Introduction
Breast cancer is the most common malignancy and is
the leading cause of cancer-related death in females worldwide
(1,2). At present, the main clinical therapy
strategies include chemotherapy, surgery and radiotherapy, but all
of these have side effects (3,4).
Mesenchymal stem cells (MSCs) are known to migrate to tumors, and
it is possible to exploit the behavior of MSCs as a tumor-targeting
method for cell-based cancer therapy. The effects of MSCs on tumor
progression remain controversial, and in particular, it is not
clear whether the clinical application of MSCs leads to unforeseen
and unwanted side effects.
Tumor development has been recognized as the result
of the interaction between tumor cells and their surrounding
supporting tissues (5). The mutual
interactions of tumor cells and stromal cells through direct
contact to various cytokines and chemokines in a paracrine manner
are thought to modulate tumor progression (6–8).
Several studies indicate that MSCs promote tumor proliferation and
metastasis (9,10), whereas other studies suggest that
MSCs display intrinsic anticancer activities (11–13).
This discrepancy requires further investigation.
The bone marrow is the main source of MSCs, but
their collection from the bone marrow is extremely difficult. The
proliferative and multilineage differentiation capacities of bone
marrow-derived MSCs (BM-MSCs) decreases with aging (14). However, umbilical cord collection is
convenient and is not associated with any ethical or legal issue
(15). Many studies have confirmed
that the proliferative and differentiation abilities of umbilical
cord MSCs (UC-MSCs) are greater than those of BM-MSCs (16). Therefore, UC-MSCs are considered a
promising source of stem cells for cancer therapy.
UC-MSCs have been found to target many primary solid
tumors and their metastases (17,18).
UC-MSCs secrete interferon-b (IFN-b), which was found to reduce the
growth of human MDA-MB-231 breast carcinoma cells by inducing
apoptosis (19). It was recently
shown that the intratumoral injection of rat umbilical cord matrix
stem cells (rUC-MSCs) caused regression of rat mammary carcinomas
(20). Human umbilical cord
Wharton's jelly stem cells (hWJSCs) have been shown to have
anti-inflammatory potential by reducing the expression of
inflammatory mediators (21). Taken
together, the results indicate that UC-MSCs exhibit an anticancer
effect. However, it remains unclear whether UC-MSCs are safe in
cancer clinical therapy.
The MDA-MB-231 cell line is a triple-negative breast
cancer cell line (22) that
exhibits stronger drug resistance and a tendency to manifest
recurrence and metastasis. The MCF-7 cell line is an estrogen
receptor-positive, hormone-dependent breast cancer cell line. In
the present study, we sought to ascertain whether UC-MSCs have the
capability to affect the migratory potential of MCF-7 cells, which
have very low metastatic potential (23), and whether UC-MSCs exert
differential effects in MDA-MB-231 and MCF-7 cells. The molecular
mechanism of UC-MSCs on cancer cells remains unclear. Thus, a
better understanding of the molecules or mechanism that regulates
the proliferative and migratory behaviors of breast cancer cells is
essential to the development of novel effective therapies. Thus,
the present study focused on the molecular mechanism underlying
these effects.
Materials and methods
Cell culture
The cells were cultured in Dulbecco's modified
Eagle's medium with low glucose (L-DMEM) supplemented with 10%
fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/ml
penicillin and streptomycin under mycoplasma-free conditions at
37°C in 5% CO2. The human breast cancer cell lines MCF-7
and MDA-MB-231 were a gift from Dr W. Zhu (Department of Medicine,
Jiangsu University, China).
Isolation and culture of human umbilical
cord MSCs (hUC-MSCs)
Fresh umbilical cords were collected from informed,
consenting mothers at the First People's Hospital of Zhenjiang
(China) and rapidly processed. Moreover, hUC-MSCs were isolated
within the optimal processing period of 6 h. The cords were rinsed
twice with phosphate-buffered saline (PBS) supplemented with
penicillin and streptomycin to remove any blood and cord vessels.
The washed cords were subsequently cut into 1 mm3
pieces, floated in L-DMEM containing 10% FBS, penicillin and
streptomycin, and incubated at 37°C with 5% CO2. The
medium was replaced every 3 days after the initial culture. When
well-developed colonies of fibroblast-like cells appeared after 10
days, the cultures were trypsinized and passaged into a new flask
for further expansion, and the medium was changed every 3 days. The
experimental protocol was approved by the Jiangsu University Ethics
Committee.
Flow cytometry
After the third passage, the cells were trypsinized
(0.25% trypsin EDTA), washed twice with PBS and stained on ice with
monoclonal antibodies against FITC-CD34, HLA-DR, PE-CD29, CD44 and
CD90 (Becton-Dickinson, San Jose, CA, USA). PE-IgG1 and FiTC-IgG1
were used as isotype controls. The stained cells were analyzed by
flow cytometry (FACSCalibur; Becton-Dickinson).
Osteogenic and adipogenic differentiation
in vitro
The differentiation of UC-MSCs was assessed in the
third passage. The cells were cultured in medium that contained
either osteogenic reagents [0.1 µM dexamethasone, 10 mM
β-glycerophosphate, 50 mg/l ascorbic acid and 4 µg/ml basic
fibroblast growth factor (bFGF)] (all from Sigma-Aldrich, St.
Louis, MO, USA) for 2 weeks or adipogenic reagents (1 µM
dexamethasone, 0.5 µM 3-isobutyl-1-methylxanthine, 5 ng/ml insulin,
60 µM indomethacin and 100 µM hydrocortisone) (all from Cyagen,
Guangzhou, China) for 3 weeks. Two or three weeks later, the degree
of osteogenic differentiation was assessed by Alizarin Red
staining, and the intracellular lipid accumulation was visualized
by Oil Red O staining.
Generation of conditioned media
UC-MSCs were plated to 70% confluency in 35-mm
plates with 10% FBS L-DMEM and allowed to adhere overnight at 37°C
in 5% CO2. The following day, the media was removed, and
the cells were washed twice with PBS. The cells were then
re-incubated with non-serum culture media. After 12 h, the
conditioned medium (CM) was collected and passed through a 0.45-µm
filter (Sigma-Aldrich). CM aliquots were frozen at -20°C until
required (not exceeding 2 weeks). To prepare different
concentrations of UC-MSC-CM (10 and 20%), 100% UC-MSC-CM was
diluted accordingly in freshly prepared L-DMEM with 10% FBS.
MTT and plate colony formation
assays
The cells were plated at a density of
2.5×103 cells/well in a 96-well plate in 180 µl of
L-DMEM and allowed to attach overnight. The cells were then treated
with 0, 10 and 20% CM for 48 h. MTT (20 µl) was added to each well
for the last 4 h. Once the reaction was terminated, the solution
was discarded, and 150 µl of dimethyl sulfoxide was added to each
well. The 96-well plate was shaken to ensure complete
solubilization of the purple formazan crystals. The absorbance at
490 nm was measured using an enzyme-linked immunosorbent assay
reader. For the colony formation assay, the cells were plated at a
density of 200 cells/plate in a 6-well plate. After culturing for
10 days, the colony units were fixed with methanol and stained with
crystal violet for 30 min before washing with water and air-drying.
The clones with >150 cells were counted with an optical
microscope, and the clone formation rate was calculated using the
following formula: Plate clone formation efficiency = (number of
clones/number of cells inoculated) × 100%. All of the experiments
were repeated 3 times, and the average values are reported.
Scratch wound assay
The cells were grown to confluence and then
scratched with a 0.2-ml pipette tip. The resulting debris was
removed by gentle washing with medium. The cells were subsequently
placed in an incubator. The cells were maintained in the presence
of 0, 10 and 20% CM for 24 h, respectively. Images of the closing
wound were acquired with an inverted microscope and analyzed using
image software (National Institutes of Health, Bethesda, MD,
USA).
Transwell migration assay
First, 0, 10 or 20% CM was added to the bottom
chambers. Then, 5×104 MCF-7 cells and 2.5×104
MDA-MB-231 cells were plated in 100 µl of non-serum L-DMEM
and added to the top of the chambers (Corning, Lowell, MA, USA),
and the plates were then incubated for 12 h at 37°C. The cells on
the top part of the filter were removed by scrubbing twice with a
cotton swab. The migrating cells were fixed in formaldehyde and
stained with crystal violet. Four low-power fields (magnification,
x200) were randomly selected from each chamber to observe the cells
and count the stained migrated cells. Each experimental group was
assessed in triplicate.
Reverse transcriptase-polymerase chain
reaction (RT-PCR)
MCF-7 and MDA-MB-231 cells were treated with CM (0,
10 or 20%) for 48 h. The total RNA was isolated with the TRIzol
reagent (invitrogen), and 1 µg of RNA was processed for cDNA
synthesis with Superscript II reverse transcriptase using Oligo-dT
primer (Toyobo, Japan). PCR was performed using 1 µg of cDNA
sample with 0.3 U of Taq polymerase (Cinnagen, Iran), 200
µM dNTPs, 10 pM of each primer, reaction buffer, and
MgCl2 (Takara, Japan) in a 25-µl volume. PCR
amplification was performed for 35 cycles using an ABI 2720 Thermal
Cycler (Applied Biosystems). The cycling conditions were 94°C for
30 sec, 60°C (primer) for 30 sec, and 72°C for 30 sec, and a final
extension at 72°C for 10 min was performed. The PCR products were
separated on a 1.5% agarose gel, stained with ethidium bromide, and
visualized under UV light. The specific primers for PCR were
designed as follows: β-actin sense, 5′-CACGAAACTACCTTCAACTC-3′ and
antisense, 5′-CATACTCCTGCTTGCTGATC-3′; E-cadherin sense,
5′-CGCATTGCCACATACACTCT-3′ and antisense, 5′-TTGG
CTGAGGATGGTGTAAG-3′; and N-cadherin sense, 5′-AGT
CAACTGCAACCGTGTCT-3′ and antisense, 5′-AGCGTTCC
TGTTCCACTCAT-3′.
Western blot assay
MCF-7 and MDA-MB-231 cells were treated with CM (0,
10 and 20%) for 48 h. The total cellular protein was extracted
using RIPA lysis buffer. Samples containing 100 µg of
protein were separated on 10% SDS-PAGE gels (Beyotime, Shanghai,
China) and transferred electrophoretically to a PVDF membrane
(Millipore Corp., Billerica, MA, USA), and the membrane was then
blocked with 5% (w/v) skim milk in TBST (20 mM Tris-HCl, 0.15 M
NaCl, and 0.05% Tween-20) for 1 h at room temperature. The
membranes were then incubated with primary antibodies at 4°C
overnight, washed in TBST and incubated for 1 h with a goat
anti-rabbit secondary antibody. The reactions were visualized using
an ECL detection system (Amersham Pharmacia Biotech, Little
Chalfont, UK). The western blot data are representatives from 3
independent experiments. The intensities of the bands obtained from
the western blot assays were quantified using the Gel Image
analysis software (Lane 1D, Beijing, China). The following primary
antibodies were used: p-ERK and T-ERK (1:1,000; Santa Cruz
Biotechnology, Santa Cruz, CA, USA), PCNA (1:1,000; Bioworld,
Minneapolis, MN, USA), E-cadherin, N-cadherin and ZEB1 (1:1,000;
Cell Signaling Technology, Beverly, MA, USA) and GAPDH and the
secondary antibody (1:2,000; Kangcheng Bio-Engineering, Shanghai,
China).
Statistical analysis
Differences between more than two groups were
analyzed by one-way ANOVA with the Newman-Keuls multiple comparison
test using the GraphPad Prism V5.0 software program (GraphPad, San
Diego, CA, USA). The results are expressed as the mean ± SD of 3
different replicates from individual assays. p<0.05, p<0.01
and p<0.001 were considered to indicate statistically
significant differences.
Results
Morphology and differentiation potential
and surface antigens of UC-MSCs
After 7 to 10 days of initial culture, the long
spindle-shaped fibroblastic cells began to form colonies and became
confluent (Fig. 1A). A multilineage
differentiation potential is the functional standard for verifying
the identity of MSCs. The differentiation of UC-MSCs was apparent
after 2 or 3 weeks of induction under specific media. At the end of
the second or third week, the UC-MSCs were capable of
differentiating into osteocytes and adipocytes, as shown by
positive staining of Oil Red O (Fig.
1B) and Alizarin Red (Fig. 1C).
The surface antigens of MSCs were positive for CD29, CD44, and CD90
but negative for CD34 and HLA-DR (Fig.
1D).
UC-MSCs enhance the proliferation of
MCF-7 and MDA-MB-231 cells
We hypothesized that various soluble factors
secreted by stem cells are capable of affecting cancer cell growth;
thus, we further investigated the effects of CM derived from
UC-MSCs on breast cancer cells. The MTT assay of MCF-7 and
MDA-MB-231 cells cultured in UC-MSC-CM (0, 10 and 20%) revealed
proliferation rates of 0.342±0.015, 0.557±0.066 and 0.534±0.047 for
MCF-7 cells and 0.143±0.017, 0.275±0.046 and 0.299±0.060 for
MDA-MB-231 cells, respectively (Fig.
2A). The increases in the proliferation rates of MCF-7 and
MDA-MB-231 cells with 10 and 20% CM were statistically significant
compared with the rates observed in the control groups (p<0.05).
After 10 days of culture, the plate clone formation rates obtained
for the control group and the 10 and 20% CM groups were
0.040±0.013, 0.088±0.013 and 0.173±0.025 for MCF-7 cells and
0.080±0.010, 0.157±0.018, and 0.128±0.013 for MDA-MB-231 cells,
respectively, and these differences were statistically significant
(p<0.05, Fig. 2B and C). Our
western blot results showed that treatment with CM increased the
PCNA protein levels in breast cancer cells (Fig. 2D). The data also revealed that the
PCNA expression level in MCF-7 and MDA-MB-231 cells treated with 10
and 20% CM presented significant differences compared with the
control groups (Fig. 2E,
p<0.05).
UC-MSCs promote the migration of MCF-7
and MDA-MB-231 cells
In this study, we sought to determine whether
UC-MSCs affect the migratory potential of the normally
non-metastatic MCF-7 cell line and the high-metastatic MDA-MB-231
cell line. In the Transwell migration assay, the mean numbers of
migrated MCF-7 cells in the lower fields after 12 h were
46.50±12.40, 77.75±6.02 and 91.00±8.52, whereas the mean numbers of
MDA-MB-231 cells were 76.50±5.97, 112.50±10.28, and 140.50±5.79,
respectively. There were significant differences between the
control group and the CM groups (p<0.05, Fig. 3A and B). Scratch wounds were
inflicted in cells pretreated with or without UC-MSC-CM for 24 h,
whereas differences were observed between the groups after 24 h of
treatment (Fig. 3C). The wound
closure ratios for MCF-7 cells were 23.18±3.73, 28.65±2.61 and
35.83±2.88% in the control, 10 and 20% CM groups, respectively. The
difference obtained with 10% CM was not significant, but that
obtained with 20% CM was statistically significant (p<0.05,
Fig. 3D). These results indicate
that MCF-7 cells have very low metastatic potential. The wound
closure ratios for the MDA-MB-231 cells were 68.00±4.16, 86.00±9.89
and 114.80±9.22 in the control and 10 and 20% CM groups,
respectively, and the values obtained for the CM groups were
significantly higher compared with that obtained for the control
group (p<0.05, Fig. 3D).
E-cadherin and N-cadherin expression
The RT-PCR and western blot results showed that
treatment with CM downregulated E-cadherin and increased N-cadherin
(Fig. 4A and B, p<0.05). The
results revealed that the mRNA and protein expression levels of
N-cadherin in breast cancer cells were significantly higher than
those in the control group. In addition, there was a significant
difference in the expression of E-cadherin in the MCF-7 and
MDA-MB-231 cells between the CM and control groups. The CM-induced
migration of UC-MSCs may be achieved by the suppression of
E-cadherin and the stimulation of N-cadherin expression.
Protein expression of UC-MSCs and the
effect of ERK inhibitor U0126 on breast cancer cells
To investigate whether CM downregulates E-cadherin
expression by modulating the transcription factor ZEB1, we examined
ZEB1 protein levels. Treatment with CM significantly increased ZEB1
expression by ~4-fold in the MCF-7 cells and 3-fold in the
MDA-MB-231 cells compared with the control groups (p<0.05,
Fig. 5Ab). These findings indicate
that the effect of UC-MSCs on breast cancer cell migration may be
achieved by EMT. We then analyzed the activation of the ERK
pathway. Treatment with 10 and 20% CM enhanced the p-ERK levels in
the MCF-7 and MDA-MB-231 cells (p<0.05, Fig. 5Aa). To determine whether the
MAPK/ERK signaling pathway is involved in the CM-induced increase
in ZEB1 protein levels, the cells were treated with the ERK
inhibitor U0126 (Promega, Madison, WI, USA) in the presence or
absence of CM. Notably, U0126 significantly decreased the ZEB1 and
p-ERK protein levels (Fig. 5Bc and
d). These results are consistent with those of previous studies
that demonstrated that MAPK/ERK is an upstream factor of ZEB1
activation in ovarian cancer cells in vitro (24) and that MAPK/ERK signaling is
required in IGF-1-induced ZEB1 expression in prostate cancer cells
(25). The ERK inhibitors
significantly decreased N-cadher in expression and increased basal
E-cadherin expression (Fig. 5Cf and
g) as well as markedly diminished but not completely abolished
the CM-induced suppression of E-cadherin expression. This finding
suggests that MAPK/ERK signaling is required for MSC-derived
CM-induced E-cadherin downregulation. Furthermore, U0126
significantly decreased the PCNA protein levels (Fig. 5Ce). Taken together, these results
indicate that the MAPK/ERK pathway is involved in CM-induced breast
cancer cell proliferation and migration.
 | Figure 5Effects of UC-MSCs and ERK inhibitor
U0126 on the protein expression levels in the breast cancer cell
lines. (A) MCF-7 and MDA-MB-231 cells were treated with 0, 10 and
20% UC-MSC-CM for 48 h. The P-ERK and ZEB1 protein levels in MCF-7
and MDA-MB-231 cells were analyzed by western blotting. T-ERK and
GAPDH protein levels were used as controls to ensure equal loading.
Three independent experiments were performed to measure the (a)
P-ERK and (b) ZEB1 protein levels. (B and C) MCF-7 and MDA-MB-231
cells were pretreated with 10 µM U0126 for 60 min before the
addition of 0 and 20% UC-MSC-CM. The P-ERK, ZEB1, E-cadherin,
N-cadherin, and PCNA protein levels were analyzed by western
blotting. Three independent experiments were used to measure the
(c) P-ERK, (d) ZEB1, (e) PCNA, (f) E-cadherin and (g) N-cadherin
protein levels. *p<0.05 and **p<0.01
compared with the control group; #p<0.05 and
##p<0.01 for the comparison between the U0126+20% CM
group and the 20% CM group. Lane 1, control; lane 2, 10% CM; lane
3, 20% CM; lane 4, U0126; lane 5, U0126+20% CM. CM, conditioned
medium; UC-MSCs, umbilical cord mesenchymal stem cells. |
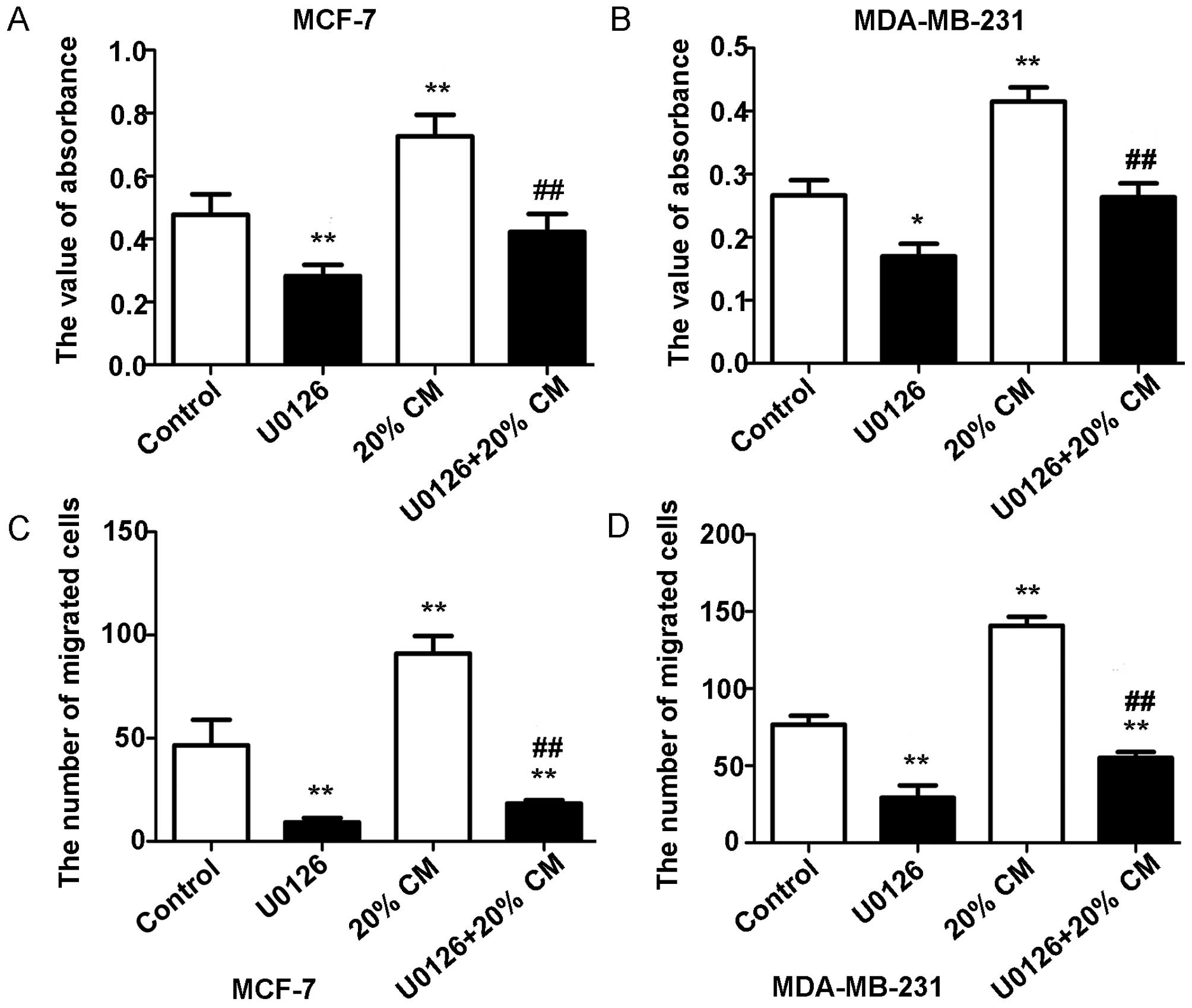
Effects of the ERK inhibitor U0126 on
CM-stimulated cell proliferation and migration
To determine whether the ERK pathway is involved in
the effects of CM on breast cancer cells, we used the ERK inhibitor
U0126 to specifically block the MAPK/ERK pathway in MCF-7 and
MDA-MB-231 cells. As shown in Fig. 6A
and B, the MTT assay results showed that U0126 treatment
significantly decreased breast cancer cell proliferation
(p<0.05). The proliferation rates obtained were 0.477±0.065,
0.2820±0.036, 0.7267±0.068 and 0.4220±0.058 for MCF-7 cells and
0.267±0.048, 0.170±0.040, 0.415±0.045, and 0.264±0.044 for
MDA-MB-231 cells. Our results also showed that CM-induced cell
migration was markedly diminished but not totally abolished by
treatment with U0126 (p<0.05). The numbers of migrated MCF-7
cells were 46.50±12.40, 9.200±2.168, 91.00±8.524 and 18.25±1.708,
whereas the numbers of migrated MDA-MB-231 cells were 78.33±4.509,
28.67±2.082, 138.7±7.572 and 54.25±5.058 (Fig. 6C and D). This finding also suggests
that other pathways may be involved in the response of breast
cancer cells to hUC-MSCs.
Discussion
In the present study, UC-MSCs from human umbilical
cord tissues showed a homogenous immunophenotype and multilineage
differentiation potential (osteoblast and adipocyte lineages). We
demonstrated that these were homogeneously positive for the
mesenchymal cell markers CD29, CD90 and CD44 but negative for CD34
and HLA-DR. These results are consistent with those of previous
studies (26,27). The data showed that UC-MSCs were
Alizarin Red-positive and Oil Red O-positive after induction. Taken
together, the findings suggest that the isolated adherent cells
from the umbilical cord were in fact MSCs.
Furthermore, we observed the effects of UC-MSCs on
the proliferation and migration of the human breast cancer cell
lines MCF-7 and MDA-MB-231 in vitro. The MTT cell
proliferation results showed that UC-MSC-CM significantly
stimulates breast cancer cell proliferation. Therefore, it was
suggested that UC-MSCs may exert certain increasing effects on the
growth of breast cancer cells in vitro. The statistical
analysis of the scratch wound and Transwell migration assay results
revealed that CM significantly promoted MCF-7 and MDA-MB-231 cell
migration. Our results are consistent with those of previous
studies (28–31).
E-cadherin functions as a cell-cell adhesion protein
and tumor-suppressor that is silenced in many malignancies
(32). E-cadherin is known to
suppress tumor invasion, and the re-expression of E-cadherin in
E-cadherin-deficient tumors reverts cells to a less invasive
phenotype (33,34). Some findings indicate that hMSCs
decrease cell-to-cell contact and decrease epithelial cell adhesion
markers (i.e., E-cadherin) in breast cancer cells (35,36).
Several transcription factors have been identified to suppress
E-cadherin, including Twist, Snail, Slug and ZEB1, via their
interaction with the E-box binding site in the E-cadherin promoter
(37,38). In the present study, we demonstrated
that CM reduced the E-cadherin protein and mRNA levels and
increased N-cadherin and ZEB1 expression via activation of the
MAPK/ERK signaling pathways. Finally, our results found that the
downregulation of E-cadherin mediated by CM enhanced the migration
of breast cancer cells.
PCNA is a well-defined regulator of DNA replication
and cell cycle control (39).
Treatment with CM significantly increased the PCNA protein levels.
An inhibitor of ERK was able to downregulate the expression of
PCNA. Furthermore, this effect was regulated through ERK nuclear
translocation, resulting in enhanced PCNA expression. These
findings suggest that UC-MSC-CM induced the proliferation of MCF-7
and MDA-MB-231 cells via the MAPK/ERK pathways.
However, our results are contrary to those of
previous studies, which suggest that hUC-MSCs inhibit the growth of
breast cancer cells (40,41). We speculate that this discrepancy
may be related to the sources and numbers of the MSCs, differences
in the culture and experimental methods, the type and site of the
carcinoma, or a combination of these factors. We believe that
UC-MSCs provide potential for cancer therapy, and further study of
UC-MSCs will offer a better understanding of the relationship
between MSCs and tumor progression and the mechanism governing this
relationship.
Acknowledgments
The present study was supported by the Foundation of
Jiangsu University for Seniors (grant no. 11JDG0089) and the
Innovation Project of Cultivating Graduates of Jiangsu Province
(grant no. CXLX13_689) and the Science Foundation of Kunshan (grant
no. KS1331).
References
|
1
|
Nelson HD, Zakher B, Cantor A, Fu R,
Griffin J, O'Meara ES, Buist DS, Kerlikowske K, van Ravesteyn NT,
Trentham-Dietz A, et al: Risk factors for breast cancer for women
aged 40 to 49 years: A systematic review and meta-analysis. Ann
intern Med. 156:635–648. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hugosson J, Stranne J and Carlsson SV:
Radical retropubic prostatectomy: A review of outcomes and
side-effects. Acta Oncol. 50(Suppl 1): 92–97. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Ruysscher D, Van Meerbeeck J,
Vandecasteele K, Oberije C, Pijls M, Dingemans AM, Reymen B, van
Baardwijk A, Wanders R, Lammering G, et al: Radiation-induced
oesophagitis in lung cancer patients. Is susceptibility for
neutropenia a risk factor? Strahlenther Onkol. 188:564–567. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mbeunkui F and Johann DJ Jr: Cancer and
the tumor microenvironment: A review of an essential relationship.
Cancer Chemother Pharmacol. 63:571–582. 2009. View Article : Google Scholar
|
|
6
|
Orimo A, Gupta PB, Sgroi DC,
Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL
and Weinberg RA: Stromal fibroblasts present in invasive human
breast carcinomas promote tumor growth and angiogenesis through
elevated SDF-1/CxCL12 secretion. Cell. 121:335–348. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whiteside TL: The tumor microenvironment
and its role in promoting tumor growth. Oncogene. 27:5904–5912.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shinagawa K, Kitadai Y, Tanaka M, Sumida
T, Kodama M, Higashi Y, Tanaka S, Yasui W and Chayama K:
Mesenchymal stem cells enhance growth and metastasis of colon
cancer. Int J Cancer. 127:2323–2333. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu JM, Jun ES, Bae YC and Jung JS:
Mesenchymal stem cells derived from human adipose tissues favor
tumor cell growth in vivo. Stem Cells Dev. 17:463–473. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khakoo AY, Pati S, Anderson SA, Reid W,
Elshal MF, Rovira II, Nguyen AT, Malide D, Combs CA, Hall G, et al:
Human mesenchymal stem cells exert potent antitumorigenic effects
in a model of Kaposi's sarcoma. J Exp Med. 203:1235–1247. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JK, Park SR, Jung BK, Jeon YK, Lee YS,
Kim MK, Kim YG, Jang JY and Kim CW: Exosomes derived from
mesenchymal stem cells suppress angiogenesis by down-regulating
VEGF expression in breast cancer cells. PLoS One. 8:e842562013.
View Article : Google Scholar
|
|
13
|
Gauthaman K, Yee FC, Cheyyatraivendran S,
Biswas A, Choolani M and Bongso A: Human umbilical cord Wharton's
jelly stem cell (hWJSC) extracts inhibit cancer cell growth in
vitro. J Cell Biochem. 113:2027–2039. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rao MS and Mattson MP: Stem cells and
aging: Expanding the possibilities. Mech Ageing Dev. 122:713–734.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Secco M, Zucconi E, Vieira NM, Fogaça LL,
Cerqueira A, Carvalho MD, Jazedje T, Okamoto OK, Muotri AR and Zatz
M: Multipotent stem cells from umbilical cord: Cord is richer than
blood! Stem Cells. 26:146–150. 2008. View Article : Google Scholar
|
|
16
|
Baksh D, Yao R and Tuan RS: Comparison of
proliferative and multilineage differentiation potential of human
mesenchymal stem cells derived from umbilical cord and bone marrow.
Stem Cells. 25:1384–1392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ayuzawa R, Doi C, Rachakatla RS, Pyle MM,
Maurya DK, Troyer D and Tamura M: Naïve human umbilical cord matrix
derived stem cells significantly attenuate growth of human breast
cancer cells in vitro and in vivo. Cancer Lett. 280:31–37. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu WT, Bian ZY, Fan QM, Li G and Tang TT:
Human mesenchymal stem cells (hMSCs) target osteosarcoma and
promote its growth and pulmonary metastasis. Cancer Lett.
281:32–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rachakatla RS, Pyle MM, Ayuzawa R, Edwards
SM, Marini FC, Weiss ML, Tamura M and Troyer D: Combination
treatment of human umbilical cord matrix stem cell-based
interferon-beta gene therapy and 5-fluorouracil significantly
reduces growth of metastatic human breast cancer in SCID mouse
lungs. Cancer Invest. 26:662–670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ganta C, Chiyo D, Ayuzawa R, Rachakatla R,
Pyle M, Andrews G, Weiss M, Tamura M and Troyer D: Rat umbilical
cord stem cells completely abolish rat mammary carcinomas with no
evidence of metastasis or recurrence 100 days post-tumor cell
inoculation. Cancer Res. 69:1815–1820. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moodley Y, Atienza D, Manuelpillai U,
Samuel CS, Tchongue J, Ilancheran S, Boyd R and Trounson A: Human
umbilical cord mesenchymal stem cells reduce fibrosis of
bleomycin-induced lung injury. Am J Pathol. 175:303–313. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Caldas-Lopes E, Cerchietti L, Ahn JH,
Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A,
Guo Y, et al: Hsp90 inhibitor PU-H71, a multimodal inhibitor of
malignancy, induces complete responses in triple-negative breast
cancer models. Proc Natl Acad Sci USA. 106:8368–8373. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soule HD, Vazguez J, Long A, Albert S and
Brennan M: A human cell line from a pleural effusion derived from a
breast carcinoma. J Natl Cancer Inst. 51:1409–1416. 1973.PubMed/NCBI
|
|
24
|
Lau MT, So WK and Leung PC: Fibroblast
growth factor 2 induces E-cadherin down-regulation via
PI3K/Akt/mTOR and MAPK/ERK signaling in ovarian cancer cells. PLoS
One. 8:e590832013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Graham TR, Zhau HE, Odero-Marah VA,
Osunkoya AO, Kimbro KS, Tighiouart M, Liu T, Simons JW and O'Regan
RM: Insulin-like growth factor-I-dependent up-regulation of ZEB1
drives epithelial-to-mesenchymal transition in human prostate
cancer cells. Cancer Res. 68:2479–2488. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qiao C, Xu W, Zhu W, Hu J, Qian H, Yin Q,
Jiang R, Yan Y, Mao F and Yang H: Human mesenchymal stem cells
isolated from the umbilical cord. Cell Biol Int. 32:8–15. 2008.
View Article : Google Scholar
|
|
27
|
Fong CY, Richards M, Manasi N, Biswas A
and Bongso A: Comparative growth behaviour and characterization of
stem cells from human Wharton's jelly. Reprod Biomed Online.
15:708–718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu W, Huang L, Li Y, Qian H, Shan X, Yan
Y, Mao F, Wu X and Xu WR: Mesenchymal stem cell-secreted soluble
signaling molecules potentiate tumor growth. Cell Cycle.
10:3198–3207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang T, Lee YW, Rui YF, Cheng TY, Jiang
XH and Li G: Bone marrow-derived mesenchymal stem cells promote
growth and angiogenesis of breast and prostate tumors. Stem Cell
Res Ther. 4:702013. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ke CC, Liu RS, Suetsugu A, Kimura H, Ho
JH, Lee OK and Hoffman RM: In vivo fluorescence imaging reveals the
promotion of mammary tumorigenesis by mesenchymal stromal cells.
PLoS One. 8:e696582013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan XL, Fu CJ, Chen L, Qin JH, Zeng Q,
Yuan HF, Nan X, Chen HX, Zhou JN, Lin YL, et al: Mesenchymal stem
cells from primary breast cancer tissue promote cancer
proliferation and enhance mammosphere formation partially via
EGF/EGFR/Akt pathway. Breast Cancer Res Treat. 132:153–164. 2012.
View Article : Google Scholar
|
|
32
|
Nollet F, Berx G and van Roy F: The role
of the E-cadherin/catenin adhesion complex in the development and
progression of cancer. Mol Cell Biol Res Commun. 2:77–85. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gottardi CJ, Wong E and Gumbiner BM:
E-cadherin suppresses cellular transformation by inhibiting
beta-catenin signaling in an adhesion-independent manner. J Cell
Biol. 153:1049–1060. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yanagisawa M and Anastasiadis PZ: p120
catenin is essential for mesenchymal cadherin-mediated regulation
of cell motility and invasiveness. J Cell Biol. 174:1087–1096.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fierro FA, Sierralta WD, Epuñan MJ and
Minguell JJ: Marrow-derived mesenchymal stem cells: Role in
epithelial tumor cell determination. Clin Exp Metastasis.
21:313–319. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Martin FT, Dwyer RM, Kelly J, Khan S,
Murphy JM, Curran C, Miller N, Hennessy E, Dockery P, Barry FP, et
al: Potential role of mesenchymal stem cells (MSCs) in the breast
tumour microenvironment: Stimulation of epithelial to mesenchymal
transition (EMT). Breast Cancer Res Treat. 124:317–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bolós V, Peinado H, Pérez-Moreno MA, Fraga
MF, Esteller M and Cano A: The transcription factor Slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: A comparison with Snail and E47 repressors. J Cell
Sci. 116:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Strzalka W and Ziemienowicz A:
Proliferating cell nuclear antigen (PCNA): A key factor in DNA
replication and cell cycle regulation. Ann Bot (Lond).
107:1127–1140. 2011. View Article : Google Scholar
|
|
40
|
Sun B, Yu KR, Bhandari DR, Jung JW, Kang
SK and Kang KS: Human umbilical cord blood mesenchymal stem
cell-derived extracellular matrix prohibits metastatic cancer cell
MDA-MB-231 proliferation. Cancer Lett. 296:178–185. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma Y, Hao X, Zhang S and Zhang J: The in
vitro and in vivo effects of human umbilical cord mesenchymal stem
cells on the growth of breast cancer cells. Breast Cancer Res
Treat. 133:473–485. 2012. View Article : Google Scholar
|