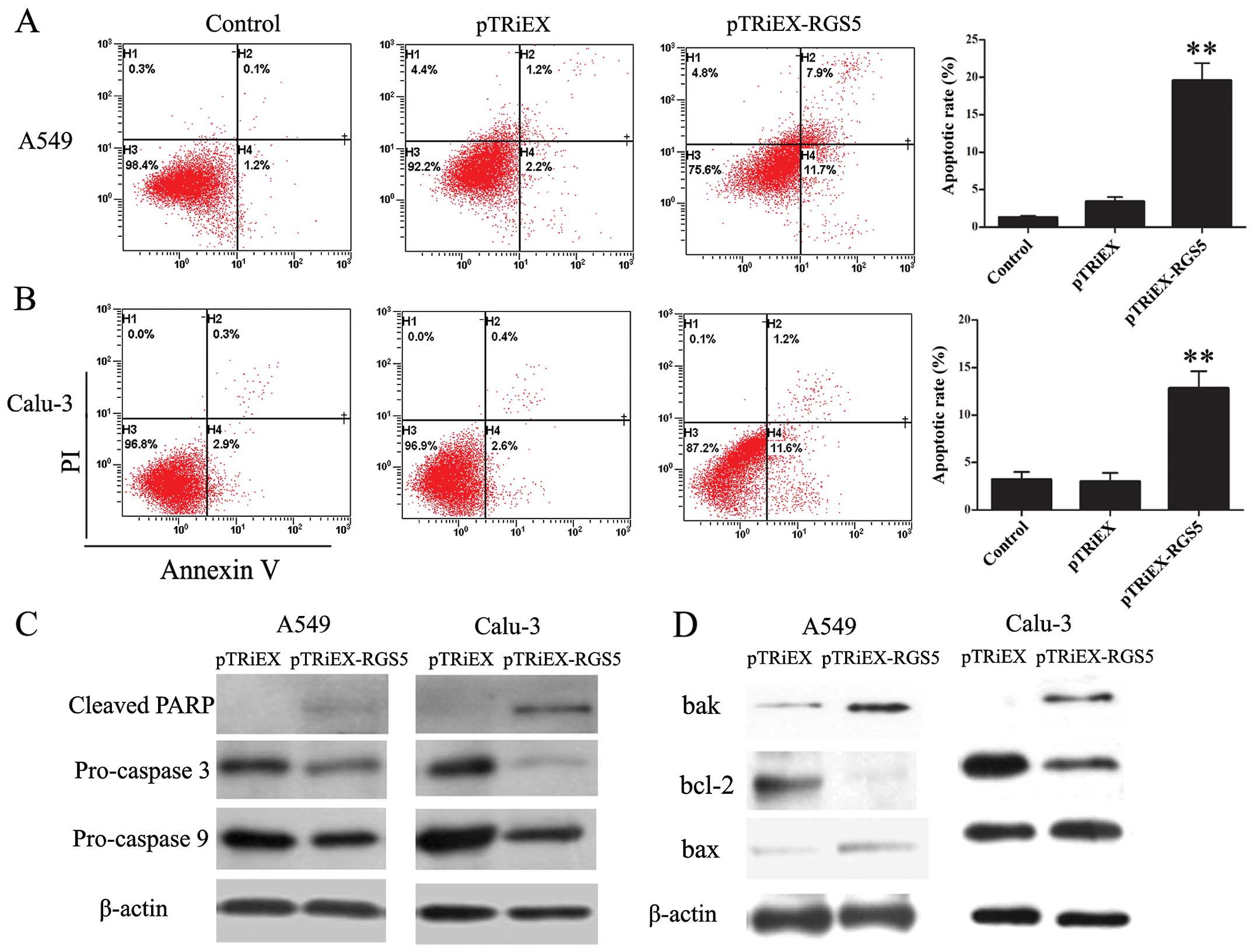
RGS5 induces lung cancer cell apoptosis
through activation of the mitochondrial pathway
To explore the possible mechanism responsible for
RGS5-mediated cell growth inhibition, cellular apoptosis was
evaluated by Annexin V-PI staining and analyzed by flow cytometry.
Both A549 and Calu-3 cells have increased populations undergoing
apoptosis when transfected with RGS5-expressing plasmid (Fig. 4A and B). The proportion of the
apoptotic cells untreated, transfected with pTRiEX or pTRiEX-RGS5
at 36 h post-transfection in A549 cells was 1.3±0.2, 3.4±0.6 and
19.6±2.3%, respectively; whereas 3.2±0.8, 3.0±0.9 and 12.8±1.8%,
respectively, in Calu-3 cells.
To determine whether caspases were involved in the
RGS5-induced apoptosis, we studied the activation of caspase 3 and
9 and the cleavage of PARP. As shown in Fig. 4C, A549 and Calu-3 cells
overexpressing RGS5 had decreased levels of pro-caspase 3 and 9,
whereas the levels of cleaved form of PARP in the same cell lines
were increased, indicating the increased activation of both
caspases.
Mitochondria play a critical role in regulating
apoptosis. Pro-apoptotic proteins Bax and Bak, as well as
anti-apoptotic protein Bcl-2 are involved in regulating the release
of mitochondrial protein cytochrome c to initiate the
apoptotic cascade. We investigated the effect of RGS5 expression on
the levels of Bax, Bak and Bcl-2 proteins. As shown in Fig. 4D, cells transfected with pTRiEX-RGS5
had increased levels of Bax and Bak protein but decreased levels of
Bcl-2. Therefore, the ratio of pro-apoptotic/anti-apoptotic was
increased when RGS5 was overexpressed in the cells. These results
indicated that RGS5 induced apoptosis via mitochondrial pathway in
human lung cancer cells.
RGS5 inhibits adhesion and migration of
lung cancer cells
Previous studies have reported that the low level of
RGS5 is associated with an increased incidence of lymph node
metastasis and more invasion in non-small cell lung cancer patients
(21). Thus, to understand whether
RGS5 is responsible for the observed phenotype, we investigated
whether the increased levels of RGS5 in human lung cancer cells
would inhibit cell adhesion or migration.
Cell adhesion was assessed on the fibronectin-coated
surface. A549 and Calu-3 cells were transfected with RGS5
expressing plasmid or vector only control, and were allowed to
adhere to the fibronectin-coated surface for 1 h. As shown in
Fig. 5A, the overexpression of RGS5
reduced A549 and Calu-3 cell adhesion to fibronectin-coated
surfaces by 45.9 and 28.9% respectively, compared to the control
cells. The differences were apparent from the microscope
images.
Similarly, the effect of RGS5 on cell migration was
assessed in the RGS5 expressing A549 and Calu-3 cells. The cells
were seeded in the upper chamber, and allowed to migrate through an
8-μm filter. Over a 12-h incubation period, the number of
cells penetrated through the filter in vector- and the
RGS5-transfected A549 and Calu-3 cells was 39.9±11.2, 10.23±4.42,
44.56±10.6 and 13.08±3.37/field, respectively (Fig. 5B). To determine the mechanisms of
the observation, expression levels of several cell migration
markers including Rock1, Rock2, cdc42 and MMP-2 were studied in
these cells by western blot analysis. As demonstrated in Fig. 5C, in A549 or Calu-3 cells
transfected with pTRiEX-RGS5, the levels of all four proteins were
decreased compared to the cells transfected with pTRiEX.
RGS5 enhances the cytotoxic effects of
X-ray irradiation on lung cancer cells
To determine whether increased levels of RGS5
increased the sensitivity of lung cancer cells to radiation, we
performed a colony-forming assay with cells expressing higher
levels of RGS5 followed by irradiation treatment. The cells
transfected with either pTRiEX-RGS5 or pTRiEX were subjected to
radiation exposure (2 Gy). Radiation of the cells expressing RGS5
resulted in further inhibition of colony formation (Fig. 6A and B). These results suggested
that RGS5 enhances the cytotoxic effect of radiation treatment.
To further explore the mechanism of colony formation
inhibition after transfection with RGS5 and radiation, the two cell
lines were transfected with pTRiEX-RGS5, irradiated at 6 Gy, and
then the cell lysates were analyzed by western blot analysis.
Combined radiation treatment with RGS5 overexpression led to
increased levels of cleaved PARP, cleaved caspase 3 and 9 in both
cell types (Fig. 6C). It is known
that radiation treatment would lead to the activation of p53
pathway (24). We found that
phosphorylation of p53 at S15 was increased upon radiation, and was
further increased when RGS5 was overexpressed in the radiated cells
(Fig. 6C). As a result, the p53
protein levels were increased in the radiated cells.
Discussion
G-protein-coupled receptors (GPCRs) are a large
family of versatile membrane proteins involved in a wide range of
physiological processes and pathological diseases (25). GPCRs classically transmit their
signal via the activation of G protein heterotrimer, which
exchanges GDP for GTP, dissociates into two parts and is released
from the receptors. Both free GTP-bound α subunit and βγ dimers are
capable of activating downstream effectors. Signaling is terminated
when α subunit hydrolyzes GTP, returns to the GDP-bound state and
re-associates with βγ dimers to form inactive heterotrimers
(26). RGS promotes the hydrolysis
of GTP, thus, reduces the life span of the active form of α
subunit. RGS proteins are emerging as important negative regulators
of G-protein-coupled receptor (GPCR) signaling. Growing evidence
has demonstrated that RGS proteins are potential drug targets in
pathologies including central nervous system diseases,
cardiovascular diseases and diabetes. Targeting RGS proteins with
small molecule modulators or inhibitors provide specific control or
treatment of pathophysiological conditions (27,28).
Despite the critical roles GPCRs and G proteins play
in tumor and metastasis (29),
studies on the function of RGS protein in cancer cells are still
lacking. However, several studies point to the roles of RGS in
regulating cancer cell proliferation. Overexpression of RGS3 in
HL-60 promoted cell apoptosis (30). Overexpression of RGS16 inhibited
epidermal growth factor (EGF)-induced proliferation and Akt
phosphorylation in MCF7 breast cancer cells (31). Altman et al (32) showed that inducible RGS5 expression
significantly reduced proliferation of HeyA8-MDR ovarian cancer
cells. Maity et al (33)
observed that RGS6 induced breast cancer cell apoptosis.
In the present study, it was demonstrated that
overexpression of RGS5 significantly reduced the proliferation of
human lung cancer cells by inducing apoptosis. Previous studies
have reported that RGS5 is upregulated by hypoxia and
overexpression of RGS5 induces apoptosis in HUVECs (34). It was demonstrated that in human
lung cancer cells, the overexpression of RGS5-induced the
expressions of pro-apoptotic proteins Bax and Bak; while it reduced
the expression of anti-apoptotic protein Bcl-2. Bax, Bak and Bcl-2
are known to be involved in the regulation of the mitochondrial
pathway of apoptosis (35). As a
consequence, activation of caspase 3 and 9 was also observed in the
RGS5 overexpressing cells.
A previous study indicates an association between
the low level of RGS5 and an increased lymph node invasion of the
human non-small cell cancer. In addition, RGS4 is found to be
involved in reducing breast cancer metastatic abilities in
vitro (36,37). We found that when overexpressed,
RGS5 decreased the adhesion and migration abilities of human lung
cancer cells. Accordingly, several cell migration markers including
Rock1, Rock2, cdc42 and MMP-2 were downregulated in RGS5 expressing
cells (38,39). Cancer cell migration is believed to
be the initial step in metastasis, in which primary tumor cells
migrate and invade neighboring tissues and enter the circulation to
establish new or secondary tumor sites during metastasis (40). Thus, inhibition of cancer cell
migration represents one of the potent strategies for cancer
treatment. To our knowledge, this is the first demonstration of the
impact of RGS5 overexpression on tumor cell migration.
Our findings partially explained why lower levels of
RGS5 are strongly associated with cancer vascular invasion and
lymph node metastasis in NSCLC and indicate RGS5 as a promising
target for cancer therapy.
RGS proteins have been shown to modulate the
responsiveness of tumor cells to chemotherapeutic agents. The
cytotoxic effects of cisplatin, vincristine and docetaxel were
reduced following RGS10 and RGS17 knockdown in SKOV-3 as well as
MDR-HEYA8 cells (41). RGS6 was
shown to mediate the activation of ATM and p53 by doxorubicin, and
genetic loss of RGS6 dramatically impaired doxorubicin-induced
activation of ATM and p53 in breast cancer cells (42). Our observation also pointed to a
possible role of RGS5 in increasing the sensitivity of the human
lung cancer cells to radiation. Colony formation of both cell lines
was reduced when RGS5 was overexpressed in the cells, and it was
further reduced when these cells were irradiated. Not surprisingly,
p53 pathway was activated upon radiation. It is known that p53
promotes apoptosis through modulating the mitochondrial apoptosis
pathway. We observed increased levels of cleaved PARP in the RGS5
expressing cells, and they were further increased when the cells
were irradiated. We found that RGS5 overexpression led to an
increased level of p53, at least in the A549 cells; whereas the
effect was less apparent in Calu-3 cells. This may be the
explanation for the changes in the levels of pro-apoptotic and
anti-apoptotic Bcl-2 proteins in the RGS5 overexpressing cells.
Although the exact underlying mechanism of these observations still
requires investigation, to our best knowledge, this is the first
study on the antitumor effects of RGS protein combined with
radiation.
The present study has noticeable limitation in that
expression of the RGS5 was transient, which may bring the question
of consistency of the transfection rate and the expression level of
each experiment. During the study, each experiment was coupled with
detection of the RGS5 expression for validation of the data. Our
study used an inducible expression system in cancer to further
analyze the function of RGS5.
In conclusion, we demonstrated the effects of RGS5
overexpression in growth inhibition, apoptosis and reduction of
cell migration in human lung cancer cells. These findings provided
evidence that RGS5 played an inhibitory role in human lung cancer
cells, which explained the pathoclinical observation that high
expression of RGS5 as a favorable prognostic factor in NSCLC
patients. Furthermore, RGS5 was shown to enhance the antitumor
effects of radiation in human lung cancer cells. Collectively, our
results implied that RGS5 may be a promising target for cancer
therapy.
Acknowledgments
The present study was supported by the National
Nature Science Foundation of China (nos. 30872974, 81172116 and
81201736), the Science and Technology Planning Project of Guangdong
Province, China (no. 2009B030801154) and the PhD Programs
Foundation of Guangdong Medical College (XB1334).
References
|
1
|
Koelle MR: A new family of G-protein
regulators - the RGS proteins. Curr Opin Cell Biol. 9:143–147.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Willars GB: Mammalian RGS proteins:
Multifunctional regulators of cellular signalling. Semin Cell Dev
Biol. 17:363–376. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bansal G, Druey KM and Xie Z: R4 RGS
proteins: Regulation of G-protein signaling and beyond. Pharmacol
Ther. 116:473–495. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bondjers C, Kalén M, Hellström M, Scheidl
SJ, Abramsson A, Renner O, Lindahl P, Cho H, Kehrl J and Betsholtz
C: Transcription profiling of platelet-derived growth
factor-B-deficient mouse embryos identifies RGS5 as a novel marker
for pericytes and vascular smooth muscle cells. Am J Pathol.
162:721–729. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho H, Kozasa T, Bondjers C, Betsholtz C
and Kehrl JH: Pericyte-specific expression of Rgs5: Implications
for PDGF and EDG receptor signaling during vascular maturation.
FASEB J. 17:440–442. 2003.PubMed/NCBI
|
|
6
|
Seki N, Sugano S, Suzuki Y, Nakagawara A,
Ohira M, Muramatsu M, Saito T and Hori T: Isolation, tissue
expression, and chromosomal assignment of human RGS5, a novel
G-protein signaling regulator gene. J Hum Genet. 43:202–205. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li H, He C, Feng J, Zhang Y, Tang Q, Bian
Z, Bai X, Zhou H, Jiang H, Heximer SP, et al: Regulator of G
protein signaling 5 protects against cardiac hypertrophy and
fibrosis during biomechanical stress of pressure overload. Proc
Natl Acad Sci USA. 107:13818–13823. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho H, Park C, Hwang IY, Han SB, Schimel
D, Despres D and Kehrl JH: Rgs5 targeting leads to chronic low
blood pressure and a lean body habitus. Mol Cell Biol.
28:2590–2597. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Adams LD, Wang X, Pabon L, Schwartz
SM, Sane DC and Geary RL: Regulator of G protein signaling 5 marks
peripheral arterial smooth muscle cells and is downregulated in
atherosclerotic plaque. J Vasc Surg. 40:519–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takata Y, Liu J, Yin F, Collins AR, Lyon
CJ, Lee CH, Atkins AR, Downes M, Barish GD, Evans RM, et al:
PPARdelta-mediated antiinflammatory mechanisms inhibit angiotensin
II-accelerated atherosclerosis. Proc Natl Acad Sci USA.
105:4277–4282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng W, Wang X, Xiao J, Chen K, Zhou H,
Shen D, Li H and Tang Q: Loss of regulator of G protein signaling 5
exacerbates obesity, hepatic steatosis, inflammation and insulin
resistance. PLoS One. 7:e302562012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Z, Balenga N, Cooper PR, Damera G,
Edwards R, Brightling CE, Panettieri RA Jr and Druey KM: Regulator
of G-protein signaling-5 inhibits bronchial smooth muscle
contraction in severe asthma. Am J Respir Cell Mol Biol.
46:823–832. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bell SE, Mavila A, Salazar R, Bayless KJ,
Kanagala S, Maxwell SA and Davis GE: Differential gene expression
during capillary morphogenesis in 3D collagen matrices: Regulated
expression of genes involved in basement membrane matrix assembly,
cell cycle progression, cellular differentiation and G-protein
signaling. J Cell Sci. 114:2755–2773. 2001.PubMed/NCBI
|
|
14
|
Berger M, Bergers G, Arnold B, Hämmerling
GJ and Ganss R: Regulator of G-protein signaling-5 induction in
pericytes coincides with active vessel remodeling during
neovascularization. Blood. 105:1094–1101. 2005. View Article : Google Scholar
|
|
15
|
Arnold C, Feldner A, Pfisterer L, Hödebeck
M, Troidl K, Genové G, Wieland T, Hecker M and Korff T: RGS5
promotes arterial growth during arteriogenesis. EMBO Mol Med.
6:1075–1089. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Furuya M, Nishiyama M, Kimura S, Suyama T,
Naya Y, Ito H, Nikaido T and Ishikura H: Expression of regulator of
G protein signalling protein 5 (RGS5) in the tumour vasculature of
human renal cell carcinoma. J Pathol. 203:551–558. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Higgins J, Cheung ST, Li R, Mason
V, Montgomery K, Fan ST, van de Rijn M and So S: Novel endothelial
cell markers in hepatocellular carcinoma. Mod Pathol. 17:1198–1210.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Silini A, Ghilardi C, Figini S, Sangalli
F, Fruscio R, Dahse R, Pedley RB, Giavazzi R and Bani M: Regulator
of G-protein signaling 5 (RGS5) protein: A novel marker of cancer
vasculature elicited and sustained by the tumor’s proangiogenic
microenvironment. Cell Mol Life Sci. 69:1167–1178. 2012. View Article : Google Scholar :
|
|
19
|
Wang JH, Huang WS, Hu CR, Guan XX, Zhou HB
and Chen LB: Relationship between RGS5 expression and
differentiation and angiogenesis of gastric carcinoma. World J
Gastroenterol. 16:5642–5646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh J, Dar M, Untch BR, Dixit D, Shi Y,
Yang Z, Adam MA, Dressman H, Wang X, Gesty-Palmer D, et al:
Regulator of G protein signaling 5 is highly expressed in
parathyroid tumors and inhibits signaling by the calcium-sensing
receptor. Mol Endocrinol. 25:867–876. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang G, Song H, Wang R, Han X and Chen L:
The relationship between RGS5 expression and cancer differentiation
and metastasis in non-small cell lung cancer. J Surg Oncol.
105:420–424. 2012. View Article : Google Scholar
|
|
22
|
Yang X, Cai W, Xu Z, Chen J, Li C, Liu S,
Yang Z, Pan Q, Li M, Ma J, et al: High efficacy and minimal peptide
required for the anti-angiogenic and anti-hepatocarcinoma
activities of plasminogen K5. J Cell Mol Med. 14:2519–2530. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Z, Fang S, Zuo Y, Zhang Y, Cheng R,
Wang Q, Yang Z, Cai W, Ma J, Yang X, et al: Combination of pigment
epithelium-derived factor with radiotherapy enhances the antitumor
effects on nasopharyngeal carcinoma by downregulating vascular
endothelial growth factor expression and angiogenesis. Cancer Sci.
102:1789–1798. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fei P and El-Deiry WS: P53 and radiation
responses. Oncogene. 22:5774–5783. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venkatakrishnan AJ, Deupi X, Lebon G, Tate
CG, Schertler GF and Babu MM: Molecular signatures of
G-protein-coupled receptors. Nature. 494:185–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karnik SS, Gogonea C, Patil S, Saad Y and
Takezako T: Activation of G-protein-coupled receptors: A common
molecular mechanism. Trends Endocrinol Metab. 14:431–437. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
O’Callaghan K, Kuliopulos A and Covic L:
Turning receptors on and off with intracellular pepducins: New
insights into G-protein-coupled receptor drug development. J Biol
Chem. 287:12787–12796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roman DL and Traynor JR: Regulators of G
protein signaling (RGS) proteins as drug targets: Modulating
G-protein-coupled receptor (GPCR) signal transduction. J Med Chem.
54:7433–7440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dorsam RT and Gutkind JS:
G-protein-coupled receptors and cancer. Nat Rev Cancer. 7:79–94.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nishiura H, Nonaka H, Revollo IS, Semba U,
Li Y, Ota Y, Irie A, Harada K, Kehrl JH and Yamamoto T: Pro- and
anti-apoptotic dual functions of the C5a receptor: Involvement of
regulator of G protein signaling 3 and extracellular
signal-regulated kinase. Lab Invest. 89:676–694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang G, Bansal G, Xie Z and Druey KM:
RGS16 inhibits breast cancer cell growth by mitigating
phosphatidylinositol 3-kinase signaling. J Biol Chem.
284:21719–21727. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Altman MK, Nguyen DT, Patel SB, Fambrough
JM, Beedle AM, Hardman WJ and Murph MM: Regulator of G-protein
signaling 5 reduces HeyA8 ovarian cancer cell proliferation and
extends survival in a murine tumor model. Biochem Res Int.
2012:5184372012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maity B, Yang J, Huang J, Askeland RW,
Bera S and Fisher RA: Regulator of G protein signaling 6 (RGS6)
induces apoptosis via a mitochondrial-dependent pathway not
involving its GTPase-activating protein activity. J Biol Chem.
286:1409–1419. 2011. View Article : Google Scholar :
|
|
34
|
Jin Y, An X, Ye Z, Cully B, Wu J and Li J:
RGS5, a hypoxia-inducible apoptotic stimulator in endothelial
cells. J Biol Chem. 284:23436–23443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View Article : Google Scholar
|
|
36
|
Mu XM, Shi W, Sun LX, Li H, Wang YR, Jiang
ZZ and Zhang LY: Pristimerin inhibits breast cancer cell migration
by up-regulating regulator of G protein signaling 4 expression.
Asian Pac J Cancer Prev. 13:1097–1104. 2012. View Article : Google Scholar
|
|
37
|
Xie Y, Wolff DW, Wei T, Wang B, Deng C,
Kirui JK, Jiang H, Qin J, Abel PW and Tu Y: Breast cancer migration
and invasion depend on proteasome degradation of regulator of
G-protein signaling 4. Cancer Res. 69:5743–5751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schofield AV and Bernard O: Rho-associated
coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem
Mol Biol. 48:301–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rane CK and Minden A: P21 activated
kinases: Structure, regulation, and functions. Small GTPases.
5:52014. View Article : Google Scholar
|
|
40
|
Luanpitpong S, Talbott SJ, Rojanasakul Y,
Nimmannit U, Pongrakhananon V, Wang L and Chanvorachote P:
Regulation of lung cancer cell migration and invasion by reactive
oxygen species and caveolin-1. J Biol Chem. 285:38832–38840. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hooks SB, Callihan P, Altman MK, Hurst JH,
Ali MW and Murph MM: Regulators of G-Protein signaling RGS10 and
RGS17 regulate chemoresistance in ovarian cancer cells. Mol Cancer.
9:2892010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang J, Yang J, Maity B, Mayuzumi D and
Fisher RA: Regulator of G protein signaling 6 mediates
doxorubicin-induced ATM and p53 activation by a reactive oxygen
species-dependent mechanism. Cancer Res. 71:6310–6319. 2011.
View Article : Google Scholar : PubMed/NCBI
|