Introduction
Large cell neuroendocrine carcinoma (LCNEC) is a
rare, aggressive malignancy most commonly arising in the lung
(1). In 1990, Hui et al
(2) described the first case of
LCNEC in the parotid gland and since then five additional cases in
the parotid and four in the submandibular gland have been reported
(2–10). Four of these patients have died of
their disease. Similar to pulmonary LCNEC, salivary gland LCNEC
shows a characteristic growth pattern, high mitotic rate and
evidence of neuroendocrine differentiation, including expression of
synaptophysin, chromogranin A and/or CD56 (11). When showing squamous, giant-cell,
spindle-cell or adenocarcinomatous features, the tumors are
designated combined LCNEC. There is no classification of salivary
LCNEC in the 2005 World Health Organization (WHO) classification of
head and neck tumors which is why the WHO criteria for pulmonary
LCNEC are applied to this type of salivary gland tumor (1,12).
Clinically, salivary gland LCNEC presents as a rapidly growing mass
with frequent cervical nodal metastases. Due to the extreme rarity
of these tumors no recommendations have been made as to its
treatment.
Little is known about the molecular pathogenesis of
salivary gland LCNEC and hence it is not known whether they are
genetically similar or different from LCNEC in other anatomical
locations, such as for example the lung where only few reports
about the genomic profile are available (13–15).
Studies of the genetics of salivary gland LCNEC may provide new
knowledge concerning potential diagnostic biomarkers and may
ultimately also lead to the identification of new treatment targets
for patients with this aggressive carcinoma. In the present study,
we report a case of LCNEC of the submandibular gland, the first
LCNEC with squamous features of the salivary glands (also known as
combined LCNEC) (1). We also
present a detailed genomic profile of the tumor based on
high-resolution array comparative genomic hybridization
(arrayCGH).
Patient and methods
Case report
A 69-year-old female was referred to the Department
of Otorhinolaryngology Head and Neck Surgery and Audiology at
Rigshospitalet (Copenhagen, Denmark) with a 3-week history of a
rapidly growing mass in the right submandibular region. Clinical
examination revealed a mobile, firm 3×7 cm mass in the
submandibular area. No palpable lymph nodes on the neck were found.
Ultrasound confirmed the submandibular origin and an inhomogeneous
echo pattern of the tumor. Fine needle aspiration cytology (FNAC)
showed carcinoma cells. PET-CT demonstrated a right-sided
submandibular mass measuring 4×5.5×4 cm without involvement of
adjacent structures (Fig. 1A and B)
as well as a 2×2×2 cm mass in the upper lobe of the left lung, both
with intense 18-FDG uptake. No enlarged or high-uptake lymph nodes
were identified. The patient underwent resection of the
submandibular gland along with selective neck dissection of level
I–III of the cervical lymph nodes. All nerves were spared and the
patient had no postoperative sequelae. The operation was
macroscopically but not microscopically radical, and the patient
received adjuvant radiotherapy (66 Gy in 33 fractions over five
weeks) and a carboplatin-etoposide chemotherapy regimen.
After completion of radiotherapy and chemotherapy,
the patient underwent lobectomy for the left-sided lung cancer
which subsequently metastasized to the right lung. The patient died
19 months after excision of the submandibular tumor due to the lung
cancer.
Histopathology and
immunohistochemistry
Formalin-fixed and paraffin-embedded (FFPE) tissues
from the submandibular and lung resected specimens were sectioned
and stained with hematoxylin and eosin according to standard
protocols. Immunohistochemistry was performed using the Ventana
BenchMark ULTRA platform (Ventana Medical Systems, Tucson, AZ, USA)
except for FLI-1, which was processed with the EnVision™ system
(Dako, Glostrup, Denmark). The following antibodies were used:
Ki-67 (clone MIB-1, code M724001, mouse anti-human; 1:100; Dako),
CD56 [clone 1B6, code NCL-CD56-1B6, mouse anti-human; 1:50;
Novocastra (Newcastle, UK)], synaptophysin [clone MRQ-50, code
760-4595, rabbit anti-human; 1:150; Roche (Mannheim, Germany)],
chromogranin A (polyclonal, code A043001, rabbit anti-human;
1:2,000; Dako), p63 (clone 4A4, code 790-4509, mouse anti-human,
ready-to-use; Roche), S-100 (polyclonal, code Z0311, rabbit
anti-human; 1:4,000), actin (clone 1A4, code M085101, mouse
anti-human; 1:2,000), vimentin (clone VIM 3B4, code M7020, mouse
anti-human; 1:400), calponin (clone CALP, code M355601, mouse
anti-human; 1:500), cytokeratin 5/6 (CK5/6) (clone D5/16 B4, code
M723701, mouse anti-human; 1:20), cytokeratin-7 (CK7) (clone OV-TL
12/30, code M701801, mouse anti-human; 1:1,000), CD99 (clone E12,
code M3601, mouse anti-human; 1:100), desmin (clone D33, code
M076001, mouse anti-human; 1:100) (all from Dako), FLI-1 [clone
G146-222, code 554266, mouse anti-human; 1:400, BD Biosciences (San
Jose, CA, USA)], CD117 (polyclonal, code A450229, rabbit
anti-human; 1:100; Dako), p16 (clone E5H4, code 825-4713, mouse
anti-human, ready-to-use; Roche), NUT [clone C52B1, code 3625,
rabbit anti-human; 1:50; Cell Signaling Technology (Danvers, MA,
USA)], napsin A (clone IP64, code NCL-L-Napsin A, mouse anti-human;
1:400) and TTF-1 (clone SPT24, code NCL-TTF-1, mouse anti-human;
1:100) (both from Novocastra). Negative control sections were
incubated identically except for the primary antibody, which was
replaced by normal rabbit serum/mouse IgG.
The Danish Data Protection Agency (REG-94-2014)
approved the investigation. Written consent from the patient's
husband was obtained. The investigation adheres to the tenets of
the Declaration of Helsinki (version 2008).
In situ hybridization (ISH)
The Epstein-Barr virus (EBER) PNA probe/flourescein
kit was used (code Y5200) and visualized with the Dako PNA ISH
Detection kit (code K5201) (both from Dako). Five-micron FFPE
sections were deparaffinized, rehydrated and processed according to
the manufacturer's instructions.
Fluorescence in situ hybridization
(FISH)
The ZytoLight SPEC EWSR1 dual-color
break-apart probe was used (ZytoVision, Bremerhaven, Germany)
according to the manufacturer's protocol using the HYBrite platform
(Abbott Molecular, Des Plaines, IL, USA). After hybridization,
nuclei were counterstained with DAPI (ZytoVision). At least 100
nuclei were counted, and only nuclei where the entire nuclear
membrane could be visualized were scored. A signal was considered
positive for EWSR1 rearrangement when the distance between
red and green signals was greater than two diameters of any
individual signal. At least 10% of nuclei should display a split
signal for a positive score.
Array comparative genomic hybridization
(arrayCGH) analysis
Genomic DNA was isolated from the FFPE LCNEC tumor
tissue using the DNeasy® Blood and Tissue kit (Qiagen
GmbH, Hilden, Germany). ArrayCGH analysis was subsequently
performed using the human genome CGH microarray 244K
oligonucleotide array (G4411B; Agilent Technologies Inc., Palo
Alto, CA, USA). The arrayCGH experiment was essentially performed
as previously described and as recommended by the manufacturer
(16,17). Slides were scanned on an Agilent
High-Resolution C Microarray Scanner, followed by data extraction
and normalization using Feature Extraction v.10.7.1 (Agilent
Technologies) with linear normalization (protocol CGH_107_Sep09).
Data analysis was carried out using Nexus Copy Number
Software® Discovery Edition v. 7.5 (BioDiscovery Inc.,
El Segundo, CA, USA) as previously described. The FASST2
segmentation algorithm was used to define non-random regions of
CNAs across the genome with a significance threshold set to
p=1.0E-8. The log2 ratio thresholds for aberration calls were set
to 1.5 for high copy number gain/amplification, 0.3 for gain, −0.3
for loss and −1.5 for homozygous deletion. Each aberration was
checked manually to confirm the accuracy of the call. Gender
chromosomes and regions partially or completely covered by a
previously reported copy number variation (Database of Genomic
Variants; http://dgvbeta.tcag.ca/dgv/app/news?ref=NCBI37/hg19)
were excluded from the analysis.
Results
Histopathology
The submandibular tumor was firm, homogeneous with a
shiny cut surface. The tumor did not grow beyond the
well-encapsulated submandibular gland. Microscopic examination of
the tumor revealed a mainly undifferentiated tumor consisting of
large sheets of medium-sized tumor cells with large, vesicular
nuclei with conspicuous nucleoli and scarce cytoplasm along with
abrupt, isolated squamous components organized in well-demarcated
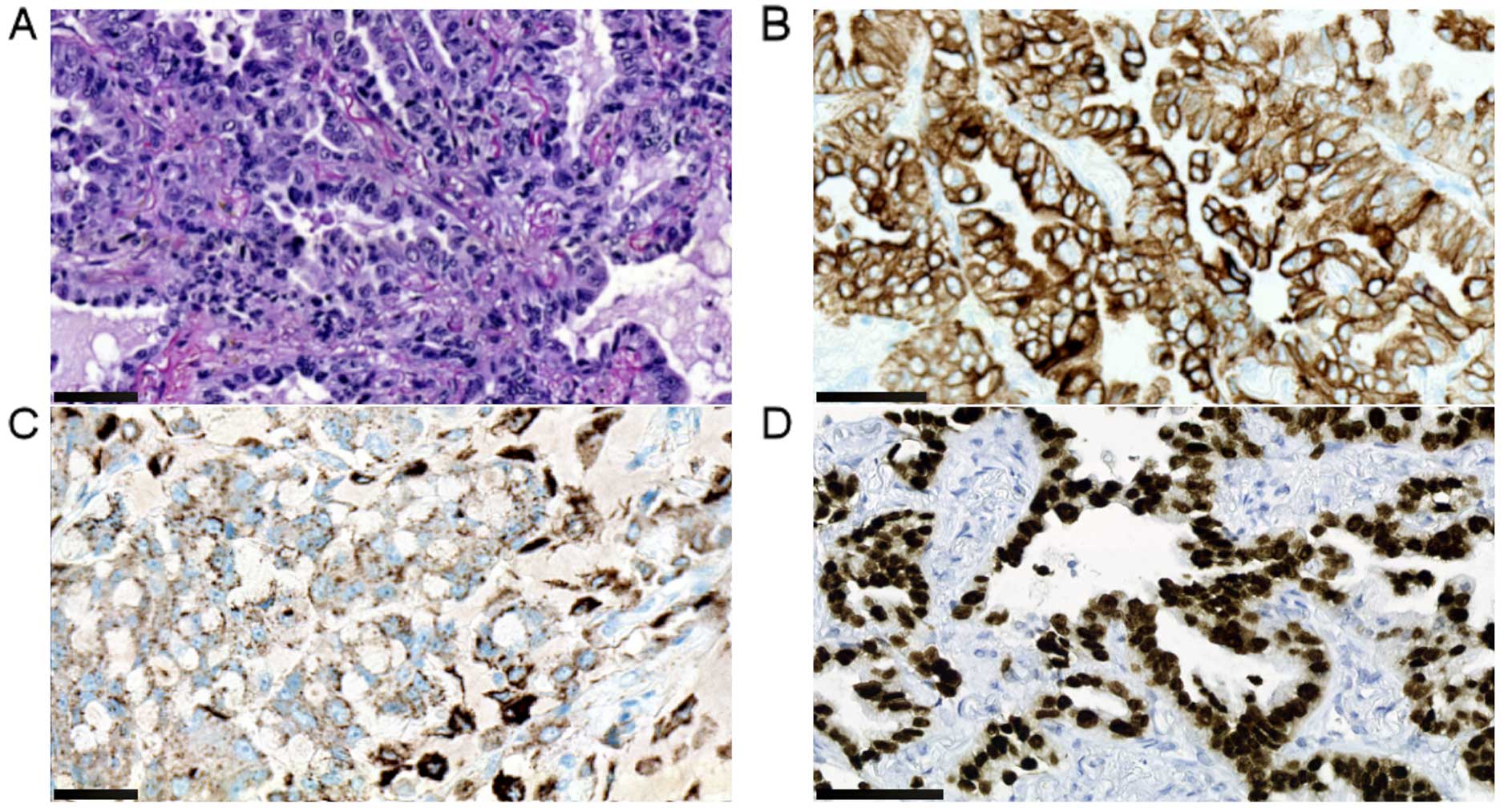
round nests. The latter made up ~30% of the tumor (Fig. 2A). Both cell types were positive for
p63 whereas primarily the squamous component expressed CK5/6
(Fig. 2B and C). The
undifferentiated component was positive for synaptophysin and CD56
whereas the squamous cells were negative for both (Fig. 2D and E). The undifferentiated cells
had a Ki-67 index of 95%, whereas the corresponding number for the
squamous cells was 30% (Fig. 2F).
Both cell populations were negative for CK7, chromogranin A, S-100,
actin, vimentin, calponin, CD117, p16, NUT, napsin A, desmin, CD99,
FLI-1 and EBV. TTF-1 showed weak reaction in 5% of undifferentiated
tumor cells. FISH analysis showed that the tumor cells were
negative for the Ewing sarcoma specific translocation t(11;22). All
lymph nodes were free of tumor cells. The final histopathologic
diagnosis was combined LCNEC of the squamous type and the patient
was staged as T3N0M0 (18). The
lung tumor was extensively sampled for neuroendocrine and squamous
differentiation but with no evidence of either of these components.
The lung tumor was also morphologically different from the
submandibular tumor and had a distinctly different
immunohistochemical profile as presented in Fig. 3 and Table I. The final diagnosis was primary
papillary adenocarcinoma of the lung.
 | Table IImmunohistochemical characteristics of
the large cell neuroendocrine carcinoma of the submandibular gland
and the papillary adenocarcinoma of the lung. |
Table I
Immunohistochemical characteristics of
the large cell neuroendocrine carcinoma of the submandibular gland
and the papillary adenocarcinoma of the lung.
| Antibody | Combined large cell
neuroendocrine carcinoma in submandibular gland
| Papillary
adenocarcinoma in lung |
|---|
| Undifferentiated
component | Squamous
component |
|---|
| Cytokeratin 5/6 | − | + | − |
| Cytokeratin 7 | − | − | + |
| p63 | + | + | − |
| TTF-1 | − | − | + |
| Napsin A | − | − | + |
| Synaptophysin | + | − | − |
| CD56 | + | − | − |
Genomic profile of the LCNEC
In order to genomically characterize the salivary
gland LCNEC we performed arrayCGH analysis using a high-resolution
244K oligonucleotide array. The results are presented in Table II and in Fig. 4A and B. The tumor had a relatively
uncomplicated genomic profile characterized by in particular copy
number losses. In total, 10 losses and 4 gains were identified.
There was no evidence of gene amplification or homozygous deletion.
Losses and gains of whole chromosomes or chromosome arms
predominated and included −3p, −4, −7q, −10 (10pter-q26) −11, −13,
−16q, +3q and +16p (Fig. 4A). In
addition, there were a few segmental gains and losses, most notably
gain of a 2.3 Mb segment in 9p23-p22.3 including the NFIB
oncogene (Fig. 4B). Notably, the
loss of 10pter-q26.2 resulted in a breakpoint in the DOCK1
gene with loss of the 5′-part of the gene (exons 1–19) and
retention of the 3′-part.
 | Table IICopy number alterations detected by
high-resolution arrayCGH analysis of the large cell neuroendocrine
carcinoma of the submandibular gland. |
Table II
Copy number alterations detected by
high-resolution arrayCGH analysis of the large cell neuroendocrine
carcinoma of the submandibular gland.
| Cytoband
location | Genomic event | Region length
(Mb) | No. of genes |
|---|
| 2q22.1-q36.3 | Gain | 86.1 | 518 |
| 3p | Loss | 90.4 | 651 |
| 3q | Gain | 104.4 | 774 |
| 4 | Loss | 188.1 | 1,120 |
| 7q | Loss | 97.4 | 837 |
| 9p23-p22.3 | Gain | 2.3 | 9 |
| 10pter-q26.2 | Loss | 128 | 971 |
| 11 | Loss | 131.0 | 1,577 |
| 13 | Loss | 85.9 | 587 |
| 16pter-p13.3 | Loss | 0.53 | 31 |
| 16p13.3-q12.1 | Gain | 24.7 | 441 |
| 16q12.1-qter | Loss | 43.2 | 468 |
|
19q13.33-q13.42 | Loss | 1.9 | 111 |
| 19q13.43 | Loss | 0.7 | 17 |
Discussion
Neuroendocrine tumors are a heterogeneous group of
lesions that may occur in all visceral subsites in the body
(11). They are, however, extremely
rare in the salivary glands. Only 10 LCNECs have been reported in
the literature, four of which were located in the submandibular
gland. In the present study, we report the first case of LCNEC with
squamous differentiation, a so-called combined LCNEC. Salivary
gland cancers are rare and constitute only 0.3% of all human
malignancies. They originate from the epithelial components of the
major and minor salivary glands (12). The origin of salivary gland LCNEC
is, however, unknown and to date no neuroendocrine cells have been
identified in the salivary glands (8). The rarity of these lesions is further
illustrated by the identification of only two cases of parotid
LCNEC in a series of 1,675 surgically resected, primary parotid
gland tumors (5). However, since a
number of different head and neck neoplasms, including those of
salivary gland origin, demonstrate focal expression of
neuroendocrine markers, the definition of what actually constitutes
a 'true' neuroendocrine tumor has been a matter of debate (19). Besides the challenge of
histopathological diagnosis of neuroendocrine neoplasms, FNAC of
LCNEC is difficult and does not contribute to diagnostic accuracy
since it often results in a suggestion of an undifferentiated
carcinoma.
Notably, in addition to the submandibular gland
LCNEC our patient also had a concurrent lung tumor. Although
several histomorphologic variants of LCNEC of the lung have been
described, thorough histopathological examination of two different
surgical specimens showed an identical image consistent with a
primary papillary adenocarcinoma of pulmonary origin. There was no
evidence of neuroendocrine differentiation or histopathologic
overlap with the submandibular LCNEC, thus excluding the
possibility of a metastatic pulmonary lesion (20,21).
The well-encapsulated appearance of the submandibular tumor along
with the lack of metastases did not call for chemotherapy and/or
radiotherapy. No recurrences or metastases of the LCNEC were
identified during the 19 months of follow-up.
Genomic profiling of the submandibular gland LCNEC
revealed a hypodiploid genome predominated by losses of whole
chromosomes or chromosome arms involving chromosomes 3p, 4, 7q, 10,
11, 13, 16q and gains of 3q and 16p. To the best of our knowledge,
a similar genomic profile has not been detected in any of the major
types of salivary gland carcinomas analyzed to date (16,22–25,
unpublished data). In addition, the present case had two
particularly interesting copy number alterations, namely gain of a
2.3-Mb segment in 9p23-p22.3, including the NFIB gene and a
breakpoint in the DOCK1 gene in 10q26.2. We previously
identified recurrent gains of NFIB in pleomorphic adenomas
and carcinoma ex pleomorphic adenomas (26). We have also shown that NFIB,
which encodes a transcription factor, is involved in recurrent gene
fusions with HMGA2 in pleomorphic adenomas and with
MYB in adenoid cystic carcinomas of the salivary glands
(27–29). These observations together with
recent arrayCGH studies demonstrating copy number
gain/amplification and overexpression of NFIB in human and
experimental small cell lung cancers and triple-negative breast
cancers further supports the notion that NFIB has oncogenic
properties in several types of human neoplasms (30,31).
We also detected a breakpoint in the DOCK1 gene in 10q26.2.
This is of special interest because gene fusions involving
DOCK1 and at least 5 different partner genes were recently
detected in breast cancer and astrocytomas (25,32).
Whether DOCK1 is involved in a gene fusion also in the
present case of LCNEC remains to be shown.
Although genomic profiling data is only available
from the present case of salivary gland LCNEC, it is interesting to
note that this case has several genomic features in common with
LCNEC of the lung, including losses of 3p, 4q, 10q, 13q and 16q and
gain involving 3q (13–15). Analyses of additional cases of
salivary gland LCNEC are, however, necessary to confirm these
genomic similarities. Notably, although LCNEC is most commonly
found in the lungs it is still a rare tumor that lacks a consensus
treatment (33). In the current WHO
classification of lung cancers, LCNEC is categorized as a non-small
cell lung cancer (non-SCLC) despite the fact that the clinical and
biological characteristics are more similar to those of small cell
lung cancer (SCLC) (33,34). In addition, several studies favor
SCLC chemotherapy regimens for lung LCNECs as compared to non-SCLC
regimens (11,34). The response of LCNEC of the salivary
glands to chemotherapy is yet to be determined but could prove to
be an attractive treatment option in patients with advanced
disease.
In conclusion, we report a rare case of LCNEC of the
salivary glands and the first case of the combined subtype. The
patient was staged as T3N0M0 and radically operated for a
well-circumscribed tumor. There was no recurrence or metastasis
during the follow-up of 19 months after which the patient died of a
concurrent papillary adenocarcinoma of the lung. Continued studies
of salivary gland LCNEC may provide new knowledge concerning
potential diagnostic biomarkers and may ultimately also lead to the
identification of new treatment targets for patients with this type
of carcinoma.
Acknowledgments
We thank Pernille Frederiksen and Sanni Pedersen for
excellent technical support with immunohistochemistry and FISH,
respectively. This study was supported in part by the Region
Zealand, the Swedish Cancer Society and BioCARE - a National
Strategic Research Program at the University of Gothenburg.
References
|
1
|
Brambilla E, Lantuejoul S, Pugatch B,
Chang YL, Geisinger K, Petersen I, Gal A, Meyerson M, Sheppard MN,
Hanash SM, et al: Large cell carcinoma. World Health Organization
Classification of Tumours, Pathology and Genetics, Tumours of the
Lung, Pleura, Thymus and Heart. Travis WD, Brambilla E,
Müller-Hermelink K and Harris CC: IARC Press; Lyon: pp. 45–50.
2004
|
|
2
|
Hui KK, Luna MA, Batsakis JG, Ordóñez NG
and Weber R: Undifferentiated carcinomas of the major salivary
glands. Oral Surg Oral Med Oral Pathol. 69:76–83. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Casas P, Bernáldez R, Patrón M,
López-Ferrer P and García-Cabezas MA: Large cell neuroendocrine
carcinoma of the parotid gland: Case report and literature review.
Auris Nasus Larynx. 32:89–93. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ueo T, Kaku N, Kashima K, Daa T, Kondo Y,
Yoshida K, Suzuki M and Yokoyama S: Carcinosarcoma of the parotid
gland: An unusual case with large-cell neuroendocrine carcinoma and
rhabdomyosarcoma. APMIS. 113:456–464. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagao T, Sugano I, Ishida Y, Tajima Y,
Munakata S, Asoh A, Yamazaki K, Muto H, Konno A, Kondo Y, et al:
Primary large-cell neuroendocrine carcinoma of the parotid gland:
Immunohistochemical and molecular analysis of two cases. Mod
Pathol. 13:554–561. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Larsson LG and Donner LR: Large cell
neuroendocrine carcinoma of the parotid gland: Fine needle
aspiration, and light microscopic and ultrastructural study. Acta
Cytol. 43:534–536. 1999.PubMed/NCBI
|
|
7
|
Yamamoto N, Minami S, Kidoguchi M, Shindo
A, Tokumaru Y and Fujii M: Large cell neuroendocrine carcinoma of
the submandibular gland: Case report and literature review. Auris
Nasus Larynx. 41:105–108. 2014. View Article : Google Scholar
|
|
8
|
Petrone G, Santoro A, Angrisani B, Novello
M, Scarano E, Rindi G and Lauriola L: Neuroendocrine tumors of the
submandibular gland: Literature review and report of a case. Int J
Surg Pathol. 21:85–88. 2013. View Article : Google Scholar
|
|
9
|
Kawaratani H, Tsujimoto T, Yoshikawa M,
Kawanami F, Shirai Y, Yoshiji H, Morita K and Fukui H: Large cell
neuroendocrine carcinoma presenting with neck swelling in the
submandibular gland: A case report. J Med Case Rep. 7:812013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sowerby LJ, Matthews TW, Khalil M and Lau
H: Primary large cell neuroendocrine carcinoma of the submandibular
gland: Unique presentation and surprising treatment response. J
Otolaryngol. 36:E65–E69. 2007. View Article : Google Scholar
|
|
11
|
Kusafuka K, Ferlito A, Lewis JS Jr,
Woolgar JA, Rinaldo A, Slootweg PJ, Gnepp DR, Devaney KO, Travis WD
and Barnes L: Large cell neuroendocrine carcinoma of the head and
neck. Oral Oncol. 48:211–215. 2012. View Article : Google Scholar
|
|
12
|
Barnes L, Eveson JW, Reichart P and
Sidransky D: Salivary Glands. World Health Organization
Classification of Tumours, Pathology and Genetics of Head and Neck
Tumours. IARC Press; Lyon: pp. 2102005
|
|
13
|
Peng WX, Shibata T, Katoh H, Kokubu A,
Matsuno Y, Asamura H, Tsuchiya R, Kanai Y, Hosoda F, Sakiyama T, et
al: Array-based comparative genomic hybridization analysis of
high-grade neuroendocrine tumors of the lung. Cancer Sci.
96:661–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ullmann R, Petzmann S, Sharma A, Cagle PT
and Popper HH: Chromosomal aberrations in a series of large-cell
neuroendocrine carcinomas: Unexpected divergence from small-cell
carcinoma of the lung. Hum Pathol. 32:1059–1063. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walch AK, Zitzelsberger HF, Aubele MM,
Mattis AE, Bauchinger M, Candidus S, Präuer HW, Werner M and Höfler
H: Typical and atypical carcinoid tumors of the lung are
characterized by 11q deletions as detected by comparative genomic
hybridization. Am J Pathol. 153:1089–1098. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Persson F, Winnes M, Andrén Y, Wedell B,
Dahlenfors R, Asp J, Mark J, Enlund F and Stenman G:
High-resolution array CGH analysis of salivary gland tumors reveals
fusion and amplification of the FGFR1 and PLAG1 genes in ring
chromosomes. Oncogene. 27:3072–3080. 2008. View Article : Google Scholar
|
|
17
|
Barrett MT, Scheffer A, Ben-Dor A, Sampas
N, Lipson D, Kincaid R, Tsang P, Curry B, Baird K, Meltzer PS, et
al: Comparative genomic hybridization using oligonucleotide
microarrays and total genomic DNA. Proc Natl Acad Sci USA.
101:17765–17770. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer-Verlag; New York: pp. 79–86. 2010
|
|
19
|
Bell D, Hanna EY, Weber RS, DeMonte F,
Triantafyllou A, Lewis JS Jr, Cardesa A, Slootweg PJ, Stenman G,
Gnepp DR, et al: Neuroendocrine neoplasms of the sinonasal region.
(Head Neck)Jun 3–2015.Epub ahead of print. View Article : Google Scholar
|
|
20
|
Travis WD, Linnoila RI, Tsokos MG,
Hitchcock CL, Cutler GB Jr, Nieman L, Chrousos G, Pass H and
Doppman J: Neuroendocrine tumors of the lung with proposed criteria
for large-cell neuroendocrine carcinoma. An ultrastructural,
immunohistochemical, and flow cytometric study of 35 cases. Am J
Surg Pathol. 15:529–553. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang SX, Kameya T, Shoji M, Dobashi Y,
Shinada J and Yoshimura H: Large cell neuroendocrine carcinoma of
the lung: A histologic and immunohistochemical study of 22 cases.
Am J Surg Pathol. 22:526–537. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Persson M, Andrén Y, Moskaluk CA, Frierson
HF Jr, Cooke SL, Futreal PA, Kling T, Nelander S, Nordkvist A,
Persson F, et al: Clinically significant copy number alterations
and complex rearrangements of MYB and NFIB in head and neck adenoid
cystic carcinoma. Genes Chromosomes Cancer. 51:805–817. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Mitani Y, Caulin C, Rao PH, Kies
MS, Saintigny P, Zhang N, Weber RS, Lippman SM and El-Naggar AK:
Detailed genome-wide SNP analysis of major salivary carcinomas
localizes subtype-specific chromosome sites and oncogenes of
potential clinical significance. Am J Pathol. 182:2048–2057. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jee KJ, Persson M, Heikinheimo K,
Passador-Santos F, Aro K, Knuutila S, Odell EW, Mäkitie A, Sundelin
K, Stenman G, et al: Genomic profiles and CRTC1-MAML2 fusion
distinguish different subtypes of mucoepidermoid carcinoma. Mod
Pathol. 26:213–222. 2013. View Article : Google Scholar
|
|
25
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations and Gene Fusions in
Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
|
|
26
|
von Holstein SL, Fehr A, Persson M,
Nickelsen M, Therkildsen MH, Prause JU, Heegaard S and Stenman G:
Lacrimal gland pleomorphic adenoma and carcinoma ex pleomorphic
adenoma: Genomic profiles, gene fusions, and clinical
characteristics. Ophthalmology. 121:1125–1133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geurts JM, Schoenmakers EF, Röijer E,
Aström AK, Stenman G and van de Ven WJ: Identification of NFIB as
recurrent translocation partner gene of HMGIC in pleomorphic
adenomas. Oncogene. 16:865–872. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Persson M, Andrén Y, Mark J, Horlings HM,
Persson F and Stenman G: Recurrent fusion of MYB and NFIB
transcription factor genes in carcinomas of the breast and head and
neck. Proc Natl Acad Sci USA. 106:18740–18744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stenman G, Persson F and Andersson MK:
Diagnostic and therapeutic implications of new molecular biomarkers
in salivary gland cancers. Oral Oncol. 50:683–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han W, Jung EM, Cho J, Lee JW, Hwang KT,
Yang SJ, Kang JJ, Bae JY, Jeon YK, Park IA, et al: DNA copy number
alterations and expression of relevant genes in triple-negative
breast cancer. Genes Chromosomes Cancer. 47:490–499. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dooley AL, Winslow MM, Chiang DY, Banerji
S, Stransky N, Dayton TL, Snyder EL, Senna S, Whittaker CA, Bronson
RT, et al: Nuclear factor I/B is an oncogene in small cell lung
cancer. Genes Dev. 25:1470–1475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoshihara K, Wang Q, Torres-Garcia W,
Zheng S, Vegesna R, Kim H and Verhaak RG: Tumours of the lung. In:
The landscape and therapeutic relevance of cancer-associated
transcript fusions. Oncogene. 34:4845–4854. 2015. View Article : Google Scholar
|
|
33
|
Iyoda A, Makino T, Koezuka S, Otsuka H and
Hata Y: Treatment options for patients with large cell
neuroendocrine carcinoma of the lung. Gen Thorac Cardiovasc Surg.
62:351–356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Travis WD, Brambilla E, Müller-Hermelink K
and Harris CC: World Health Organization Classification of Tumours,
Pathology and Genetics, Tumours of the Lung, Pleura, Thymus and
Heart. IARC Press; Lyon: pp. 102004
|