Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common type of cancer worldwide and the third leading cause of
cancer-related deaths, with more than 700,000 cases being diagnosed
yearly (1–3). The diagnosis and management of HCC
have changed greatly within the past decade, but postoperative
recurrence occurs frequently and the 5-year survival rate of HCC
patients remains quite low (2,4).
Hepatocarcinogenesis is a complex and multistep process in which
many signalling factors are altered, leading to a multifarious
molecular profile (5–8). Although much effort has been made to
identify key molecules involved in the development and progression
of HCC, our understanding of the molecular pathogenesis of this
disease remains elusive. Hence, there is an urgent need to develop
novel strategies for the diagnosis, treatment and prognosis of
HCC.
microRNAs (miRNAs) are endogenous, ~22-nucleotide
long, non-coding RNAs that negatively regulate the expression of
multiple target genes at the post-transcriptional level by binding
to the 3′-untranslated region (3′-UTR) of target mRNAs, resulting
in mRNA degradation or blockade of mRNA translation (9). Growing evidence indicates that miRNAs
play an important role in diverse biological processes, and
aberrant expression of specific miRNAs is involved in a wide range
of human cancers, functioning as classical oncogenes or
tumor-suppressor genes (10,11).
Deregulation of miRNAs which have been associated with HCC patient
clinicopathological features, can also contribute to HCC
development by influencing cell growth, apoptosis, migration or
invasion (12–15). Hence, more extensive investigations
are needed to identify miRNAs in order to reveal the underlying
mechanisms of HCC carcinogenesis and progression, and to facilitate
targeted therapy and improve the prognosis of HCC patients.
Currently, gene expression profiling is employed to
investigate the spectrum of differentially expressed genes in HCC
cells and clinical specimens. Numerous candidate genes potentially
involved in HCC developmental processes, such as proliferation,
apoptosis, angiogenesis and invasion have been identified. Our
previous microarray profiling showed that miR-128-3p is
downregulated in HCC tissues. The role of miR-128-3p in HCC
carcinogenesis and progression, however, remains unknown. In the
present study, the roles of miR-128-3p in HCC development were
investigated. We found that low miR-128-3p expression in HCC
tissues was correlated with a worse prognosis for HCC patients.
Additionally, we also found that miR-128-3p downregulated PIK3R1 to
inhibit the PI3K-AKT pathway and thereby suppress HCC progression.
Therefore, these findings demonstrate that miR-128-3p is a
prognostic predictor for HCC patients, and provide new insights for
the study of the molecular mechanisms of HCC and subsequent
treatment.
Materials and methods
Patients and tissue samples
Surgically resected paired HCC and adjacent
noncancerous tissues were collected from 72 primary HCC patients at
The Affiliated Tumor Hospital of Guangxi Medical University between
March 2011 and May 2013. Tissue samples were immediately frozen in
liquid nitrogen until analysis. The cases selected were based on a
clear pathological diagnosis, follow-up data, and had first
undergone radical resection of HCC, and had not received
preoperative adjuvant chemotherapy, radiotherapy, targeted therapy
or immunotherapy. Informed consent was obtained from each patient,
and the study was approved by the Ethics Committee of Guangxi
Medical University, Nanning, China. The investigations were
conducted according to the Declaration of Helsinki Principles.
RNA extraction and quantitative
RT-PCR
Total RNA, including miRNA, was extracted using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. cDNAs were synthesized using ReverTra
Ace qPCR RT kit (FSQ-101; Toyobo, Kagoshima, Japan). microRNA was
reversely transcribed using First Strand cDNA Synthesis kit
ReverTra Ace-α-(FSK-100; Toyobo). Real-time PCR analyses were
performed with Thunderbird SYBR qPCR mix (QPS-201; Toyobo) on an
MxPro Mx3000P Sequence Detection system (Stratagene, La Jolla, CA,
USA). U6 small nuclear RNA or β-actin was used as an internal
normalized reference, and fold changes were calculated by relative
quantification (2−ΔΔCt). The primers used were:
miR-128-3p specific stem-loop reverse transcription primers,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACT GGATACGACAAAGAG-3′;
miR-128-3p forward, 5′-GGTC ACAGTGAACCGGTC-3′ and reverse,
5′-GTGCAGGGTCC GAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; PIK3R1 forward,
5′-AAGAAGTTGAACGAGTGGTTGG-3′ and reverse,
5′-GCCCTGTTTACTGCTCTCCC-3′; β-actin forward,
5′-AGTGTGACGTTGACATCCGT-3′ and reverse, 5′-GCAGCTCAGTAACAGTCCGC-3′.
All samples were amplified in triplicate.
Cell culture and transfection
Cells were obtained from the Institute of
Biochemistry and Cell Biology of the Chinese Academy of Sciences
(Shanghai, China). Human HCC cell lines (QGY-7703, SK-hep1,
QGY-7404, SMMC-7721, Huh7 and HepG2) and human normal liver cells
(HL-7702) were maintained in RPMI-1640 medium with 10% fetal bovine
serum (FBS; Gibco, USA) at 37°C in a humidified incubator
containing 5% CO2. miR-128-3p duplex mimics and a
negative control (NC) were obtained from Genepharma (Shanghai,
China). Cells were transfected with RNAs using INTERFERin
Transfection reagent (Polyplus Transfection, Illkirch, France) at a
final concentration of 100 nM according to the manufacturer’s
instructions.
Cell proliferation and colony formation
assays
Cells were seeded into 96-well plates
(5×103/well) and transfected with miR-128-3p mimics or
the NC. The cell proliferation of HCC cell lines was determined
using WST-8 staining with the Cell Counting Kit-8 (Dojindo, Japan)
at the indicated time-points (24, 48, 72 and 96 h) according to the
manufacturer’s instructions. For the colony formation assay, cells
were seeded into 6-well plates at a low density (1×103
cells/well) and cultured for 10 days. Then, cells were fixed with
4% paraformaldehyde for 30 min and surviving colonies (>50
cells/colony) were counted after staining with 1% crystal violet.
The experiments were carried out in triplicate wells for at least 3
times.
Cell cycle distribution
Forty-eight hours after transfection in 6-well
plates, the QGY-7703 or SK-hep1 cells were harvested and washed
with cold 1X PBS. Then, cells were fixed in 70% ethanol at 4°C
overnight, and washed with PBS twice, resuspended with 100
μl RNase A, and incubated at 37°C for 30 min. Staining for
DNA content was performed using 400 μl propidium iodide
(KeyGen, Nanjing, China) at 4°C for 30 min in the dark, and
analyzed using an Epics XL flow cytometer (Beckman Coulter, Brea,
CA, USA).
In vitro migration assay
Migration assays were performed using the 24-Well
Cell Migration assay with an 8-μm pore size polycarbonate
membrane (Corning, New York, NY, USA), according to the
manufacturer’s instructions. Briefly, 24 h after the transfection,
5×104 QGY-7703 cells or 1×104 SK-hep1 cells were
resuspended in 200 μl serum-free medium and plated in the
top chamber. The lower chambers were filled with 0.6 ml of medium
containing 10% FBS. Medium with 10% FBS was added to the lower
chamber as a chemoattractant. After a 24-h incubation at 37°C, the
cells on the upper surface of the membrane were removed, and the
cells on the lower surface were fixed, stained, photographed, and
counted under a microscope in five fields.
Western blot analysis
Antibodies for p85, p-AKT (Ser473), p-mTOR, p-p70S6K
and β-actin were purchased from Cell Signaling Technology, and all
the antibodies were rabbit anti-human. Cells were harvested and
then lysed with RIPA buffer supplemented with 1 mmol/l PMSF (both
from Boster, Wuhan, China), and then centrifuged at 15,000 rpm at
4°C for 10 min. Protein concentrations of the extracts were
measured using the bicinchoninic acid (BCA) protein assay kit
(KeyGen). Equal amounts of the proteins were concentrated and
separated through SDS-PAGE, and then transferred to polyvinylidene
difluoride (PVDF) membranes (Boster). After blocking in TBST
(Tris-buffered saline with Tween-20) which contained 5% non-fat
milk for 60 min, the membranes were incubated with the primary
antibody (1:1,000 dilution; β-actin, as a loading control, 1:2,500
dilution) overnight at 4°C. The membranes were incubated with the
secondary antibodies (mouse anti-rabbit and HRP-linked antibody,
1:5,000 dilution; Cell Signaling Technology). After incubating in
enhanced chemiluminescence solution (Boster), the proteins on the
membranes were detected using Bio-Rad Universal Hood III, and
analyzed by Image Lab™ software 2.0 (Bio-Rad).
miRNA target predictions
Predicted targets of miR-128-3p and its sites were
analyzed using TargetScan (http://www.targetscan.org/).
Statistical analysis
Data are presented as the mean ± standard deviation
(SD) of one representative experiment. The χ2 test and
Fisher’s exact test was used to analyze the relationship between
the expression level of miR-128-3p and the clinicopathological
characteristics. Unless otherwise noted, the differences between
the groups were analyzed by one-way analysis of variance (ANOVA)
when there were more than two groups. Differences in miR-128-3p
expression between the HCC and noncancerous tissues of the human
subjects were calculated using a two-tailed independent sample
Student’s t-test. Disease-free survival (DFS) was displayed by
Kaplan-Meier survival curves, and DFS of the different groups was
compared by log-rank test. The relationship between the expression
level of miR-128-3p and P85 was measured by Pearson’s correlation
coefficient analysis. In all cases, differences were considered
statistically significant at P<0.05. All analyses were performed
using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA).
Results
miR-128-3p is downregulated in the HCC
tissues as well as in the HCC cell lines
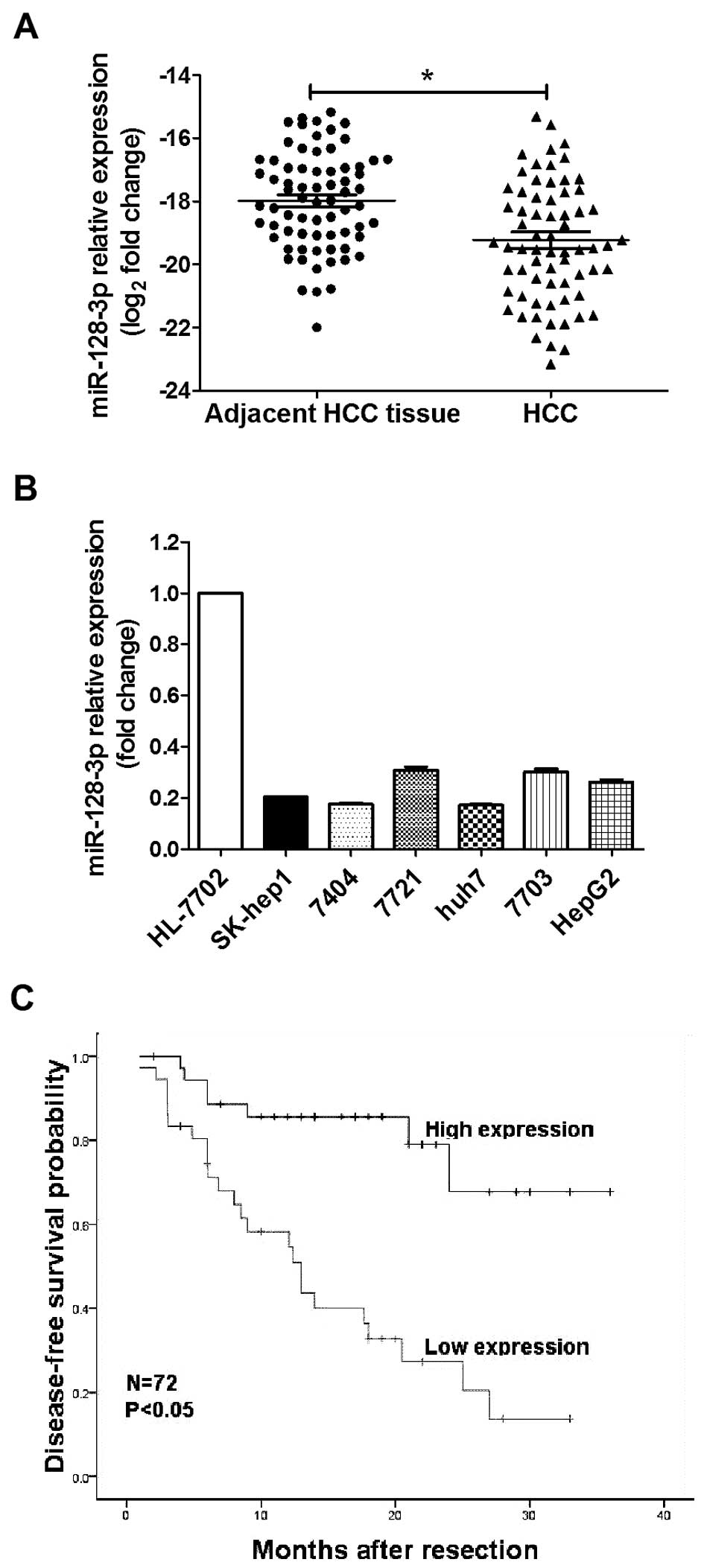
In order to investigate the expression of miR-128-3p
in HCC, the levels of miR-128-3p in 72 paired HCC tissues and 6 HCC
cell lines and human normal liver cells were tested by qRT-PCR. As
shown in Fig. 1A, miR-128-3p
expression was significantly downregulated in 65.30% (47 of 72) of
the HCC samples compared to their matched controls. The median
miR-128-3p expression level in all the HCC tissues was ~2.5-fold
lower than that in the matched controls (P<0.001). In addition,
the expression of miR-128-3p in the HCC cell lines (including
SK-hep1, QGY-7404, SMMC-7721, Huh7, QGY-7703, HepG2) was lower than
that in the human normal liver cell line HL-7702 (Fig. 1B). These findings indicate that
miR-128-3p was consistently decreased in HCC, which may contribute
to HCC pathogenesis.
Correlation between miR-128-3p expression
and the clinicopathological features of the HCC patients
To further understand the relationship between the
miR-128-3p expression levels and clinicopathological factors, 72
HCC patients who underwent radical resection and relapsed after a
3-year follow-up, were divided into a high or low miR-128-3p
expression group according to the 50th percentile (median) of
relative miR-128-3p expression, as analyzed by the χ2
test (Table I). We found that low
expression of miR-128-3p was strongly correlated with tumor-node
metastasis (TNM) and tumor size (P<0.05). To exclude the
confounder effect, we further performed Kaplan-Meier survival
analysis and Cox proportional hazards regression analysis.
Strikingly, the Kaplan-Meier survival analysis showed that low
miR-128-3p expression was correlated with a shorter DFS (P<0.05)
(Fig. 1C) in the HCC patients.
Multivariate analysis further confirmed that a reduced miR-128-3p
level is an independent predictor for a short DFS of HCC patients
(P<0.05) (Table II). These
results indicate that miR-128-3p may be involved in the tumor
development and progression of HCC.
 | Table IAssociations between the expression
of miR-128-3p and the clinicopathological features of the HCC
patients. |
Table I
Associations between the expression
of miR-128-3p and the clinicopathological features of the HCC
patients.
|
Characteristics | No. of
patients | miR-128-3p
| P-value | χ2 |
|---|
| High, n (%) | Low, n (%) |
|---|
| 72 | 36 (50.0) | 36 (50.0) | | |
| Gender |
| Male | 66 | 32 (48.5) | 34 (51.5) | 0.674 | 0.727 |
| Female | 6 | 4 (66.7) | 2 (33.3) | | |
| Age (years) |
| ≥50 | 25 | 14 (56.0) | 11 (44.0) | 0.311 | 0.551 |
| <50 | 47 | 22 (46.8) | 25 (53.2) | | |
| Hepatitis B |
| Positive (+) | 64 | 32 (55.6) | 32 (44.4) | 1.000 | 0.000 |
| Negative (−) | 8 | 4 (49.2) | 4 (50.8) | | |
| Tumor size
(cm) |
| <5 | 29 | 20 (69.0) | 9 (31.0) | 0.008 | 6.986 |
| ≥5 | 43 | 16 (37.2) | 27 (62.8) | | |
| Tumor number |
| Solitary | 59 | 30 (50.8) | 29 (49.2) | 1.000 | 0.094 |
| Multiple | 13 | 6 (46.2) | 7 (53.8) | | |
| Tumor capsule |
| Void or
particle | 31 | 17 (54.8) | 14 (45.2) | 0.634 | 0.510 |
| Intact | 41 | 19 (46.3) | 22 (53.7) | | |
| AFP (ng/ml) |
| ≤20 | 21 | 13 (62.5) | 8 (37.5) | 0.150 | 1.681 |
| >20 | 51 | 23 (43.8) | 28 (56.3) | | |
| TNM |
| I+II | 56 | 32 (57.1) | 24 (42.9) | 0.045 | 5.143 |
| III+IV | 16 | 4 (25.0) | 12 (75.0) | | |
 | Table IIMultivariate Cox regression analyses
of overall survival in the 72 patients with HCC. |
Table II
Multivariate Cox regression analyses
of overall survival in the 72 patients with HCC.
| Tumor
characteristics | Relative risk (95%
CI) | P-value |
|---|
| Tumor size (>5
cm) | 0.259
(0.046–1.440) | 0.123 |
| Age (years) | 1.442
(0.590–3.521) | 0.422 |
| Tumor number | 0.580
(0.146–2.301) | 0.439 |
| Tumor capsule | 0.747
(0.316–1.766) | 0.507 |
| Hepatitis B | 3.387
(0.447–25.676) | 0.238 |
| AFP (>20
ng/ml) | 0.757
(0.282–2.029) | 0.579 |
| TNM (III+IV) | 0.620
(0.000–9.632E72) | 0.996 |
| miR-128-3p
(low) | 0.323
(0.121–0.864) | 0.024 |
miR-128-3p overexpression suppresses HCC
cell proliferation and clonogenicity
Following the finding of decreased expression of
miR-128-3p in HCC, it was then determined whether miR-128-3p
functions as a tumor suppressor. The effect of miR-128-3p on HCC
cell growth was observed. Firstly, two of the HCC cell lines were
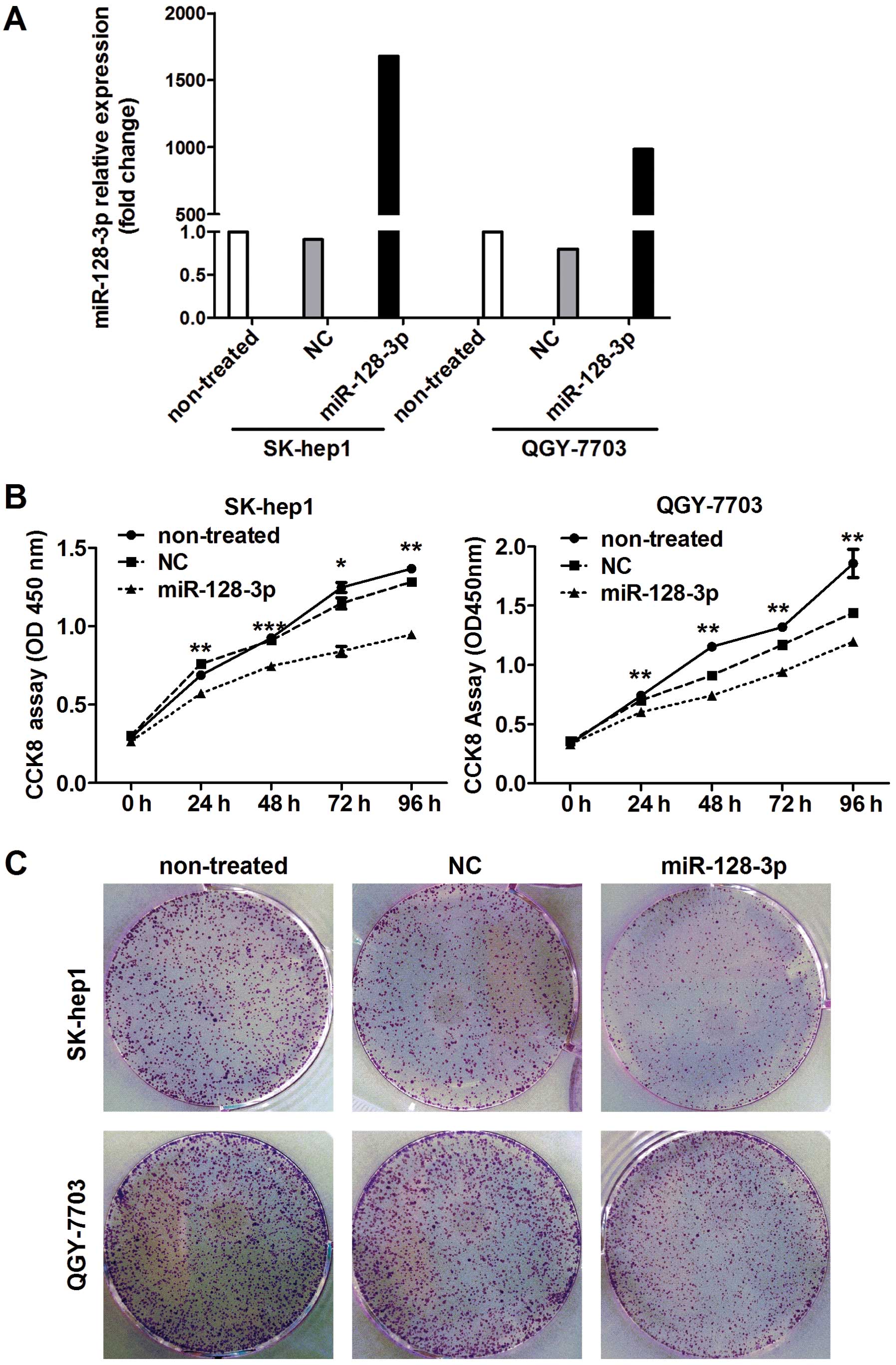
randomized to do follow-up experiments. The transfection efficiency
of miR-128-3p mimics in the SK-hep1 and QGY-7703 HCC cells was
assessed by qRT-PCR after transfection with the miR-128-3p mimics
and NC after 24 h. As shown in Fig.
2A, the expression of miR-128-3p in the HCC cells was
significantly increased after transfection with the miRNA mimics.
Then, we evaluated the effect of miR-128-3p overexpression on the
proliferation of the HCC cells. The results showed that
proliferation of the HCC cells was suppressed by miR-128-3p
overexpression (Fig. 2B). We also
further investigated the effect of miR-128-3p overexpression on the
clonogenicity of the SK-hep1 and QGY-7703 cells, which were
transfected with the miR-128-3p mimics or the NC. Compared with the
NC transfectants, HCC cells transfected with the miR-128-3p mimics
displayed notably fewer colonies (Fig.
2C).
miR-128-3p inhibits HCC cell cycle
progression to suppress tumor growth
Since overexpression of miR-128-3p inhibited HCC
cell proliferation, we ascertained whether the effect of miR-128-3p
is relevant to the cell cycle. Cell cycle analysis of the HCC
SK-hep1 and QGY-7703 cell lines indicated that miR-128-3p inhibited
cell cycle progression, most likely due to G0–G1 phase arrest
(P<0.05) (Fig. 3). These results
indicate that miR-128-3p can inhibit HCC cell growth through
inhibition of cell cycle progression.
miR-128-3p inhibits HCC cell
migration
The role of miR-128-3p in HCC cell migration was
then investigated. As shown in Fig.
4, HCC SK-hep-1 and QGY-7703 cells transfected with the
miR-128-3p mimics had significantly weaker migratory ability when
compared to that of the control cells. These observations imply
that miR-128-3p may inhibit HCC metastasis.
miR-128-3p inhibits PI3K/AKT pathway
activation by downregulating p85α expression
To elucidate the underlying molecular mechanisms of
miR-128-3p in proliferation and migration, 1,047 putatively
conserved gene targets of miR-128-3p in TargetScan (http://www.targetscan.org) were subjected to
enrichment analysis of the cell signaling pathways using the Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway database
(http://www.genome.jp/kegg/). It was
found that the signaling pathway in cancer (map05200) and PI3K/AKT
(Table III) were the most
significantly enriched pathways compared to the other signaling
pathways. As shown in Fig. 5A, the
human PIK3R1, encoding p85α and involving 28.77% of the 212
miR-128-3p-related pathways, is a member of the pathway in cancer
and the PI3K-AKT signaling pathway, which is known to be involved
in cancer development (16–19), and has one miR-128-3p binding site
in its 3′-UTR (Fig. 5A). Therefore,
to verify whether miR-128-3p regulates PIK3R1, the cellular mRNA
expression of p85α was detected by qRT-PCR after treatment with the
miRNA mimics for 24 h. Compared with the NC duplex, the p85 mRNA
expression in the HCC cells was extremely suppressed by the
miR-128-3p mimics (Fig. 5B). In
addition, p85α was found to be related to the activation of the
PI3K/AKT pathway. Therefore, whether miR-128-3p influences the
activation of the PI3K pathway was also studied. Phosphorylation of
the essential molecules in the PI3K pathway was analyzed by western
blot analysis. As shown in Fig. 5C and
D, the protein levels of p85α, phosphorylated AKT, mammalian
target of rapamycin (p-mTOR) and p-p70S6K were inhibited by
miR-128-3p overexpression in the HCC SK-hep1 and QGY-7703
cells.
 | Table IIIEnrichment analysis of predicted
miR-128-3p targets in the KEGG signaling pathway database. |
Table III
Enrichment analysis of predicted
miR-128-3p targets in the KEGG signaling pathway database.
| Pathway | Count |
|---|
| Pathways in
cancer | 32 |
| PI3K-Akt signaling
pathways | 30 |
| MAPK signaling
pathways | 29 |
| Proteoglycans in
cancer | 26 |
| Rap1 signaling
pathway | 25 |
| Ras signaling
pathway | 24 |
| Transcriptional
misregulation in cancer | 23 |
| Focal adhesion
kinase | 21 |
To confirm the relevance of our in vitro
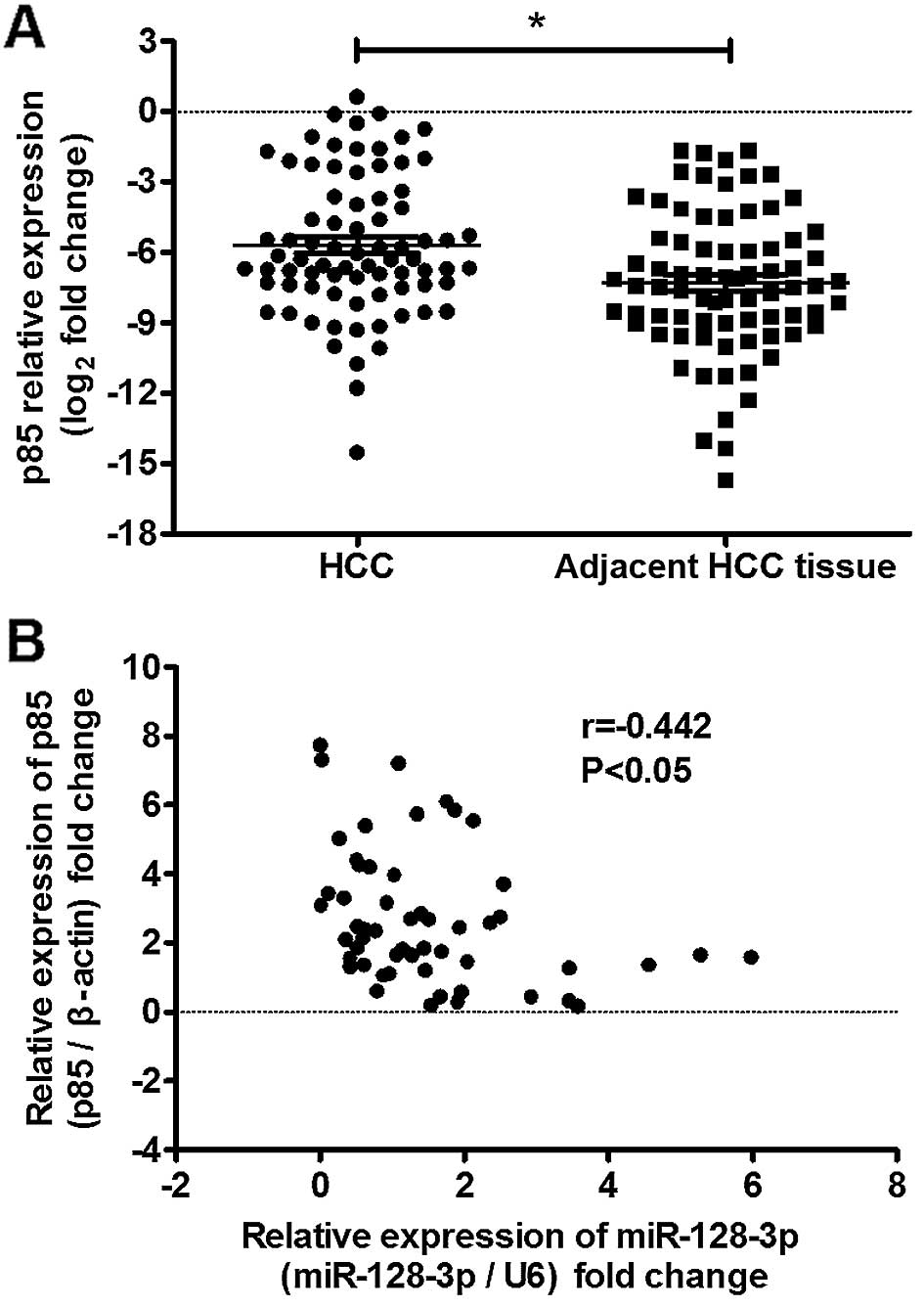
findings, p85α expression was assessed in the same 72 HCC samples.
As shown in Fig. 6, p85α expression
was upregulated in 68.06% (49 of 72) of the HCC samples and showed
an inverse correlation with miR-128-3p expression in the HCC
samples.
These findings further suggest that decreased
expression of miR-128-3p triggered upregulation of p85α partially
in HCC, and demonstrate that miR-128-3p can inhibit HCC progression
by downregulating PIK3R1 expression and repressing PI3K-AKT pathway
activation.
Only the top 8 enriched cell signaling pathways of
predicted miR-128-3p targets in KEGG database are shown. HCC,
hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and
Genomes.
Discussion
In recent years, growing evidence suggests that
aberrant expression of miRNAs contributes to tumorigenesis. Changes
in miRNA profiling are implicated in almost all aspects of cancer
biology, including cell proliferation, apoptosis, migration and
angiogenesis. Thus, miRNAs are increasingly viewed as potential
diagnostic and therapeutic tools. In the present study, miR-128-3p
was found to be markedly decreased in HCC. Low expression of
miR-128-3p was significantly associated with a worse prognosis for
HCC patients. Moreover, miR-128-3p may function as a tumor
suppressor, as it was found to be involved in the development and
progression of HCC through repressing PI3K/AKT pathway activation
by regulating p85α. These results suggest that miR-128-3p may be a
new prognostic predictor as well as a potential therapeutic target
for HCC.
Concerning the roles of miR-128 in tumorigenesis and
development, research has demonstrated that miR-128 can regulate
proliferation, differentiation and apoptosis of various types of
tumor cells. For example, P70S6k1 is known as one of the key
downstream targets of mTOR and is involved in tumor angiogenesis.
Shi et al (20) found that
miR-128 over-expression acted as a tumor suppressor by targeting
p70S6K1 consequently attenuating tumor growth and angiogenesis in
glioma. miR-128 also suppressed prostate and breast cancer by
inhibiting BIM-1 in tumor-initiating cells consequently influencing
the self-renewal and malignant transformation of tumor stem cells
(21,22). Moreover, Zhu et al (23) found that reduced miR-128 induced
chemotherapeutic resistance via influencing multidrug resistance
associated protein (ABCC5, MRP5). Research indicated that miR-128
overexpression can inhibit Reelin and doublecortin (DCX) expression
consequently reducing neuroblastoma cell motility and invasiveness
(24). In the present study,
overexpression of miR-128-3p in the HCC cells not only inhibited
HCC proliferation by arresting the cell cycle at the G1 phase, but
also suppressed HCC cell colony formation and migration. These
results indicate that miR-128-3p can act as a tumor suppressor and
is involved in the tumor development and progression of HCC.
Identifying the molecular markers correlated with
the survival of cancer patients has attracted much research
interest. Hence, the suppressor role of miR-128-3p motivated us to
detect the relationship between the miR-128-3p expression levels
and clinicopathological factors. Firstly, the expression of
miR-128-3p was found to be markedly decreased in HCC. This result
was similar to previous studies that miR-128-3p is repressed in
ovarian cancer, non-small cell lung cancer, glioma progression, and
in acute myeloid leukemia cells (25–28).
Conversely, miR-128 expression was reported to be high in acute
leukemia, and in undifferentiated gastric and prostate cancer
(22,29,30).
Thus, miR-128-3p is a tissue-specific gene. Furthermore, we
analyzed the relationship between miR-128-3p expression and the
clinicopathological features of HCC and found that low expression
of miR-128-3p was strongly correlated with TNM and was correlated
with a shorter DFS in the HCC patients. These results are
consistent with previous reports (22,31)
that the level of miR-128-3p is an independent predictor for
reduced DFS of HCC patients.
Before further discussing the antitumor molecular
mechanisms of miR-128-3p in HCC, it is important to note that the
mechanisms responsible for miR-128-3p downregulation in cancers are
largely unknown. According to previous studies, there are several
possible reasons for the downregulation of miR-128-3p in HCC
tissues. Firstly, miR-128-3p (known as miR-128) is the same major
mature microRNA of miR-128-1 and miR-128-2. miR-128-1 and miR-128-2
are located on chromosomes 2q and 3q, respectively (26). This location contains multiple
tumor-suppressor genes and is one which commonly presents with loss
of heterozygosity in various types of tumors, such as HCC (32), clear cell renal carcinoma (33) and lung cancer (34). Allelic loss of the genomic region
may be responsible for the downregulation of miR-128-3p. Secondly,
epigenetic alteration through DNA methylation also causes
miR-128-3p downregulation (28,31,35).
Additionally, the expression of miR-128 can be regulated by a
transcriptional factor. Snail and p53 directly bind to the promoter
region of miR-128 consequently influencing the expression of
miR-128 (27,36,37).
Further studies are required to evaluate the cause of miR-128-3p
deregulation in HCC development.
To elucidate the antitumor mechanism of miR-128-3p
in HCC, the target genes of miR-128-3p were investigated. One such
target gene was p85α, a well-accepted regulatory subunit of Class
1A PI3K. To investigate the role of p85α in tumorigenesis, research
on gain-of-function mutations in the nSH2 and/or iSH2 domain of
p85α have revealed that these mutations can relieve the repression
on p110 catalytic activity and enhance PI3K signaling (38–41).
Moreover, depletion of PI3K p85α can decrease the expression of
cyclin D1, CDK4 and p27/kip1 and induce tumor cell apoptosis in
colorectal cancer by negatively regulating the activity of Forkhead
family transcription factors (42).
Regarding metastasis, Hong et al (43) reported that the activation of
non-smad TGF-β signaling can promote mesenchymal transition
depending upon focal adhesion kinase (FAK) binding with p85α.
Hence, p85 can act as an oncogene in tumorigenesis. In the present
study, p85α was found to be upregulated in the HCC tissues, and
correlation analysis showed an inverse correlation linking
miR-128-3p and p85α expression. Restoring miR-128-3p significantly
repressed the PI3K/AKT pathway activation by downregulating p85α,
thereby explaining why miR-128-3p suppresses HCC cell proliferation
and metastasis.
It has been acknowledged that a single miRNA can
regulate the expression of multiple genes by targeting different
mRNAs (44), indicating that there
may be other molecules or signaling pathways also targeted by
miR-128-3p. This presumption requires future research to reveal the
complete function of miR-128-3p in HCC carcinogenesis and
progression.
In summary, we demonstrated that miR-128-3p is
commonly downregulated in HCC, and is closely associated with the
prognosis of HCC patients. miR-128-3p acts as a tumor-suppressor by
silencing PI3KR1 to regulate the PI3K/AKT signaling pathway.
Further investigation is required to fully reveal the molecular
mechanisms of miR-128-3p and to determine whether this miRNA is a
potential therapeutic target for the treatment of HCC.
Acknowledgments
The present study was supported by grants from the
Key Research Project of the Health Department of Guangxi Zhuang
Autonomous Region (no. s201301-10), and the National Natural
Science Foundation of China (no. 81360315).
Abbreviations:
|
TGF-β
|
transforming growth factor-β
|
|
3′-UTR
|
3′-untranslated region
|
|
HCC
|
hepatocellular carcinoma
|
|
DFS
|
disease-free survival
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
|
mTOR
|
mammalian target of rapamycin
|
|
FAK
|
focal adhesion kinase
|
|
ABCC5
|
ATP-binding cassette, sub-family C
(CFTR/MRP), member 5
|
|
DCX
|
doublecortin
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
4
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar
|
|
7
|
Moeini A, Cornellà H and Villanueva A:
Emerging signaling pathways in hepatocellular carcinoma. Liver
Cancer. 1:83–93. 2012. View Article : Google Scholar
|
|
8
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ,
Liu LL, Yi C, Fu J, Hu W, Wen JM, et al: miR-625 suppresses tumour
migration and invasion by targeting IGF2BP1 in hepatocellular
carcinoma. Oncogene. 34:965–977. 2015. View Article : Google Scholar
|
|
16
|
Gedaly R, Angulo P, Hundley J, Daily MF,
Chen C, Koch A and Evers BM: PI-103 and sorafenib inhibit
hepatocellular carcinoma cell proliferation by blocking
Ras/Raf/MAPK and PI3K-AKT-mTOR pathways. Anticancer Res.
30:4951–4958. 2010.PubMed/NCBI
|
|
17
|
Chen M, Gu J, Delclos GL, Killary AM, Fan
Z, Hildebrandt MA, Chamberlain RM, Grossman HB, Dinney CP and Wu X:
Genetic variations of the PI3K-AKT-mTOR pathway and clinical
outcome in muscle invasive and metastatic bladder cancer patients.
Carcinogenesis. 31:1387–1391. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao L and Vogt PK: Class I PI3K in
oncogenic cellular transformation. Oncogene. 27:5486–5496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian
X, Yin Y, Zhao P, Wang YY, Wang XF, et al: MiR-128 inhibits tumor
growth and angiogenesis by targeting p70S6K1. PLoS One.
7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jin M, Zhang T, Liu C, Badeaux MA, Liu B,
Liu R, Jeter C, Chen X, Vlassov AV and Tang DG: miRNA-128
suppresses prostate cancer by inhibiting BMI-1 to inhibit
tumor-initiating cells. Cancer Res. 74:4183–4195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu YD, Wang L, Sun C, Fan L, Zhu DX, Fang
C, Wang YH, Zou ZJ, Zhang SJ, Li JY, et al: Distinctive microRNA
signature is associated with the diagnosis and prognosis of acute
leukemia. Med Oncol. 29:2323–2331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao
H, Gong C, Chen J, Su F, Zhang Y, et al: Reduced miR-128 in breast
tumor-initiating cells induces chemotherapeutic resistance via
Bmi-1 and ABCC5. Clin Cancer Res. 17:7105–7115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li M, Fu W, Wo L, Shu X, Liu F and Li C:
miR-128 and its target genes in tumorigenesis and metastasis. Exp
Cell Res. 319:3059–3064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li B, Chen H, Wu N, Zhang WJ and Shang LX:
Deregulation of miR-128 in ovarian cancer promotes cisplatin
resistance. Int J Gynecol Cancer. 24:1381–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G,
Fu S, Zhang Y, Feng K and Feng Y: microRNA-128 plays a critical
role in human non-small cell lung cancer tumourigenesis,
angiogenesis and lymphangiogenesis by directly targeting vascular
endothelial growth factor-C. Eur J Cancer. 50:2336–2350. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong Q, Cai N, Tao T, Zhang R, Yan W, Li
R, Zhang J, Luo H, Shi Y, Luan W, et al: An axis involving SNAI1,
microRNA-128 and SP1 modulates glioma progression. PLoS One.
9:e986512014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seca H, Lima RT, Almeida GM,
Sobrinho-Simoes M, Bergantim R, Guimaraes JE and Vasconcelos MH:
Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia
cells. Curr Pharm Biotechnol. 15:492–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khan AP, Poisson LM, Bhat VB, Fermin D,
Zhao R, Kalyana-Sundaram S, Michailidis G, Nesvizhskii AI, Omenn
GS, Chinnaiyan AM, et al: Quantitative proteomic profiling of
prostate cancer reveals a role for miR-128 in prostate cancer. Mol
Cell Proteomics. 9:298–312. 2010. View Article : Google Scholar :
|
|
30
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
31
|
Takahashi Y, Iwaya T, Sawada G, Kurashige
J, Matsumura T, Uchi R, Ueo H, Takano Y, Eguchi H, Sudo T, et al:
Up-regulation of NEK2 by microRNA-128 methylation is associated
with poor prognosis in colorectal cancer. Ann Surg Oncol.
21:205–212. 2014. View Article : Google Scholar
|
|
32
|
Zhang X, Li HM, Liu Z, Zhou G, Zhang Q,
Zhang T, Zhang J and Zhang C: Loss of heterozygosity and
methylation of multiple tumor suppressor genes on chromosome 3 in
hepatocellular carcinoma. J Gastroenterol. 48:132–143. 2013.
View Article : Google Scholar
|
|
33
|
Gatto F, Nookaew I and Nielsen J:
Chromosome 3p loss of heterozygosity is associated with a unique
metabolic network in clear cell renal carcinoma. Proc Natl Acad Sci
USA. 111:E866–E875. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saint-Georges F, Garçon G, Escande F,
Abbas I, Verdin A, Gosset P, Mulliez P and Shirali P: Role of air
pollution Particulate Matter (PM(2.5)) in the occurrence of loss of
heterozygosity in multiple critical regions of 3p chromosome in
human epithelial lung cells (L132). Toxicol Lett. 187:172–179.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly
MB, Wang Y, Qian Z, Jin J, Zhang Y, et al: MicroRNA expression
signatures accurately discriminate acute lymphoblastic leukemia
from acute myeloid leukemia. Proc Natl Acad Sci USA.
104:19971–19976. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tao T, Li G, Dong Q, Liu D, Liu C, Han D,
Huang Y, Chen S, Xu B and Chen M: Loss of SNAIL inhibits cellular
growth and metabolism through the miR-128-mediated
RPS6KB1/HIF-1α/PKM2 signaling pathway in prostate cancer cells.
Tumour Biol. 35:8543–8550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Adlakha YK and Saini N: miR-128 exerts
pro-apoptotic effect in a p53 transcription-dependent and
-independent manner via PUMA-Bak axis. Cell Death Dis. 4:e5422013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheung LW, Hennessy BT, Li J, Yu S, Myers
AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al:
High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer
elucidates a novel mechanism for regulation of PTEN protein
stability. Cancer Discov. 1:170–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jaiswal BS, Janakiraman V, Kljavin NM,
Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring
P, et al: Somatic mutations in p85α promote tumorigenesis through
class IA PI3K activation. Cancer Cell. 16:463–474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun M, Hillmann P, Hofmann BT, Hart JR and
Vogt PK: Cancer-derived mutations in the regulatory subunit p85α of
phosphoinositide 3-kinase function through the catalytic subunit
p110α. Proc Natl Acad Sci USA. 107:15547–15552. 2010. View Article : Google Scholar
|
|
41
|
Urick ME, Rudd ML, Godwin AK, Sgroi D,
Merino M and Bell DW: PIK3R1 (p85α) is somatically mutated at high
frequency in primary endometrial cancer. Cancer Res. 71:4061–4067.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun Y, Zhao S, Tian H, Xie X, Xiao F, Li K
and Song Y: Depletion of PI3K p85α induces cell cycle arrest and
apoptosis in colorectal cancer cells. Oncol Rep. 22:1435–1441.
2009.PubMed/NCBI
|
|
43
|
Hong M, Wilkes MC, Penheiter SG, Gupta SK,
Edens M and Leof EB: Non-Smad transforming growth factor-β
signaling regulated by focal adhesion kinase binding the p85
subunit of phosphatidylinositol 3-kinase. J Biol Chem.
286:17841–17850. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|