Introduction
Nasopharyngeal carcinoma (NPC) is the most common
type of head and neck cancer with significant regional differences
(1). The annual incidence of NPC in
regions of Southeast Asian is much higher than that of other
regions worldwide (2,3). Genetic factors, EB virus infection,
environmental factors and diet are all possible related etiologies
of NPC, while the molecular characteristics and biological
behaviors of NPC remain elusive (4–7).
Radiation therapy, surgical treatment and chemotherapy are all
conventional treatment options for NPC patients and the majority of
early stage NPC patients achieve long-term survival with
standardized radiation therapy (8,9).
However, more than 15% of NPC patients suffer from recurrence due
to radioresistance while the exact mechanism behind this remains
elusive (8,10).
Histone modification, a post-translational
modification process including methylation, phosphorylation,
acetylation, ubiquitination and ADP-ribosylation, can alter the
chromatin state or alter the binding affinity of transcription
factors to DNA and thus affect gene expression (11,12).
Both arginine and lysine residues can be methylated. Methylated
lysines are well known marks of the histone code, and the
methylation status of specific lysines is able to determine gene
expression states (13).
Methylation of lysines H3K4 and H3K36 can transcriptionally
activate gene expression while methylation of H3-K9, H3-K27 and
H4-K20 always inhibits gene transcription (14–16).
In regards to arginine methylation, two categories of protein
N-arginine methyltransferases (PRMTs) have been identified; protein
arginine methyltransferase 5 (PRMT5) is responsible for the mono-
and symmetric-dimethylation of arginine and all other PRMTs (PRMT1,
PRMT3, RMT1/HMT and PRMT4/CAMR1) mainly account for the asymmetric
dimethylation of arginine (17,18).
Recently, the role of PRMT5 in controlling cell proliferation and
signaling transduction has been identified. Knockout of PRMT5 in
mice was found to induce early embryonic lethality (19). Furthermore, PRMT5 regulates cell
apoptosis by affecting the activity of p53, a well-established
tumor suppressor involved in multiple biological processes
(20). In human lung
adenocarcinoma, PRMT5 was found to be correlated with vessel
invasion and regulated epithelial-mesenchymal transition of lung
cancer cells (21). Silencing of
PRMT5 in colorectal cancer cells was able to decrease the arginine
methylation of eIF4E and fibroblast growth factor receptor 3
(FGFR3) and thus inhibit colorectal cancer growth (22). Although PRMT5 has been implicated in
various human tumors, the expression level of PRMT5 in human NPC
cell lines and tissue samples has not been previously reprted. In
the present study, we aimed to explore the clinical significance of
PRMT5 in NPC tissues and the biological role, particularly
radioresistance-related mechanisms of PRMT5 in NPC cells.
Materials and methods
Patients and follow-up
All specimens were obtained following the informed
consent of patients prior to surgery. All human and animal studies
were approved by the Ethics Committee of The First Affiliated
Hospital of Zhengzhou University, Zhengzhou University School of
Medicine, and were therefore performed in accordance with the
ethical standards laid down in the 1964 Declaration of Helsinki and
its later amendments. One hundred and twelve NPC patients were
enrolled in the present study, between December 2008 and February
2010. Follow-up data were summarized in 2015, and the patients were
evaluated every 2–3 months during the first 2 years and every 3–6
months thereafter. The overall survival (OS) was calculated from
the date of resection to the date of death or last follow-up.
One hundred and twelve NPCs of different stages were
used to construct a tissue microarray. The tissue microarray was
stained for PRMT5 (Abcam, USA) expression. Scoring was performed by
two researchers independently. The number of stained cells was
counted and scored as 0 (no staining), 1 (1–25%), 2 (26–50%) and 3
(>50%) accordingly. Staining intensity was scored as 0 (no
staining), 1 (weak staining), 2 (moderate staining) and 3 (strong
staining). The final score was obtained by multiplying the score
for the number of stained cells with the staining intensity score
providing a final range of 0–9. Specimens scoring 0–1 were
considered as having low expression, 2 and 3 represent moderate
expression and scoring of 4–9 was regarded as high expression.
Cell culture and lentivirus
production
Human NPC cell lines TW06, CNE1, 5-8F and CNE2, and
the immortalized nasopharyngeal epithelial cell line NP69 were
obtained from the Cell Bank of the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China) and conserved in our
laboratory. All cells were cultured in Dulbecco's modified Eagle's
medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco,
Australia) at 37°C with 5% CO2. Lentiviruses expressing
shPRMT5, shFGFR3 or shNC were purchased from GeneChem (Shanghai,
China). The effective short hairpin RNA (shRNA) targeted against
the sequence in the coding region of the human PRMT5 gene was:
5′-GGATAA AGCTGTATGCTGT-3′. 5-8F and CNE2 cells (1×105)
were plated in 6-well plates and transfected with the lentiviruses
according to the manufacturer's protocol. Stable cell lines were
obtained by continuous selection by puromycin. The sequences of the
shRNAs are presented in Table
I.
 | Table IList of oligonucleotides used in the
present study. |
Table I
List of oligonucleotides used in the
present study.
| Name | Sequences
(5′–3′) | Procedure |
|---|
| PRMT5-Forward |
5′-GATGGCGGCGATGGCA-3′ | RT-PCR |
| PRMT5-Reverse |
5′-CTGTGTGTGTAGTCGG-3′ | RT-PCR |
| FGFR3-Forward |
5′-GCAGGAGCAGTTGGTCTTC-3′ | RT-PCR |
| FGFR3-Reverse |
5′-TCCTCGTGGGAGGCATTCAG-3′ | RT-PCR |
| GAPDH-Forward |
5′-AGCCACATCGCTCAGACAC-3′ | RT-PCR |
| GAPDH-Reverse |
5′-GCCCAATACGACCAAATCC-3′ | RT-PCR |
| shPRMT5-Upper |
5′-CCGGGGATAAAGCTGTATGCTGTCTCGAGACAGCATACAGCTTTATCCTTTTTG-3′ | shRNA cloning |
| shPRMT5-Lower |
5′-AATTCAAAAAGGATAAAGCTGTATGCTGTCTCGAGACAGCATACAGCTTTATCC-3′ | shRNA cloning |
| shFGFR3-Upper |
5′-GATCCCCAATGCCTCCCACGAGGACTCCTTCAAGAGAGGAGTCCTCGTGGGAGGCATTTTTTTGGAA-3′ | shRNA cloning |
| shFGFR3-Lower |
5′-AGCTTTTCCAAAAAAATGCCTCCCACGAGGACTCCTCTCTTGAAGGAGTCCTCGTGGGAGGCATTGGG-3′ | shRNA cloning |
Quantitative RT-PCR
Total RNA was extracted by TRIzol reagent
(Invitrogen). Reverse transcription was performed using the
ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan) according to the
manufacturer's protocol. In regards to the real-time PCR analysis,
aliquots of double-stranded cDNA were amplified using
SYBR®-Green Real-Time PCR Master Mix (Toyobo). The
relative expression levels (fold-change) of the target genes were
determined by the 2−ΔΔCt (ΔCt = Cttarget -
CtGAPDH; ΔΔCt = ΔCtexpressing vector -
ΔCtcontrol vector) method. The primer sequences are
listed in Table I.
Transient transfection
In regards to the transient transfection, cells were
plated into 6-well plates at a density of 3×105
cells/well and transfected with pcDNA3.1-FGFR3 or control plasmids
and Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer's instructions. Quantitative RT-PCR and
immunoblotting were used to ensure effective transfection
efficiency. All functional studies were performed after
transfection for 48 h.
Cell proliferation assay
6-MV X-rays from a linear accelerator
(Varian-2300EX; Varian, Palo Alto, CA, USA) were used at a dose
rate of 3 Gy/min. Cell Counting Kit-8 (CCK-8) (Dojindo) was
employed for cell proliferation assay according to the
manufacturer's instructions. Cells were plated in 96-well cell
plates at a density of 1,500 cells/well and incubated for 12 h.
Then, cells of each group were treated with the indicated doses of
radiation according to the aim of the experiment. Optical density
(OD) values at 450 nm were recorded by a microplate reader (Epoch;
BioTek), and each experiment was performed in triplicate and then
averaged.
Colony formation assay
The plate colony formation assay was performed
according to previous studies (23,24).
Logarithmic growth phase cells in each group were incubated in
6-well plates at a density of 1,000 cells/well for 12 h.
Subsequently the cells were exposed to ionizing radiation (IR) with
a dose of 6 Gy and incubated for another 14 days. The surviving
colonies (colonies containing >50 cells) were counted by
microscopic inspection in three independent experiments.
Immunoblotting assay
For the immunoblotting analysis, NPC tissues/cells
were digested in RIPA (Solarbio, China) buffer in the presence of
protease inhibitor cocktail (Sigma, USA). Protein concentration was
detected by the Bradford assay according to the manufacturer's
protocol. Then, total protein was transferred to PVDF membranes
(Millipore, Billerica, MA, USA). The membranes were blocked in 5%
milk for 2 h at room temperature and incubated with the antibody
overnight at 4°C. The primary antibodies used were rabbit
anti-PRMT5, rabbit anti-FGFR3 (both from Abcam) and rabbit
anti-GAPDH, and the secondary antibodies were horseradish
peroxidase (HRP)-conjugated secondary anti-rabbit antibody (both
from Cell Signaling Technology, Danvers, MA, USA). Pierce ECL Plus
Substrate (Thermo Scientific, USA) was employed for the western
blotting visualization.
Statistical analysis
The software Microsoft Excel and SPSS 18.0 (SPSS
Inc., Chicago, IL, USA) were used for statistical analysis. t-test
and one-way ANOVA analysis were used for analyzing the difference
in measured data while the appropriate. χ2 test was used
for evaluating the difference in categorical data. Survival curves
(Kaplan-Meier) and log-rank test were used to assess the survival
status of the different groups.
Results
Elevated PRMT5 expression in NPC patients
is correlated with a poor prognosis
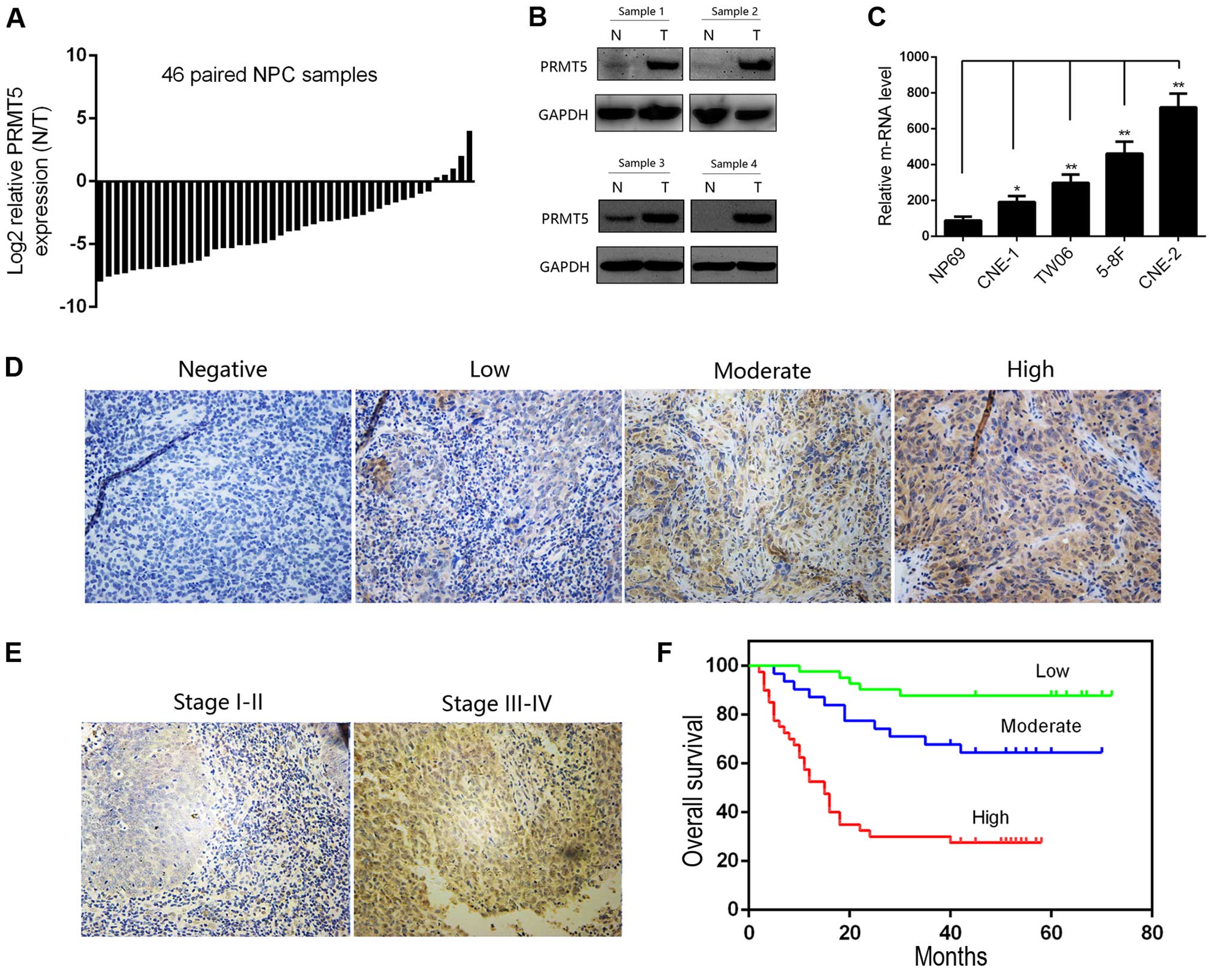
We first evaluated the mRNA level of PRMT5 in 46
paired NPC and corresponding normal tissues by quantitative RT-PCR.
The expression level of PRMT5 in the NPC tissues was significantly
higher than that in the adjacent non-tumor tissues, which indicated
the potential oncogenic role of PRMT5 in NPC patients (Fig. 1A). Furthermore, we confirmed this
difference by immunoblotting analysis of 4 pairs of NPC cases
(Fig. 1B). We then investigated the
expression of PRMT5 in the NPC cell lines at the RNA level.
Notably, PRMT5 was also upregulated in a panel of cancer cell lines
compared with its expression in the NP69 cells, an immortalized
nasopharyngeal epithelial cell line (Fig. 1C). We next evaluated the clinical
role of PRMT5 in NPC. A tissue microarray containing 112 cases was
conducted for evaluating the relationship between the PRMT5 level
and clinicopathological parameters of NPC patients. Representative
images are presented in Fig. 1D and
E. By scoring the results of our tissue microarray, we found
that the nuclear but not the cytoplasmic PRMT5 level was correlated
with a poorer tumor stage and lymph nodal status, and the
relationship between PRMT5 expression and the clinicopathological
parameters of the NPC patients is summarized in Table II. The 5-year OS (Fig. 1F) indicated that a high nuclear
PRMT5 expression level in the NPC patients predicted poorer
outcomes when compared to the outcome of patients with low
expression. All of these results revealed the clinical significance
of PRMT5 in NPC patients.
 | Table IIAssociation between expression of
PRMT5 and clinicopathological features of the NPC cases
(n=112). |
Table II
Association between expression of
PRMT5 and clinicopathological features of the NPC cases
(n=112).
| Parameters | n | PRMT5 expression (%)
| P-value |
|---|
| Low | Moderate | High |
|---|
| Age (years) | | | | | |
| ≥50 | 55 | 19 | 14 | 22 | 0.4398 |
| <50 | 57 | 22 | 17 | 18 | |
| Gender | | | | | |
| Male | 77 | 28 | 22 | 27 | 0.9405 |
| Female | 35 | 13 | 9 | 13 | |
| Histological
type | | | | | |
| DNKC | 17 | 7 | 6 | 4 | 0.3799 |
| UDC | 95 | 34 | 25 | 36 | |
| Clinical stage | | | | | |
| I–II | 54 | 14 | 13 | 27 | 0.0214 |
| III–IV | 58 | 27 | 18 | 13 | |
| T
classification | | | | | |
| T1–T2 | 81 | 29 | 21 | 31 | 0.5008 |
| T3–T4 | 31 | 12 | 10 | 9 | |
| N
classification | | | | | |
| N0–N1 | 56 | 28 | 14 | 14 | 0.0028 |
| N2–N3 | 56 | 13 | 17 | 26 | |
| Distant
metastasis | | | | | |
| Yes | 9 | 2 | 2 | 5 | 0.2212 |
| No | 103 | 39 | 29 | 35 | |
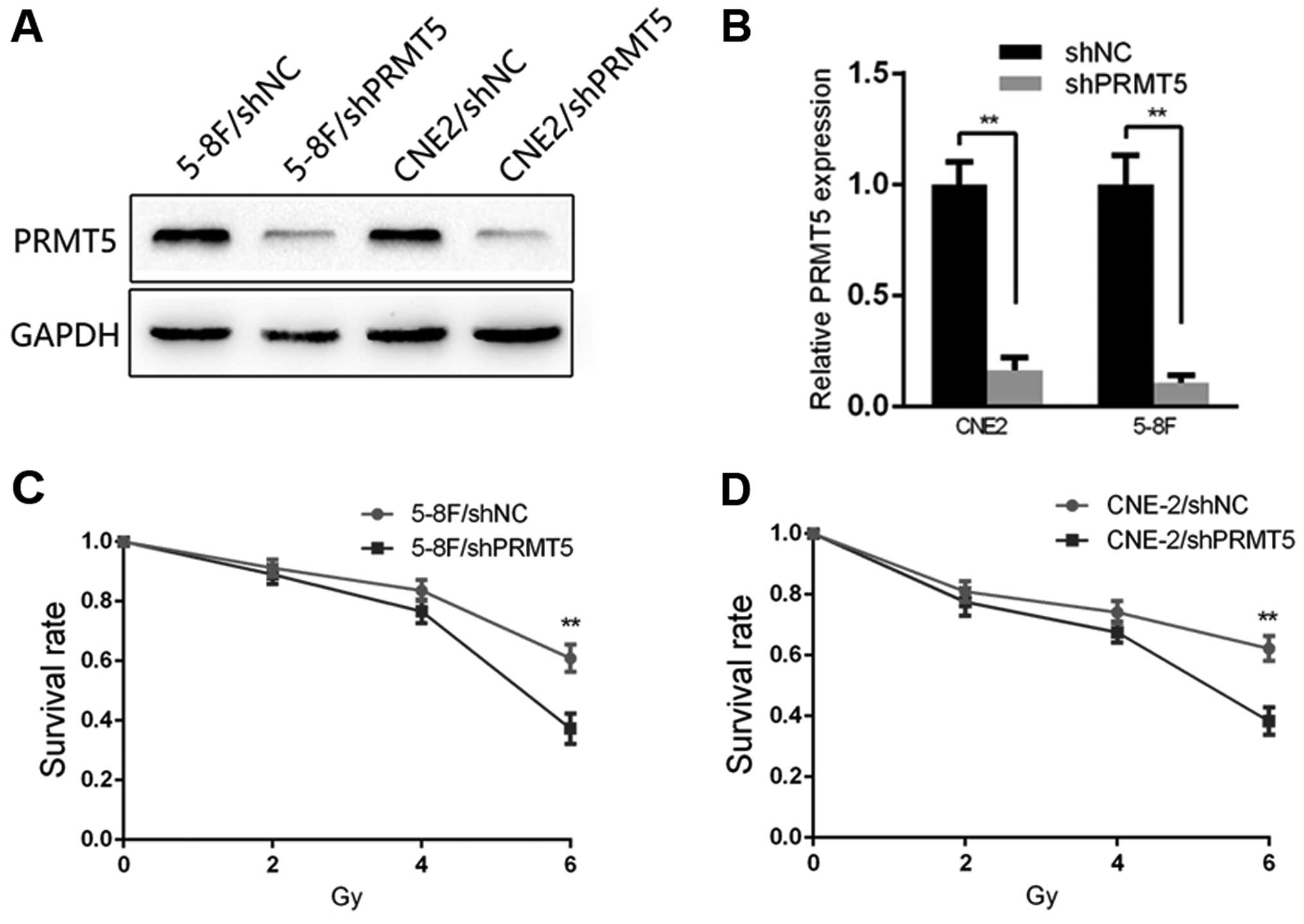
Silencing of PRMT5 in NPC cell lines
increases radiosensitivity
To gain a more comprehensive understanding of the
effect of PRMT5 on radiosensitivity, we generated 5-8F and CNE2
cell lines expressing shPRMT5 (5-8F/shPRMT5 and CNE2/shPRMT5) and
the corresponding negative control cells (5-8F/shNC and CNE2/shNC).
The transfection efficiency of 5-8F and CNE2 was confirmed by
qRT-PCR and immunoblotting, respectively. The result of qRT-PCR
showed a significant interference effect at the RNA level in both
cell lines, and the result of immunoblotting showed an effective
interference effect at the protein level (Fig. 2A and B). Based on the clinical role
and differential expression of PRMT5 in the NPC cells, we explored
the potential significance of PRMT5 in NPC radioresistance. The
results of the CCK-8 assay revealed that following 2, 4 and 6 Gy
irradiation stimulation, the survival rate of the 5-8F/shPRMT5
cells was decreased compared with that of the 5-8F/shNC cells
(Fig. 2C). A similar result was
observed in the CNE2 cells (Fig.
2D).
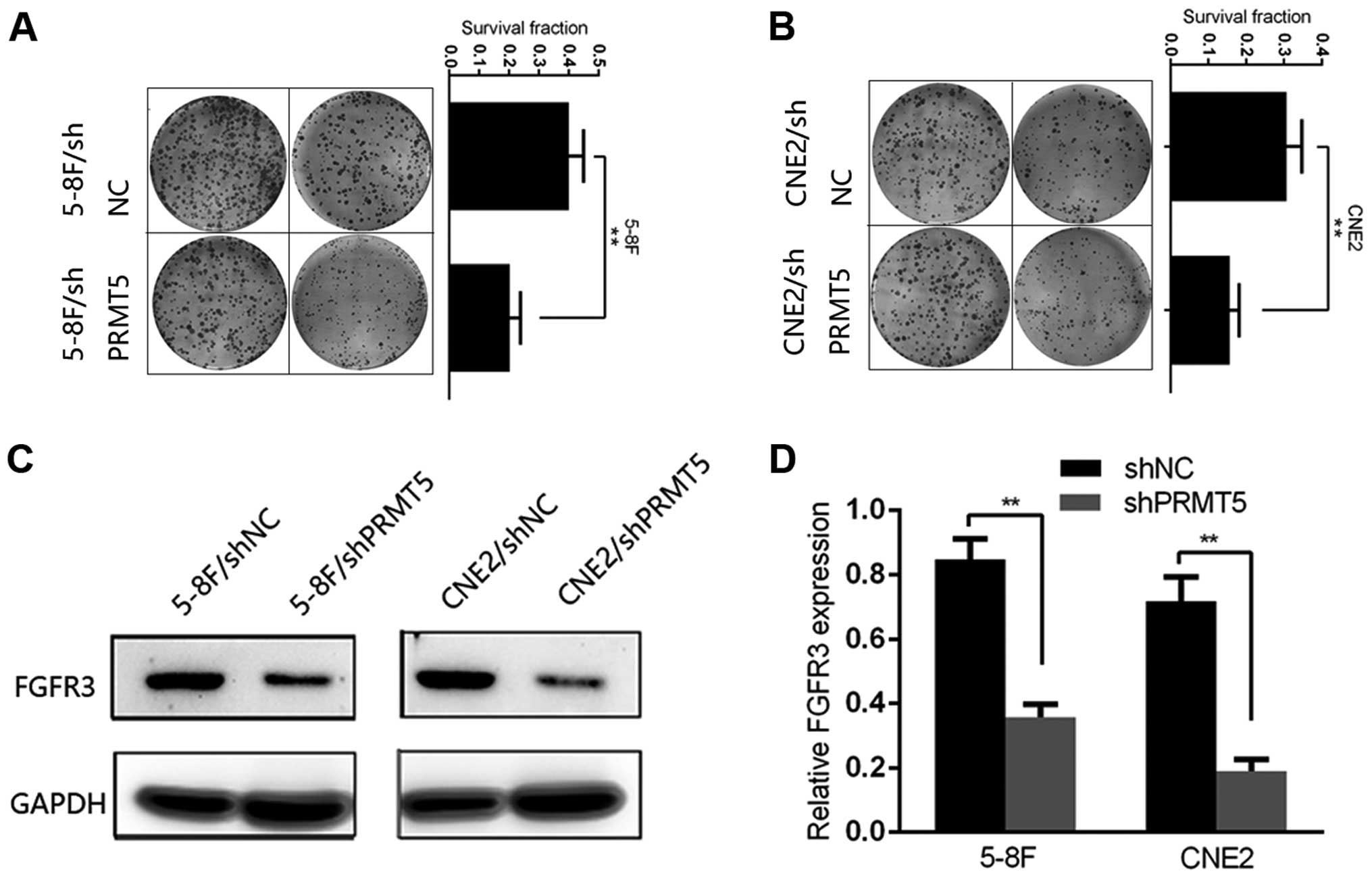
FGFR3 is a major downstream effector of
PRMT5 in NPC cell lines
We next conducted plate colony formation assay to
investigate the colony formation ability of each group. 5-8F and
CNE2 cells of each group were plated in a 6-well plate and
incubated for 14 days after irradiation exposure at a dose of 6 Gy.
Cells transfected with shNC showed a significant colony formation
advantage over the shPRMT5 group and the survival fraction was
decreased in the 5-8F/shPRMT5 and CNE2/shPRMT5 cells compared with
that noted in the 5-8F/shNC and CNE2/shNC cells, respectively
(Fig. 3A and B). Then, we explored
the potential mechanisms of PRMT5-induced radioresistance. FGFR3
has been identified as a major downstream effector through which
PRMT5 regulates tumor progression in lung and colorectal cancer. In
the present study, we tentatively examined the protein level of
FGFR3 in the shPRMT5 and shNC cells. Notably, we found that the
protein level of FGFR3 was significantly decreased after silencing
of PRMT5 in the 5-8F and CNE2 cells (Fig. 3C). To better understand the
relationship between PRMT5 and FGFR3 in the NPC cells, we detected
the mRNA levels and luciferase activity of FGFR3 in the two group
of 5-8F and CNE2 cells. As a result, the change in the mRNA level
of FGFR3 was in accordance with the protein level in both cell
lines (Fig. 3D).
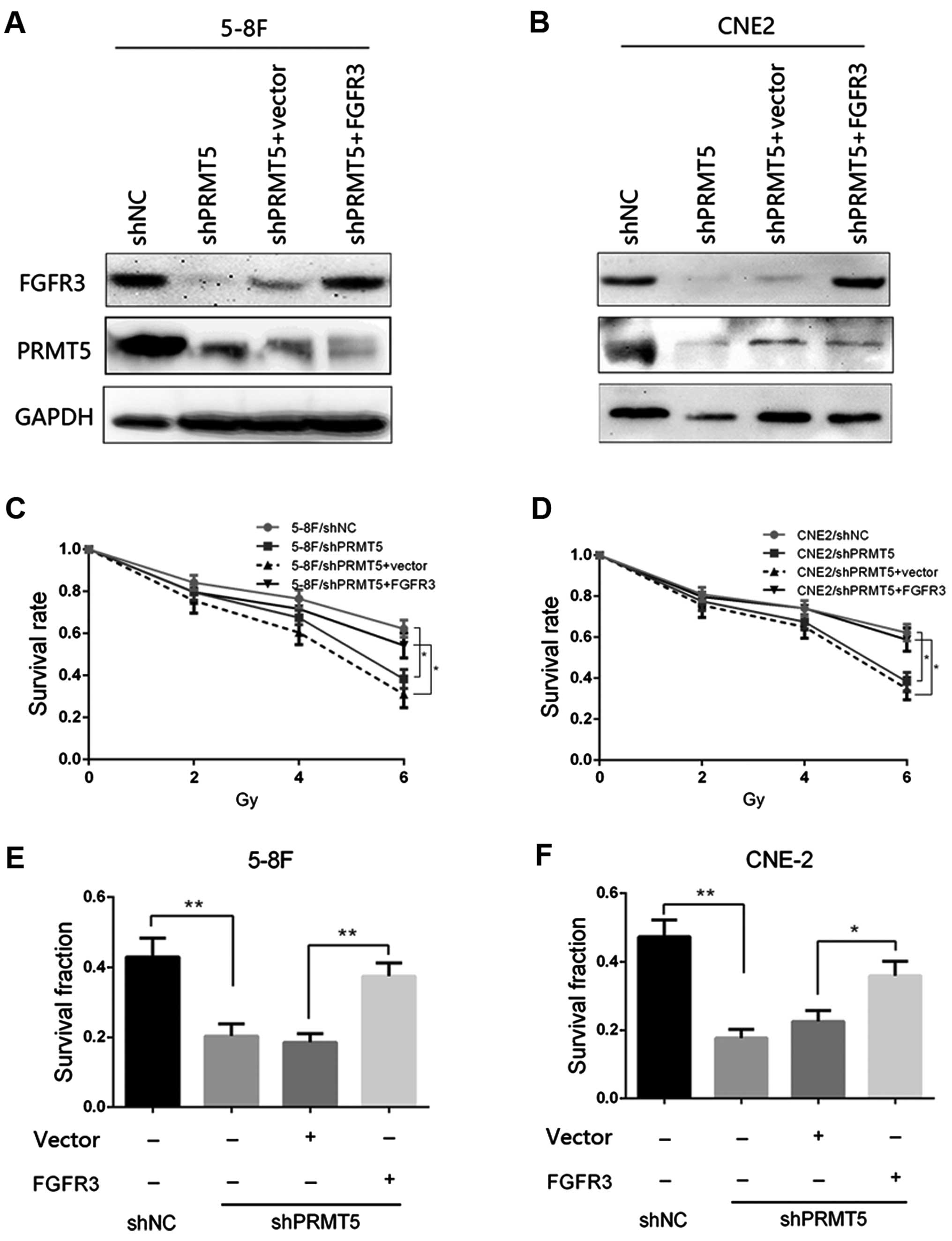
Overexpression of FGFR3 restores PRMT5
silenced-induced radiosensitivity
Given that FGFR3 is a downstream target of PRMT5 in
NPC cells, we then investigated whether FGFR3 restoration in the
5-8F/shPRMT5 and CNE2/shPRMT5 cells could reverse the phenotypes
induced by PRMT5 silencing. The FGFR3-overexpressing vector
pcDNA3.1-FGFR3 and corresponding control plasmids were transfected
into the 5-8F/shPRMT5 and CNE2/shPRMT5 cells, respectively. The
transfection effect was confirmed by immunoblotting and the results
are presented in Fig. 4A and B. We
found that re-expression of FGFR3 in the 5-8F/shPRMT5 and
CNE2/shPRMT5 cells increased the survival rate of both cell lines
to similar levels as observed in the 5-8F/shNC and CNE2/shNC cells
as determined by CCK-8 analysis (Fig.
4C and D). Furthermore, FGFR3 restoration in the 5-8F/shPRMT5
and CNE2/shPRMT5 cells increased the survival fraction by colony
formation analysis compared with that of the 5-8F/shPRMT5 and
CNE2/shPRMT5 cells transfected with the vector plasmids (Fig. 4E and F). Taken together, these
results suggest that FGFR3 expression could restore the
radiosensitivity induced by PRMT5 silencing in the 5-8F and CNE2
cells.
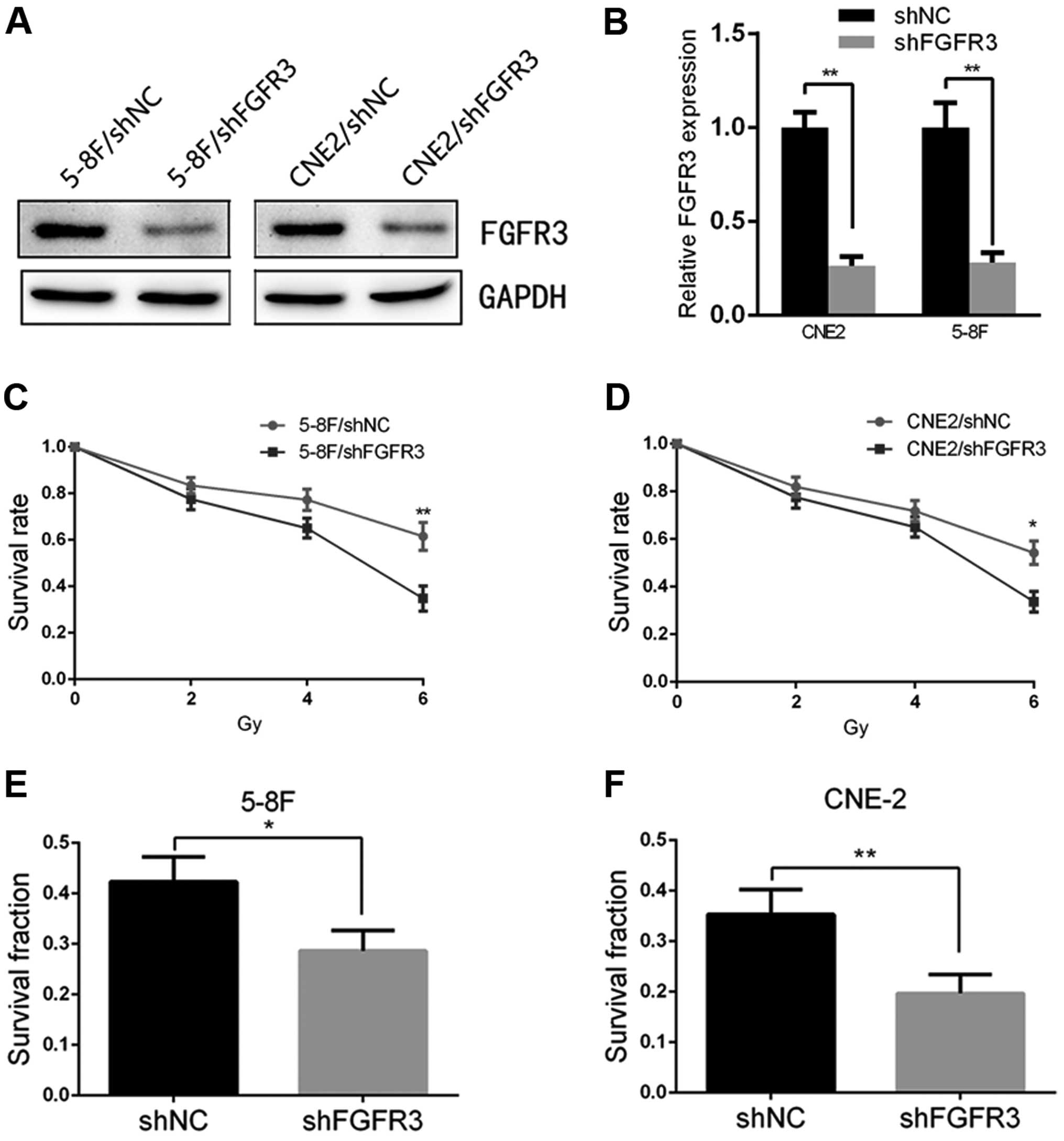
Silencing of FGFR3 in the NPC cell lines
increases radiosensitivity
Finally, to demonstrate the potential of the
FGFR3-mediated regulation of cell radiosensitivity, we transfected
shRNAs targeting FGFR3 and a negative control into 5-8F and CNE2
cells, respectively. The results of the qRT-PCR and immunoblotting
showed that the 5-8F and CNE2 cells transfected with shFGFR3
yielded a significant downregulation of FGFR3 compared with the
5-8F/shNC and CNE2/shNC cells (Fig. 5A
and B). We observed that the silencing of FGFR3 in the 5-8F and
CNE2 cells significantly decreased the survival rate to similar
levels as observed in the 5-8F/shPRMT5 and CNE2/shPRMT5 cells as
determined by CCK-8 analysis (Fig. 5C
and D). In addition, inhibition of FGFR3 also reduced the
survival fraction of both cell lines by colony formation assay
(Fig. 5E and F). These results were
similar to the effects of silencing PRMT5 in NPC cells.
Discussion
Radioresistance is a major cause for the poor
prognosis of NPC patients (25).
Recently, biological and clinical studies have focused on the
molecular characteristics and biological behaviors of NPC,
particularly on the mechanisms of radioresistance: i) genetic
changes including chromosome alteration and gene mutation (4,26); ii)
ectopic expression of protein kinases, epigenetic-related enzymes
and DNA repair proteins (27,28);
iii) overactivation of signaling pathways including ERK, PI3K/AKT,
Wnt and NF-kB (23,29–32).
All of these changes may finally induce proliferation, apoptosis,
angiogenesis and autophagy. Although many efforts have been made to
investigate the mechanisms of radioresistance in previous studies,
there is still an urgent need for screening novel molecular targets
attibuted to the radioresistance of NPC. Recently, a relationship
between the aggressive behaviors of NPC and the nucleosome
remodeling complex SWI/SNF (SWItch/sucrose non-fermentable) has
attacted much attention. The overexpression of Rsf-1, a chromatin
remodeling protein, was correlated with poor therapeutic response
and adverse outcomes of NPC patients (33). Furthermore, a mutational landscape
of 128 cases conducted by Lin et al provided a comprehensive
understanding of the genetic changes of NPC (26). These studies revealed a preliminary
relationship between the remodeling complex SWI/SNF and the NPC
malignant phenotype, which warrants further research.
In the present study, we found that PRMT5, a member
of the SWI/SNF complex, was overexpressed in NPC tissues and cell
lines compared with its levels in adjacent non-tumor tissues and
the immortalized nasopharyngeal epithelial NP69 cell line, which
indicated the potential oncogenic role of PRMT5 in NPC patients.
Next, we conducted a tissue microarray containing 112 NPC cases to
evaluate the clinical significance of PRMT5 in NPC tissues. By
scoring the staining result and Kaplan-Meier survival analysis, we
found that high expression of PRMT5 in NPC tissues predicted a
poorer survival rate of the NPC patients. Furthermore, a higher
level of PRMT5 was correlated with a more advanced tumor stage and
adverse lymph nodal status of NPC patients. Given the clinical
significance of PRMT5 in NPC tissues, we next explored the
biological role of PRMT5 in NPC cells and the potential mechanisms.
We generated stable cell lines transfected with shPRMT5 and shNC to
compare the radiosensitivity of the two cell groups in the 5-8F and
CNE2 cells. The results of the CCK-8 assay showed that the
silencing of PRMT5 in the 5-8F and CNE2 cells could increase the
radiosensitivity and the result was confirmed by a colony formation
analysis. These results indicated that PRMT5-mediated
radioresistance may be responsible for the poor prognosis of some
NPC patients.
We next explored the mechanisms by which PRMT5
regulates the radiosensitivity of NPC cells. FGFR3 was identified
as a target of PRMT5 in lung and colorectal cancer (22,34).
In the present study, the protein level of FGFR3 was decreased
after silencing of PRMT5 in the 5-8F and CNE2 cells, which was
verified at the mRNA level and by luciferase activity change. The
chromatin immunoprecipitation (ChIP) results of previous studies
revealed that FGFR3 and eIF4E are two critical downstream genes
regulated by PRMT5. Our present study also identified that PRMT5
could transcriptionally activate FGFR3 expression. Furthermore,
silencing of FGFR3 induced similar phenotypes as PRMT5 inhibition,
and re-expression of FGFR3 in the 5-8F/shPRMT5 and CNE2/shPRMT5
cells increased the radiosensitivity of cells as determined by
CCK-8 and colony formation assays. Taken together, PRMT5 regulates
the apoptosis-dependent radiosensitivity of NPC cells via targeting
FGFR3.
In conclusion, the present study demonstrated the
clinical significance of PRMT5 expression in NPC tissues. PRMT5
plays an important role in the enhancement of radiosensitivity in
NPC cells, and FGFR3 is a downstream effector of PRMT5-induced
radioresistance. The PRMT5-FGFR3 axis may be a potential
therapeutic target for NPC patients.
Acknowledgments
The present study was supported by the Second Batch
of Science and Technology Projects of Henan Province
(142102310055).
References
|
1
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo P, Huang ZL, Yu P and Li K: Trends in
cancer mortality in China: An update. Ann Oncol. 23:2755–2762.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lo KW, Chung GT and To KF: Acquired
genetic and epigenetic alterations in nasopharyngeal carcinoma.
Nasopharyngeal Carcinoma. Busson P: Springer; New York: pp. 61–81.
2013, View Article : Google Scholar
|
|
5
|
Hildesheim A and Wang CP: Genetic
predisposition factors and nasopharyngeal carcinoma risk: A review
of epidemiological association studies, 2000–2011: Rosetta Stone
for NPC: Genetics, viral infection, and other environmental
factors. Semin Cancer Biol. 22:107–116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
West RB: Fingerprints of Epstein-Barr
virus in nasopharyngeal carcinoma. Nat Genet. 46:809–810. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polesel J, Serraino D, Negri E, Barzan L,
Vaccher E, Montella M, Zucchetto A, Garavello W, Franceschi S, La
Vecchia C, et al: Consumption of fruit, vegetables, and other food
groups and the risk of nasopharyngeal carcinoma. Cancer Causes
Control. 24:1157–1165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshizaki T, Ito M, Murono S, Wakisaka N,
Kondo S and Endo K: Current understanding and management of
nasopharyngeal carcinoma. Auris Nasus Larynx. 39:137–144. 2012.
View Article : Google Scholar
|
|
9
|
Bensouda Y, Kaikani W, Ahbeddou N, Rahhali
R, Jabri M, Mrabti H, Boussen H and Errihani H: Treatment for
metastatic nasopharyngeal carcinoma. Eur Ann Otorhinolaryngol Head
Neck Dis. 128:79–85. 2011. View Article : Google Scholar
|
|
10
|
Yu F and Loh KS: Cancer stem cells in
nasopharyngeal carcinoma: Current evidence. JNPC. 1:e62014.
View Article : Google Scholar
|
|
11
|
Zentner GE and Henikoff S: Regulation of
nucleosome dynamics by histone modifications. Nat Struct Mol Biol.
20:259–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Waldmann T and Schneider R: Targeting
histone modifications - epigenetics in cancer. Curr Opin Cell Biol.
25:184–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chung S, Sundar I, Rahman I and Yao H:
Cigarette smoke causes a residue-specific histone methylation and
its cross-talk with histone acetylation in human lung epithelial
cells. Am J Respir Crit Care Med. 185:A34742012.
|
|
15
|
Eram MS, Bustos SP, Lima-Fernandes E,
Siarheyeva A, Senisterra G, Hajian T, Chau I, Duan S, Wu H,
Dombrovski L, et al: Trimethylation of histone H3 lysine 36 by
human methyltransferase PRDM9 protein. J Biol Chem.
289:12177–12188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang H, Fang H, Yin E, Brasier AR, Sowers
LC and Zhang K: Multiplexed parallel reaction monitoring targeting
histone modifications on the QExactive mass spectrometer. Anal
Chem. 86:5526–5534. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Di Lorenzo A and Bedford MT: Histone
arginine methylation. FEBS Lett. 585:2024–2031. 2011. View Article : Google Scholar
|
|
18
|
Berrens RV and Reik W: Prmt5: A guardian
of the germline protects future generations. EMBO J. 34:689–690.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim S, Günesdogan U, Zylicz JJ, Hackett
JA, Cougot D, Bao S, Lee C, Dietmann S, Allen GE, Sengupta R, et
al: PRMT5 protects genomic integrity during global DNA
demethylation in primordial germ cells and preimplantation embryos.
Mol Cell. 56:564–579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chu Z, Niu B, Zhu H, He X, Bai C, Li G and
Hua J: PRMT5 enhances generation of induced pluripotent stem cells
from dairy goat embryonic fibroblasts via down-regulation of p53.
Cell Prolif. 48:29–38. 2015. View Article : Google Scholar
|
|
21
|
Ibrahim R, Matsubara D, Osman W, Morikawa
T, Goto A, Morita S, Ishikawa S, Aburatani H, Takai D, Nakajima J,
et al: Expression of PRMT5 in lung adenocarcinoma and its
significance in epithelial-mesenchymal transition. Hum Pathol.
45:1397–1405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y,
Zhao Q, Shao X, Bu Q, Li H, et al: Targeting protein arginine
methyltransferase 5 inhibits colorectal cancer growth by decreasing
arginine methylation of eIF4E and FGFR3. Oncotarget. 6:22799–22811.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y
and Qiu Y: MicroRNA-324-3p regulates nasopharyngeal carcinoma
radioresistance by directly targeting WNT2B. Eur J Cancer.
49:2596–2607. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S,
Deng T, Huang D, Tian Y and Qiu Y: miR-185-3p regulates
nasopharyngeal carcinoma radioresistance by targeting WNT2B in
vitro. Cancer Sci. 105:1560–1568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chua DT, Nicholls JM, Sham JS and Au GK:
Prognostic value of epidermal growth factor receptor expression in
patients with advanced stage nasopharyngeal carcinoma treated with
induction chemotherapy and radiotherapy. Int J Radiat Oncol Biol
Phys. 59:11–20. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin DC, Meng X, Hazawa M, Nagata Y, Varela
AM, Xu L, Sato Y, Liu LZ, Ding LW, Sharma A, et al: The genomic
landscape of nasopharyngeal carcinoma. Nat Genet. 46:866–871. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang F, Qian X-J, Qin W, Deng R, Wu XQ,
Qin J, Feng GK and Zhu XF: Dual phosphoinositide 3-kinase/mammalian
target of rapamycin inhibitor NVP-BEZ235 has a therapeutic
potential and sensitizes cisplatin in nasopharyngeal carcinoma.
PLoS One. 8:e598792013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan Y, Zhang Q, Atsaves V, Yang H and
Claret FX: Suppression of Jab1/CSN5 induces radio- and
chemo-sensitivity in nasopharyngeal carcinoma through changes to
the DNA damage and repair pathways. Oncogene. 32:2756–2766. 2013.
View Article : Google Scholar :
|
|
29
|
Agaoglu FY, Dizdar Y, Dogan O, Alatli C,
Ayan I, Savci N, Tas S, Dalay N and Altun M: p53 overexpression in
nasopharyngeal carcinoma. In Vivo. 18:555–560. 2004.PubMed/NCBI
|
|
30
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-kB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi D, Guo W, Chen W, Fu L, Wang J, Tian
Y, Xiao X, Kang T, Huang W and Deng W: Nicotine promotes
proliferation of human nasopharyngeal carcinoma cells by regulating
α7AChR, ERK, HIF-1α and VEGF/PEDF signaling. PLoS One.
7:e438982012. View Article : Google Scholar
|
|
32
|
Liu Y, Chen LH, Yuan YW, Li QS, Sun AM and
Guan J: Activation of AKT is associated with metastasis of
nasopharyngeal carcinoma. Tumour Biol. 33:241–245. 2012. View Article : Google Scholar
|
|
33
|
Tai HC, Huang HY, Lee SW, Lin CY, Sheu MJ,
Chang SL, Wu LC, Shiue YL, Wu WR, Lin CM, et al: Associations of
Rsf-1 overexpression with poor therapeutic response and worse
survival in patients with nasopharyngeal carcinoma. J Clin Pathol.
65:248–253. 2012. View Article : Google Scholar
|
|
34
|
Gu Z, Gao S, Zhang F and Wang Z, Ma W,
Davis RE and Wang Z: Protein arginine methyltransferase 5 is
essential for growth of lung cancer cells. Biochem J. 446:235–241.
2012. View Article : Google Scholar : PubMed/NCBI
|