Introduction
Hepatocellular carcinoma (HCC) is one of the most
common human cancers worldwide and ranks third in cancer-related
death (1). Although remarkable
achievements have been made (2),
the long-term prognosis of HCC patients remains poor, particularly
for those in advanced stages, with the 5-year survival rate lower
than 5% (3). Local and systemic
metastasis are the critical reasons for the unsatisfactory survival
of HCC patients in advanced stages (4). Elucidating the molecular mechanism of
the metastasis of HCC is critical for identifying novel therapeutic
targets and may significantly improve the prognosis of HCC
patients.
MicroRNAs (miRNAs) are a group of short non-conding
RNAs and serve as important post-transcriptional modulator of gene
expression by binding to 3′ untranslated region (3′-UTR) of
targeted mRNAs and resulting in the suppression of protein
translation or the degradation of the target mRNAs (5,6).
Numerous studies have demonstrated that miRNAs are involved in
various cellular activities (7–9)
including proliferation, apoptosis, differentiation and movement.
Aberrant expression and function of miRNAs has been found to play
fundamental roles in human malignancies (7,10–12).
In addition, miRNAs have become promising diagnostic and prognostic
biomarkers and attractive therapeutic targets of human cancers
(13,14).
Among numerous miRNAs, microRNA-616 (miR-616) has
been found to be a novel cancer-associated miRNA. miRNA profiling
of human gastric cancer showed that miR-616 was overexpressed in
gastric cancer tissues (15). In
addition, studies of lung (16) and
prostate cancer (17) demonstrated
that the level of miR-616 was significantly elevated in the serum
of patients. Furthermore, miR-616 was found to play an important
role in the development and maintenance of androgen-independent
prostate cancer by regulating the expression of TFPI-2 (17). However, the expression and clinical
significance of miR-616 in HCC tissues, and its functional role and
the underlying mechanisms in HCC cells, are still undefined.
In the present study, we demonstrated that miR-616
was significantly upregulated in HCC tissues. Elevated expression
of miR-616 was observed in patients with metastasis and recurrence.
Additionally, elevated miR-616 expression was associated with
adverse clinicopathological features and poor prognosis of HCC
patients. In vitro studies showed that miR-616 promoted the
invasive behavior and epithelial-mesenchymal transition (EMT) of
HCC cells. Moreover, phosphatase and tensin homolog (PTEN) was
identified to be the direct downstream target of miR-616 in HCC.
miR-616 contributed to the metastasis and EMT of HCC by inhibiting
the expression of PTEN.
Materials and methods
Human tissue specimens and cell
lines
The paired HCC and adjacent non-tumor tissues were
collected from 80 HCC patients who received surgical resection of
primary HCC in the Department of General Surgery at the Second
Affiliated Hospital of Xi'an Jiaotong University from January 2006
to December 2009. No local or systemic treatments had been
performed before the surgical resection. Informed consent was
obtained from each patient before collecting the clinical samples.
The demographic and clinicopathological data are presented in
Table I. All protocols in the
present study were approved by the Xi'an Jiaotong University Ethics
Committee according to the Declaration of Helsinki (as revised in
Tokyo, 2004).
 | Table IThe association between the
clinicopathological features and miR-616 expression in HCC. |
Table I
The association between the
clinicopathological features and miR-616 expression in HCC.
| Clinicopathological
features | Total no. of
pts. | No. of patients
| P-value |
|---|
|
miR-616High |
miR-616Low |
|---|
| Gender |
| Male | 59 | 31 | 28 | 0.446 |
| Female | 21 | 9 | 12 | |
| Age (years) |
| <50 | 34 | 18 | 16 | 0.651 |
| ≥50 | 46 | 22 | 24 | |
| HBV infection |
| Absent | 28 | 15 | 13 | 0.639 |
| Present | 52 | 25 | 27 | |
| Cirrhosis |
| Absent | 33 | 18 | 15 | 0.496 |
| Present | 47 | 22 | 25 | |
| Diameter of tumor
(cm) |
| <5 | 34 | 19 | 15 | 0.366 |
| ≥5 | 46 | 21 | 25 | |
| No. of tumor
nodules |
| 1 | 44 | 24 | 20 | 0.369 |
| >1 | 36 | 16 | 20 | |
| Venous
infiltration |
| Absent | 57 | 24 | 33 |
0.026a |
| Present | 23 | 16 | 7 | |
| Edmondson-Steiner
grading |
| I+II | 60 | 25 | 35 |
0.001a |
| III+IV | 20 | 15 | 5 | |
| TNM stage |
| I+II | 58 | 22 | 36 |
<0.001a |
| III+IV | 22 | 18 | 4 | |
Six HCC cell lines, Hep3B, HepG2, Huh7, MHCC97L
HCCLM3 and MHCC97H, and the human immortalized normal hepatocyte
cell line, LO2 (The Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences, Shanghai, China) were maintained in
complete Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS, HyClone,
Thermo Fisher Scientific, Australia) with 100 U/ml penicillin and
100 µg/ml streptomycin (Sigma, USA) in a humidified incubator
containing of 5% CO2 at 37°C.
Quantitative real-time PCR (qPCR)
analysis
Total RNA from tissues or cells was extracted using
TRIzol (Life Technologies) according to the manufacturer's
instructions. The PCR amplification for the quantification of the
miR-616 was performed using the TaqMan microRNA assay kit (Applied
Biosystems, Foster City, CA, USA). U6 was used as endogenous
controls. The relative expression of miR-616 was normalized to that
of U6.
Cell transfection
miR-616 expression vector (HmiR0186-MR04), the
control vector for miR-616 (CmiR0001-MR04), miR-616 inhibitor
(HmiR-AN0721-AM03) and the negative control for the miR-616
inhibitor (CmiR-AN0001-AM03), were purchased from GeneCopoeia
(Guangzhou, China). Plasmids carrying wt SMAD7 (pCMV5-SMAD7) were
obtained from Addgene (Cambridge, MA, USA). The vectors mentioned
above were transfected into the HCC cells using Lipofectamine 2000
(Invitrogen, USA) following the manufacturer's protocol.
Migration and invasion assays
Cell migration and invasion assays were evaluated in
chambers of 8-µm with Transwell inserts (Millipore, USA) according
to the manufacturer's instructions. Cells (5×104) were
seeded into the top chamber of each insert in serum-free medium and
serum-containing medium was used in the lower chamber as the
attractant. Specifically for the invasion assay, before seeding the
HCC cells, each chamber was coated with mixture of DMEM and
Matrigel (Becton-Dickinson Labware, USA) at a ratio of 6:1. After
incubated at 37°C for 24 h, the cells adherent to the upper surface
of the filter were removed using a cotton swab. The migrated or
invaded cells were fixed and stained with 0.1% crystal violet,
air-dried and photographed. Three independent experiments were
performed.
Western blotting
HCC cells were washed twice with phosphate-buffered
saline (PBS) and lysed on the culture dishes using RIPA lysis
buffer (BioMed, China). Protein concentration was measured using
the BCA kit (Pierce, USA) and 30 µg of each sample were separated
by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to PVDF membrane. Non-specific binding sites were
blocked by incubating with TBST containing 5% skimmed milk/TBST for
2 h at room temperature. The blots were then incubated overnight at
4°C with the following primary antibodies: PTEN (1:1,000), AKT
(1:1,000) (both from Santa Cruz, USA), p-AKT (1:1,000), E-cadherin
(1:1,000), vimentin (1:1,000) and β-actin (1:1500) (all from Cell
Signaling, USA). After washing the membranes with TBST, blots were
incubated with horseradish peroxidase-conjugated goat anti-mouse or
anti-rabbit secondary antibodies (1:10,000; Bio-Rad, USA), and were
detected using the Bio-Rad Gel imaging system.
Luciferase reporter assay
The wild-type 3′-UTR sequence of PTEN (wt
PTEN-3′-UTR) predicted to interact with miR-616 or the mutated
sequence (mt PTEN-3′-UTR) within the predicted target sites was
synthesized and inserted into the pGL3 control vector (Promega,
USA). For the reporter assay, Hep3B cells were seeded into 24-well
plates and were transfected with the above constructs and miR-616
expressing vector, miR-616 inhibitor, control vector or negative
control. After 48 h, the cells were harvested and luciferase
activity was measured using the Dual-Luciferase Reporter Assay
system (Promega) according to the manufacturer's instructions.
Results were obtained from three independent experiments performed
in duplicate.
Immunohistochemical staining
After being deparaffinized in xylene and rehydrated
in a graded alcohol series and distilled water, the paraffin
sections were heated for antigen retrieval in sodium citrate buffer
for 10 min, and were quenched for endogenous peroxidase activity in
3% hydrogen peroxide for 15 min. They were incubated with PTEN
(1:100) or p-AKT (1:50) antibody at 4°C overnight. Then, sections
were incubated with biotinylated secondary antibodies from
Zhongshan Golden Bridge Biotechnology (Beijing, China) for 2 h.
Sections were stained with the avidin-biotin-peroxidase complex
(SABC) method and visualized with diaminobenzidine and
counter-stained with hematoxylin.
The staining results for PTEN and p-AKT were
semi-quantitatively calculated by multiplying the staining
intensity and the percentage of positive staining. Staining
intensity was assessed as four grades: 0, none; 1, weak; 2,
moderate; and 3, strong. The percentage of positive staining was
assessed as the following grades: 0, <5%; 1, 6–25%; 2, 26–50%;
3, 51–75%; and 4, >75%. Ten independent high magnification
(×400) fields were assayed for each section. Two experienced
pathologists independently evaluated the sections, and obtained the
average scores of 10 fields.
Statistical analysis
Results are presented as mean ± SEM. The SPSS
statistical package for Windows version 13 (SPSS, Inc., Chicago,
IL, USA) and GraphPad Prism 5 software (GraphPad Software, Inc.,
USA) were employed to perform the statistical analysis. The
Pearson's Chi-square test, the Spearman's rank correlation
coefficient, the two-tailed Student's t-test, the Kaplan-Meier
plot, the log-rank test or ANOVA was used in appropriate situation.
Difference was considered to indicate a statistically significant
result, at P<0.05.
Results
miR-616 is upregulated in HCC and is
associated with the metastasis and recurrence of HCC
To determine the level of miR-616 expression in HCC
tissues, the quantitative RT-PCR assay was performed in 40 pairs of
HCC and adjacent non-tumor tissues. The expression level of miR-616
in HCC tissues was significantly elevated compared with that in the
adjacent non-tumor liver tissues (P<0.01, Fig. 1A). Moreover, in the recurrent cases,
the level of miR-616 was significantly higher than that in the
non-recurrent cases (P<0.01, Fig.
1B). In addition, patients with metastasis had obviously
increased level of miR-616 than those without metastasis
(P<0.01, Fig. 1C). These results
demonstrated that the expression level of miR-616 is significantly
upregulated in HCC tissues and is associated with the recurrence
and metastasis of HCC.
Then, we examined the expression level of miR-616 in
a non-transformed hepatic cell line (LO2) and six types of HCC cell
lines (Hep3B, HepG2, Huh7, MHCC97L, HCCLM3 and MHCC97H). Compared
with LO2, all HCC cell lines had significantly higher expression
level of miR-616 (P<0.01, Fig.
1D). Moreover, it was interesting to find that miR-616
expression in HCCLM3 and MHCC97H, two highly metastatic HCC cell
lines, was obviously higher than that in the low metastatic HCC
cell lines including Hep3B, HepG2, Huh7 and MHCC97L (P<0.01,
Fig. 1D). These data indicated that
miR-616 can probably potentiate the invasive ability of HCC
cells.
Elevated expression of miR-616 was
correlated with adverse clinicopathological features and poor
prognosis of HCC patients
The obvious elevation of miR-616 in HCC tissues and
its association with HCC recurrence and metastasis prompted us to
examine the clinical significance of miR-616 in HCC. The expression
level of miR-616 was assessed either low (n=40) or high (n=40)
based on the cut-off value which was defined as the median level of
miR-616 in the 80 patients cohort. As presented in Table II, elevated expression level of
miR-616 in HCC patient was associated with the venous infiltration
(P=0.026), high Edmondson-Steiner grading (P=0.001) and advanced
TNM stage (P<0.001). Furthermore, Kaplan-Meier analysis was
performed to evaluate the prognostic value of miR-616 in HCC.
Patients with high-miR-616 expression level had significantly
decreased overall survival (P<0.01, Fig. 2A) and disease-free survival
(P<0.01, Fig. 2B). These data
suggest miR-616 can serve as a prognostic predictor for HCC
patients.
 | Table IIMultivariate Cox regression analysis
of 5-year overall and disease-free survival. |
Table II
Multivariate Cox regression analysis
of 5-year overall and disease-free survival.
| Variables | Overall survival
| Disease-free
survival
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-616 | 1.808 | 1.044–3.134 |
0.035a | 1.724 | 1.014–2.932 |
0.044a |
| Venous
infiltration | 1.941 | 1.012–3.725 |
0.046a | 2.164 | 1.132–4.139 |
0.020a |
| Edmondson-Steiner
grading | 1.717 | 1.018–2.897 |
0.043a | 1.730 | 1.019–2.939 |
0.042a |
| TNM stage | 2.294 | 1.126–4.671 |
0.022a | 2.116 | 1.040–4.303 |
0.038a |
miR-616 promotes the migration, invasion
and EMT of HCC cells
Next, we further explored the functional role of
miR-616 in HCC cells. We transfected Hep3B cells with miR-616
mimics, and miR-616 expression level was significantly increased
after transfection, as suggested by the qRT-PCR (P<0.01,
Fig. 3A). Functionally, as
suggested by the Transwell assay, Hep3B cells overexpressing
miR-616 (Hep3B-miR-616 cells) showed increased ability of migration
(P<0.01, Fig. 3B) and invasion
(P<0.05, Fig. 3B). In contrast,
miR-616 inhibitor significantly downregulated the expression level
of miR-616 in MHCC97H cells (P<0.01, Fig. 3C), and resulted in an obviously
reduced number of migrated (P<0.01, Fig. 3D) and invaded (P<0.01, Fig. 3D) MHCC97H cells. These results
demonstrated that miR-616 can promote the invasive behavior of HCC
cells.
Since the EMT process, which is regarded as a
hallmark of metastasis (18),
increased the migratory and invasive ability of HCC cells (19), we next investigated whether miR-616
could modulate EMT phenotype of HCC cells. Overexpression of
miR-616 in Hep3B cells resulted in decreased level of E-cadherin
(P<0.01, Fig. 3E) and increased
expression of vimentin (P<0.01, Fig.
3E). In addition, inhibition of miR-616 in MHCC97H cells led to
increased level of E-cadherin (P<0.01, Fig. 3E) and decreased expression of
vimentin (P<0.01, Fig. 3E).
These results suggested that miR-616 can promote EMT of HCC
cells.
PTEN is a downstream target of miR-616 in
HCC cells
To clarify the underlying mechanisms by which
miR-616 regulates the invasive behavior and EMT of HCC cells, we
used two publicly available databases (TargetScan 6.2 and miRanda)
to search potential downstream target of miR-616. PTEN, which is a
well-recognized tumor suppressor and an important regulator of HCC
metastasis (20,21), was predicted as a downstream target
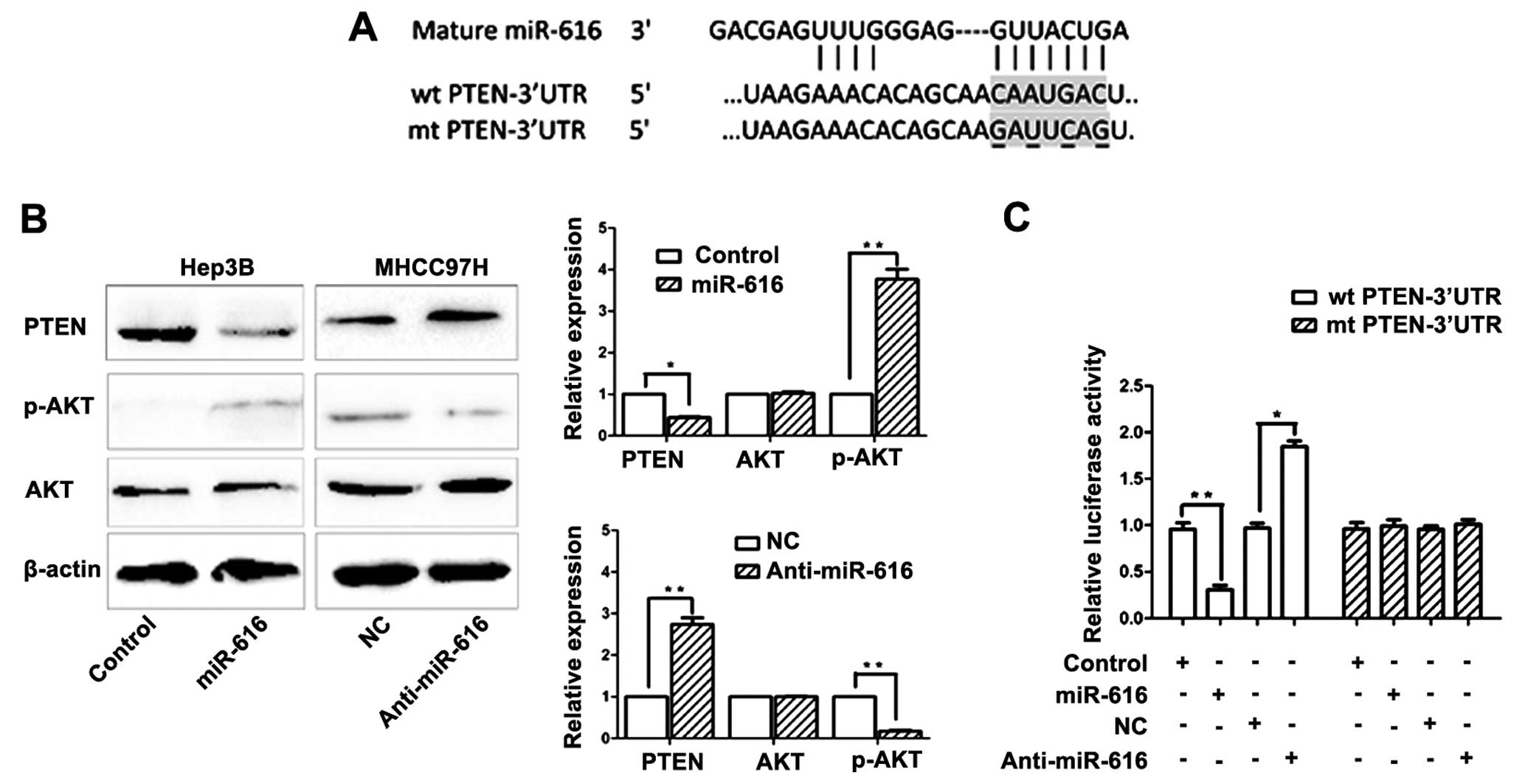
of miR-616. As shown in Fig. 4A,
the 3′-UTR of PTEN mRNA was found to contain the complementary
sequence of miR-616. This suggests that miR-616 can potentially
regulate the expression of PTEN by binding to the 3′-UTR of PTEN.
To confirm this prediction, Hep3B cells transfected with miR-616
expression vector were subjected to western blotting.
Overexpression of miR-616 in Hep3B cells resulted in significantly
reduced expression of PTEN (P<0.05, Fig. 4B). It is well recognized that
inhibiting PTEN expression can lead to increased phosphorylation of
AKT (22). Accordingly, the
phosphorylation of AKT was significantly increased after
overexpressing miR-616 in Hep3B cells (P<0.05, Fig. 4B). Conversely, inhibiting the
expression of miR-616 in MHCC97H cells resulted in significantly
increased expression of PTEN (P<0.01, Fig. 4B) and reduced phosphorylation of AKT
(P<0.01, Fig. 4B). Next, we
performed the dual-luciferase reporter assay to elucidate whether
miR-616 regulated PTEN expression by directly binding to its
3′-UTR. Overexpressing miR-616 significantly inhibited the
luciferase activity of PTEN with a wild-type (wt) 3′-UTR
(P<0.01, Fig. 4C), but had no
influence on the activity of PTEN with a mutant (mt) 3′-UTR.
Additionally, when anti-miR-616 was transfected, the luciferase
activity of wt PTEN 3′-UTR obviously increased (P<0.01, Fig. 4C) while the luciferase activity of
mt PTEN 3′-UTR remained unchanged.
To further validate the regulatory effect of miR-616
on PTEN expression, we compared the expression of PTEN and p-AKT in
HCC tissues with low-miR-616 expression level with that of
high-miR-616 expression. The IHC results showed that the PTEN
expression in high-miR-616 expression group was significantly
decreased when compared with that in the low-miR-616 expression
group (P<0.01, Fig. 5A).
Accordingly, the expression of p-AKT was significantly increased in
high-miR-616 expression group compared to the low-miR-616
expression group (P<0.01, Fig.
5B). In all, these data demonstrated that PTEN is a direct
downstream target of miR-616 and miR-616 can inhibit the expression
of PTEN by binding to its 3′-UTR.
miR-616 promotes EMT and metastatic
ability of HCC cells through inhibiting PTEN
To further clarify the functional significance of
PTEN in miR-616-mediated EMT and metastatic ability of HCC cells,
we transfected pCMV5-PTEN plasmids into Hep3B cells overexpressing
miR-616 (Hep3B-miR-616 cells). The results of western blotting
showed that transfection of pCMV5-PTEN plasmids resulted in
significant increase of PTEN level in Hep3B-miR-616 cells
(P<0.01, Fig. 6A), and led to
upregulation of E-cadherin (P<0.05, Fig. 6A) and reduced expression of vimentin
(P<0.05, Fig. 6A), indicating
that restoring PTEN expression can reverse the EMT phenotype of
Hep3B cells induced by miR-616 overexpression. Furthermore, forced
expression of PTEN partly abrogated the promoting effects of
miR-616 on the migration (P<0.05, Fig. 6B) and invasion (P<0.05, Fig. 6B) of HCC cells. These data indicated
that miR-616 promotes EMT and metastatic ability of HCC cells by
targeting PTEN.
Discussion
The initiation and progression of HCC is a complex
process in which various molecules, and numerous signaling pathways
are involved (23,24). Emerging studies have confirmed that
abnormal expression and function of miRNAs play a critical role in
this process (25,26). miR-616 is a novel cancer-associated
miRNA which has been found to be abnormally expressed in gastric
(15), lung (16) and prostate cancer (17,27).
In the present study, we initially examined the expression status
of miR-616 in clinical specimens of HCC. We confirmed for the first
time that compared with adjacent non-tumor tissues, the expression
level of miR-616 was significantly elevated in HCC tissues. In
addition, patients with recurrence and metastasis had significantly
higher miR-616 levels than those without recurrence and metastasis.
The clinical analysis further demonstrated that increased
expression level of miR-616 was associated with adverse
clinicopathological features including venous infiltration, high
Edmondson-Steiner grading and advanced TNM tumor stage.
Importantly, Kaplan-Meier analysis showed that increased expression
of miR-616 was associated with poorer prognosis of HCC patients
including overall and disease-free survival. Moreover, the
expression level of miR-616 in HCC cell lines, particularly in
those with high metastatic ability (HCCLM3 and MHCC97H), was
significantly higher than that in the non-transformed hepatic cell
line LO2. Taken together, these results indicated that miR-616 is
an important participant in the development and progression of HCC,
and can serve as a prognostic marker of HCC patients.
The obvious elevation of miR-616 expression in HCC
prompted us to examine the functional role of miR-616 in HCC. Since
our results showed that the expression of miR-616 was associated
with the recurrence and metastasis of HCC patients, we speculated
that miR-616 probably influenced the migration and invasion of HCC
cells. Transwell assay showed that over-expression of miR-616
promoted the migration and invasion of HCC cells while
downregulation of miR-616 inhibited the metastatic behavior of HCC
cells. These data suggested that miR-616 can play an active role in
HCC by promoting the migration and invasion of HCC cells. EMT, a
process in which epithelial cells lose cell polarity and
intracellular junctions and acquire mesenchymal features (19,28),
can potentiate the migratory and invasive ability of HCC cells
(19,29). Increasing evidence has demonstrated
that miRNAs, which could regulate the expression of EMT-related
genes, played a critical role in the EMT process (28,30,31).
In the present study, we found that overexpression of miR-616
reduced the expression of E-cadherin, the epithelial marker and
increased the expression of vimentin, the mesenchymal marker.
Downregulation of miR-616 resulted in elevated expression of
E-cadherin and reduced expression of vimentin. These results
suggested that miR-661 promotes the EMT of HCC cells. Altogether,
miR-616 may increase the invasive ability of HCC cells by inducing
EMT.
Phosphatase and tensin homolog (PTEN) is a
well-recognized tumor suppressor which is frequently mutated or
deleted in human cancers (22).
However, instead of the mutation or the deletion of PTEN, abnormal
expression of PTEN was confirmed in HCC (32) and was found to promote the
progression of HCC (20). However,
the molecular mechanisms responsible for the abnormal expression of
PTEN remains unclear. PTEN has been found to be the downstream
target of several miRNAs including miR-21 (20,33),
miR-216a and 217 (21), miR-221 and
222 (34), miR29a (35) and miR-32 (36). In the present study, we confirmed
that PTEN was a direct downstream target of miR-616 in HCC.
Firstly, putative binding sequences of miR-616 were found in the
3′-UTR of PTEN after searching two publicly available databases
(TargetScan and miRanda). Secondly, transfecting miR-616 mimics
into Hep3B cells resulted in decreased expression of PTEN while
downregulation of miR-616 in MHCC97H cells led to increased PTEN
expression. The phosphorylation of Akt, which is inhibited by PTEN
(37), was reduced after miR-616
overexpression and was increased after miR-616 suppression,
supporting that miR-616 regulated the expression of PTEN.
Additionally, the expression of PTEN in HCC tissues with
high-miR-616 level was significantly lower than that in those with
low-miR-616 level. This further validates the regulatory effect of
miR-616 on PTEN expression. Furthermore, altering the expression
level of miR-616 in Hep3B cells significantly influenced the
luciferase activity of wt 3′-UTR of PTEN, while had no obvious
influence on the luciferase activity of mt 3′-UTR of PTEN,
suggesting that miR-616 can regulate the expression of PTEN by
directly interacting with the 3′-UTR of PTEN.
Functionally, PTEN was able to influence the
migration of glioma cells by modulating its downstream PI3K/Akt
pathway (38). Therefore, we
performed rescue experiments in Hep3B-miR-616 cells to clarify
whether PTEN was involved in the promoting effect of miR-616 on the
metastatic ability and EMT phenotype of HCC cells. We found that
restoring the expression of PTEN abrogated the promoting effects of
miR-616 on EMT and metastatic abilities of HCC cells. Therefore,
our results suggest that PTEN is not only a direct target of
miR-616 but also a functional mediator downstream of miR-616 in
HCC. Of interest is that a study of prostate cancer (17) found TFPI-2 was a direct downstream
target of miR-616, and miR-616 induced androgen-independent growth
of prostate cancer cells by suppressing TFPI-2 expression.
Therefore, the downstream targets and the functional significance
of miRNAs in human cancers seem to be cancer-type specific.
In summary, our results demonstrated for the first
time that miR-616 expression is elevated in HCC tissues, and its
increased expression is associated with adverse prognostic features
of HCC patients. Moreover, miR-616 is a valuable biomarker in
predicting the prognosis of HCC patients. In vitro
experiments demonstrated that miR-616 can promote the migration,
invasion and EMT of HCC cells. Mechanistically, we found that PTEN
is a downstream target of miR-616 in HCC and miR-616 exerts its
functional significance on HCC cells by suppressing PTEN. Taken
together, the findings from the present study indicated that
miR-616 can serve as a valuable clinical biomarker, and may
potentially be a promising therapeutic target in HCC.
References
|
1
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127(Suppl 1): S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment, and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014.PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastro-enterology. 132:2557–2576. 2007. View Article : Google Scholar
|
|
4
|
Roberts LR: Sorafenib in liver cancer -
just the beginning. N Engl J Med. 359:420–422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosa A and Brivanlou AH: MicroRNAs in
early vertebrate development. Cell Cycle. 8:3513–3520. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang RM, Yang H, Fang F, Xu JF and Yang
LY: MicroRNA-331-3p promotes proliferation and metastasis of
hepatocellular carcinoma by targeting PH domain and leucine-rich
repeat protein phosphatase. Hepatology. 60:1251–1263. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar
|
|
14
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar
|
|
15
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
16
|
Rani S, Gately K, Crown J, O'Byrne K and
O'Driscoll L: Global analysis of serum microRNAs as potential
biomarkers for lung adenocarcinoma. Cancer Biol Ther. 14:1104–1112.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma S, Chan YP, Kwan PS, Lee TK, Yan M,
Tang KH, Ling MT, Vielkind JR, Guan XY and Chan KW: MicroRNA-616
induces androgen-independent growth of prostate cancer cells by
suppressing expression of tissue factor pathway inhibitor TFPI-2.
Cancer Res. 71:583–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Książkiewicz M, Markiewicz A and Zaczek
AJ: Epithelial-mesenchymal transition: A hallmark in metastasis
formation linking circulating tumor cells and cancer stem cells.
Pathobiology. 79:195–208. 2012. View Article : Google Scholar
|
|
19
|
Reichl P, Haider C, Grubinger M and
Mikulits W: TGF-β in epithelial to mesenchymal transition and
metastasis of liver carcinoma. Curr Pharm Des. 18:4135–4147. 2012.
View Article : Google Scholar
|
|
20
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012.PubMed/NCBI
|
|
23
|
Roberts LR and Gores GJ: Hepatocellular
carcinoma: Molecular pathways and new therapeutic targets. Semin
Liver Dis. 25:212–225. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of microRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar
|
|
27
|
Haldrup C, Kosaka N, Ochiya T, Borre M,
Høyer S, Orntoft TF and Sorensen KD: Profiling of circulating
microRNAs for prostate cancer biomarker discovery. Drug Deliv
Transl Res. 4:19–30. 2014. View Article : Google Scholar
|
|
28
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi SS and Diehl AM:
Epithelial-to-mesenchymal transitions in the liver. Hepatology.
50:2007–2013. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bullock MD, Sayan AE, Packham GK and
Mirnezami AH: MicroRNAs: Critical regulators of epithelial to
mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in
cancer progression. Biol Cell. 104:3–12. 2012. View Article : Google Scholar
|
|
31
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan XW, Jiang M, Cao HF, He YQ, Liu SQ,
Qiu XH, Wu MC and Wang HY: The alteration of PTEN tumor suppressor
expression and its association with the histopathological features
of human primary hepatocellular carcinoma. J Cancer Res Clin Oncol.
129:100–106. 2003.PubMed/NCBI
|
|
33
|
Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G,
Zeng Y, Sun B, Qian H, Chen L, et al: MicroRNA-21 suppresses PTEN
and hSulf-1 expression and promotes hepatocellular carcinoma
progression through AKT/ERK pathways. Cancer Lett. 337:226–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Garofalo M, Di Leva G, Romano G, Nuovo G,
Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P,
et al: miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kong G, Zhang J, Zhang S, Shan C, Ye L and
Zhang X: Upregulated microRNA-29a by hepatitis B virus X protein
enhances hepatoma cell migration by targeting PTEN in cell culture
model. PLoS One. 6:e195182011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hers I, Vincent EE and Tavaré JM: Akt
signalling in health and disease. Cell Signal. 23:1515–1527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dasari VR, Kaur K, Velpula KK, Gujrati M,
Fassett D, Klopfenstein JD, Dinh DH and Rao JS: Upregulation of
PTEN in glioma cells by cord blood mesenchymal stem cells inhibits
migration via downregulation of the PI3K/Akt pathway. PLoS One.
5:e103502010. View Article : Google Scholar : PubMed/NCBI
|