Introduction
Esophageal cancer is one of the most common cancers
of the upper gastrointestinal tract worldwide with a variable
geographic distribution. Esophageal squamous cell carcinoma (ESCC)
and esophageal adenocarcinoma (EAC) have been identified as the two
main histological types in regards to different etiologic and
pathologic characteristics. EAC is common in Western countries
while ESCC is frequently diagnosed in east Asia, particularly in
China whose incidence in the high-risk northern and central China
exceeds 100 cases/100,000 people/year (1). Despite remarkable improvements in the
diagnosis and treatment of this cancer, the overall 5-year survival
rate for advanced and metastatic esophageal cancer is less than 20%
after surgery in China (1). Thus,
an understanding of the molecular mechanisms of the pathogenesis of
esophageal cancer is important for identifying more tumor-specific
biomarkers and therapeutic targets for early diagnosis and
treatment.
miRNAs are an endogenous class of highly-conserved
20–25 nucleotide single-stranded non-coding RNAs that regulate gene
expression at the post-transcriptional level by binding to the
3′-untranslated region (3′-UTR) of mRNA which subsequently leads to
mRNA degradation and translation repression. Emerging evidence
demonstrates that a single gene may be targeted by different
miRNAs, while a single miRNA can modulate multiple gene expression
due to the imperfect complementarity with target mRNAs, which
allows miRNAs to control a variety of physiological processes such
as cell proliferation, differentiation, apoptosis and angiogenesis
(2). Thus, aberrant expression of
miRNAs are thought to play critical roles in cancer initiation and
progression via targeting various oncogenes and tumor-suppressor
genes (3,4).
Aberrant miRNA expression patterns have been
reported in esophageal cancer compared to corresponding
non-malignant tissues (5,6). Several miRNAs were found to be
significantly downregulated in ESCC such as miR-100, miR-203,
miR-205, miR-145, miR-27b, miR-375, miR-125b and let-7c (7,8),
suggesting that they may exert tumor-suppressive effects. We
previously demonstrated that miR-203 promoted apoptosis and
inhibited the proliferation and migration of esophageal cancer
cells through downregulation of Ran expression (9). miR-100 is located on chromosome 11 at
11q24.1 (Gene ID: 406892) (10). A
previous study showed that overexpression of miR-100 induced
apoptosis of esophageal cancer cells by targeting mTOR (11). However, further studies are required
to provide additional insights into the molecular mechanisms
underlying the regulatory activities of miR-100 in esophageal
cancer. In the present study, the regulatory effects of miR-100 on
the proliferation, migration and tumor growth of esophageal cancer
cells and its molecular mechanisms were characterized.
Materials and methods
Plasmid construction
The hsa-miR-100 (miR-100) precursor was synthesized
by Songon Tech (Beijing, China) and subcloned into the
pcDNA6.2-GW/EmGFP-miR vector (referred to as pcDNA6.2/miR-100)
according to the manufacturer's instructions (Invitrogen, Carlsbad,
CA, USA). Scramble miRNA was cloned into the same vector and used
as a negative control (named pcDNA6.2/miR-NC). The sequence of the
control miRNA was: 5′-AGGTACGAAACGCTAAGAAT-3′. To construct the
luciferase reporter vector, the human CXCR7 3′-UTR containing
putative binding sites for miR-100 was amplified by PCR and cloned
in pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega,
Madison, WI, USA). The construct was named pmiR-GLO-CXCR7. The
primers were as follows: forward primer,
5′-GGGAGCTCTCTGCCCTGGAGAGGCTCTG-3′ and reverse primer,
5′-CGTCTAGACAAAACTGAAGTCACGCTA-3′. The accuracy of all cloning was
confirmed by sequencing.
Cell transfection
Human esophageal squamous cancer cell line (Ec-109)
(kindly provided by Dr Ling Zhou from the Institute for Viral
Disease Control and Prevention, Chinese Center for Disease Control
and Prevention) was maintained in RPMI-1640 medium supplemented
with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100
µg/ml streptomycin in a humidified incubator with 5%
CO2 at 37°C. All cell culture materials were obtained
from Gibco, USA. Lipofectamine 2000 (Invitrogen) was used for cell
transfection. Briefly, cells at ~60% confluency in 6-well plates
were transfected with the plasmid pcDNA6.2/miR-100 or control
vector pcDNA6.2/miR-NC. Blasticidin (4 µg/ml) was applied to
select stable miR-100-overexpressing clones. The expression of
miR-100 was confirmed by real-time PCR after 3 weeks of selection
from individual clones.
Quantitative real-time PCR (RT-qPCR)
analysis
The expression of mature miR-100 and the primary
transcript of CXCR7 was analyzed by RT-qPCR. Total RNA was
extracted from cultured cells by TRIzol reagent (Invitrogen) based
on the provided protocols. Total RNA (1 µg) was applied for
reverse transcription to synthesize miRNA cDNA using NCode™ VILO™
miRNA cDNA synthesis kit (Invitrogen). RT-qPCR analysis was
performed using EXPRESS SYBR®-GreenER™ miRNA RT-qPCR kit
(Invitrogen) based on instructions. Reverse transcription mix (20
µl) was amplified by PCR with denaturation at 95°C for 2 min
and 40 cycles at 95°C for 10 sec and 60°C for 1 min. U6 was applied
as an endogenous control for normalization. Each sample was
analyzed in triplicate. The forward primers that target miR-100 or
U6 were purchased from Songon Tech.
To analyze the expression of CXCR7, 1 µg of
total RNA was reverse transcribed in a final volume of 20 µl
to synthesize first-strand cDNA using ImProm-II™ reverse
transcription system (Promega) based on the instructions of the
manufacturer. The transcript levels were detected using
Brilliant® II SYBR®-Green qPCR Master Mix
(Stratagene, USA) to monitor amplification. β-actin served as an
endogenous control for normalization. PCR reactions (25 µl)
in triplicate were carried out by an initial denaturation at 95°C
for 10 min followed by 40 cycles, each consisting of 30 sec at
95°C, 30 sec at 58°C, 30 sec at 72°C, and then 1 cycle for melting
curve consisting of 1 min at 95°C, 30 sec at 55°C, 30 sec at 95°C.
Primer sequences used were as follows: 5′-GGCTATGACACGCACTGCTACA-3′
(forward primer for CXCR7) and 5′-TGGTTGTGCTGCACGAGACT-3′ (reverse
primer for CXCR7); 5′-AGAAAATCTGG CACCACACC-3′ (forward primer for
β-actin) and 5′-TAGCACAGCCTGGATAGCAA-3′ (reverse primer for
β-actin). The 2−ΔΔCt method for relative quantification
of gene expression was used to determine miRNA and CXCR7 mRNA
expression levels. RT-qPCR primers were designed using Primer
Express software (version 5.0; Applied Biosystems).
Cell proliferation assay
Cells (2,000 cells/well) were passaged in 96-well
plates in RPMI-1640 culture medium containing 10
µg/µl blasticidin, supplemented with 10 or 7% FBS.
Cell proliferation were measured by the OD values using Cell
Counting Kit-8 (CCK-8) (Dojindo, Japan). The absorbance was
measured at 450 nm on a microplate reader (PerkinElmer, USA), and
the reference light was 650 nm. Every sample was measured for three
times.
Cell invasion and migration assays
Transwell insert chambers (Corning, Corning, NY,
USA) with 8-µm pores were used to conduct these assays. For
the migration assay, 1×105 cells were seeded into the
upper chamber in serum-free medium in triplicate. Medium containing
20% FBS in the lower chamber served as the chemoattractant. After
incubation for 24 h at 37°C in a 5% CO2 humidified
incubator, cells in the upper chambers were removed by wiping with
a cotton swab and cells that had migrated to the lower surface of
the filter were fixed with 4% formaldehyde overnight at 4°C and
stained with 0.2% crystal violet for 10 min. Cell migration was
scored by counting five random fields/filter under a light
microscope. For the invasion assay, 3×105 cells were
plated into upper chambers precoated with Matrigel (BD, USA) in
serum-free medium in triplicate. Medium with 20% FBS was added to
the lower chamber to serve as the chemoattractant. After incubation
for 24 h at 37°C, non-invading cells on the upper surface of the
filter were removed by wiping with a cotton swab and invading cells
that had migrated to the lower surface of the filter were fixed,
stained and scored as described above.
Soft agar assay for colony formation
Cells (2,000) were seeded in 1.5 ml of standard
growth medium containing 0.33% low-melting-temperature soft agar
(SeaKem; FMC Corporation) and plated onto a 3 ml layer of
solidified 0.66% soft agar in the same medium in a 35-mm tissue
culture dish. Cultures were fed once a week with 0.5 ml of complete
medium. Each cell clone tested was seeded in triplicate. The cell
colonies becoming visible microscopically after 3±4 weeks were
scored weekly, and photographed with a Olympus microscope
(Japan).
In vivo tumor formation assay
Female athymic BALB/c nude mice ~4–6 weeks old were
purchased from the Laboratory Animal Center of the Army Research
Center (Beijing, China), and carefully fostered under the
guidelines for the care and use of laboratory animals of the local
government. All mice were housed under a pathogen-free condition.
All experiments were performed with guidelines provided by the
National Cancer Insitute (NIH; USA). For xenografts, 10 mice were
randomly separated into 2 groups. Cells (2×106) that
stably expressed miR-100 or miR-NC were suspended in serum-free
RPMI-1640 medium and injected subcutaneously into the flank,
respectively. Five weeks post-injection, the mice were sacrificed
and photographed. Tumor volumes (cm3) were calculated
using the following standard formula: [length × width] × [(length +
width)/2].
Luciferase target gene reportor
assay
Two groups of Ec-109 cells (stably overexpressing
miR-100 or miR-NC) were seeded in 6-well plates for ~24 h to reach
60% confluency, and then transiently transfected with the
pmiR-GLO-CXCR7 gene report vector for 12 h. Empty vector
(pmiR-GLO-NC) was used as a control. Luciferase activity was
analyzed using the Dual-Glo® Luciferase Assay system
(Promega), which was analyzed by three independent experiments
performed in triplicate.
Western blotting
Proteins were extracted from the miR-100-expressing
or the control cells using the Membrane and Cytosol Protein
Extraction kit (Beyotime, China). Protein concentrations were
determined by the BCA protein assay kit (Tiangen, China). Protein
samples were denatured by boiling for 5 min and were loaded onto
SDS-PAGE (10% polyacrylamide gel) for electrophoresis, and then
transferred onto methanol-activated polyvinylidene fluoride (PVDF)
membranes (Bio-Rad Laboratories, USA). Non-specific reactivity on
the membranes was blocked by 5% skim milk [non-fat dry milk in
Tris-buffered saline with Tween-20 (TBST)] overnight at 4°C, and
then washed with TBST. Primary antibodies for human β-actin (Cell
Signaling Technology, Danvers, MA, USA) or CXCR7 (Bioss, China)
were incubated with the membranes overnight at 4°C, respectively.
The membranes were then incubated with the secondary antibody,
conjugated with fluorescent dyes: IRDye 800CW (KPL, Gaithersburg,
MD, USA) for 1 h. After washing, the membranes were developed under
the Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln,
NE, USA) to obtain the blotting images.
Statistical analysis
Data are presented as mean ± SD of at least three
independent experiments. The differences between group were
analyzed using the Student's t-test. A p-value of <0.05 was
considered statistically significant for all tests. Analyses were
performed using statistical analysis software SPSS 19.
Results
miR-100 inhibits cell proliferation,
migration and invasion of esophageal cancer cells
In the present study, the miR-100 precursor and
scramble miRNAs were exogenously expressed in esophageal cancer
cells and stable miRNA-expressing cells were acquired by
blasticidin. The ectopic expression of mature miR-100 in the
transfectants was initially confirmed by qRT-PCR (Fig. 1A). To investigate the potential
regulatory effects of miR-100 on esophageal cancer cells, CCK-8 and
Transwell assays were used to evaluate the proliferation, migration
and invasion of the esophageal cancer cells upon miR-100
transfection, respectively. The data showed that miR-100
overexpression significantly inhibited cell growth when cells were
cultured in 7% of FBS compared to the scramble miRNA (control)
(Fig. 1B). However, no obvious
inhibition in cell growth was observed when cultured in 10% of FBS
(Fig. 1C). Moreover, compared with
the control, ectopic expression of miR-100 was able to suppress the
migration (Fig. 2A and B) and
invasion (Fig. 2C and D) of the
esophageal cancer cells, implying a tumor-suppressor role of
miR-100 in esophageal cancer.
miR-100 suppresses colony formation and
the tumor growth of esophageal cancer cells
An anchorage-independent growth assay and xenograft
transplantation experiment were further conducted to examine the
effects of miR-100 on tumorigenesis in esophageal cancer cells. The
results showed that large cell colonies were generated in the
scramble miRNA-transfected cells (Fig.
3A). In contrast, overexpression of miR-100 inhibited colony
formation and significantly reduced colony numbers in vitro
when compared to the control (Fig. 3B
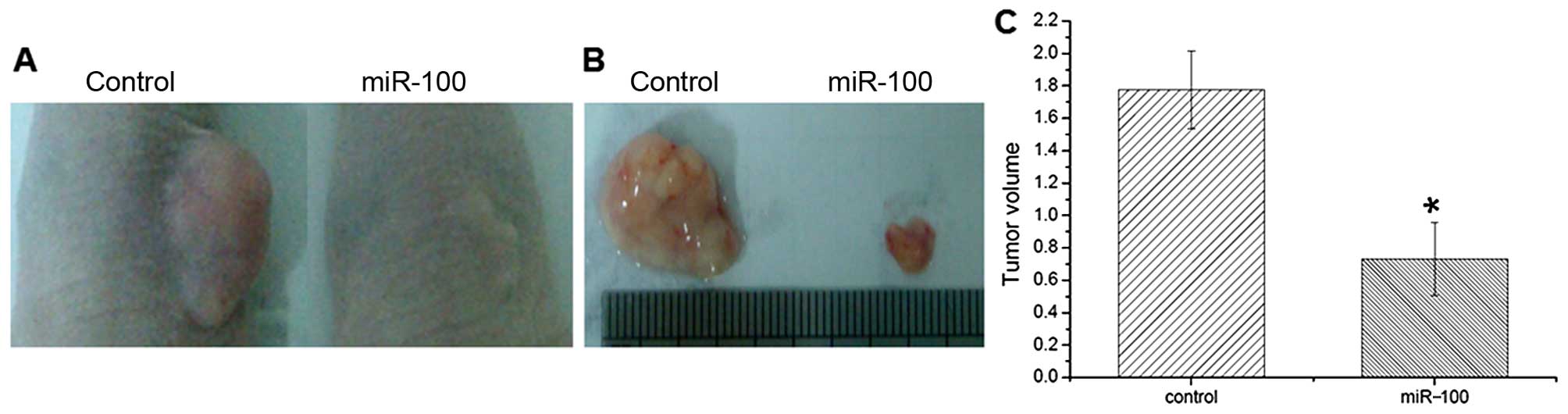
and C). In vivo xenograft transplantation analysis
further showed that miR-100 overexpression markedly suppressed
tumor growth in nude mice (Fig. 4A and
B). The tumor volume was much smaller than that of the controls
(Fig. 4C), which was consistent
with the in vitro colony formation assay. The obtained data
suggest that miR-100 plays a suppressive role in the tumorigenesis
of esophageal cancer.
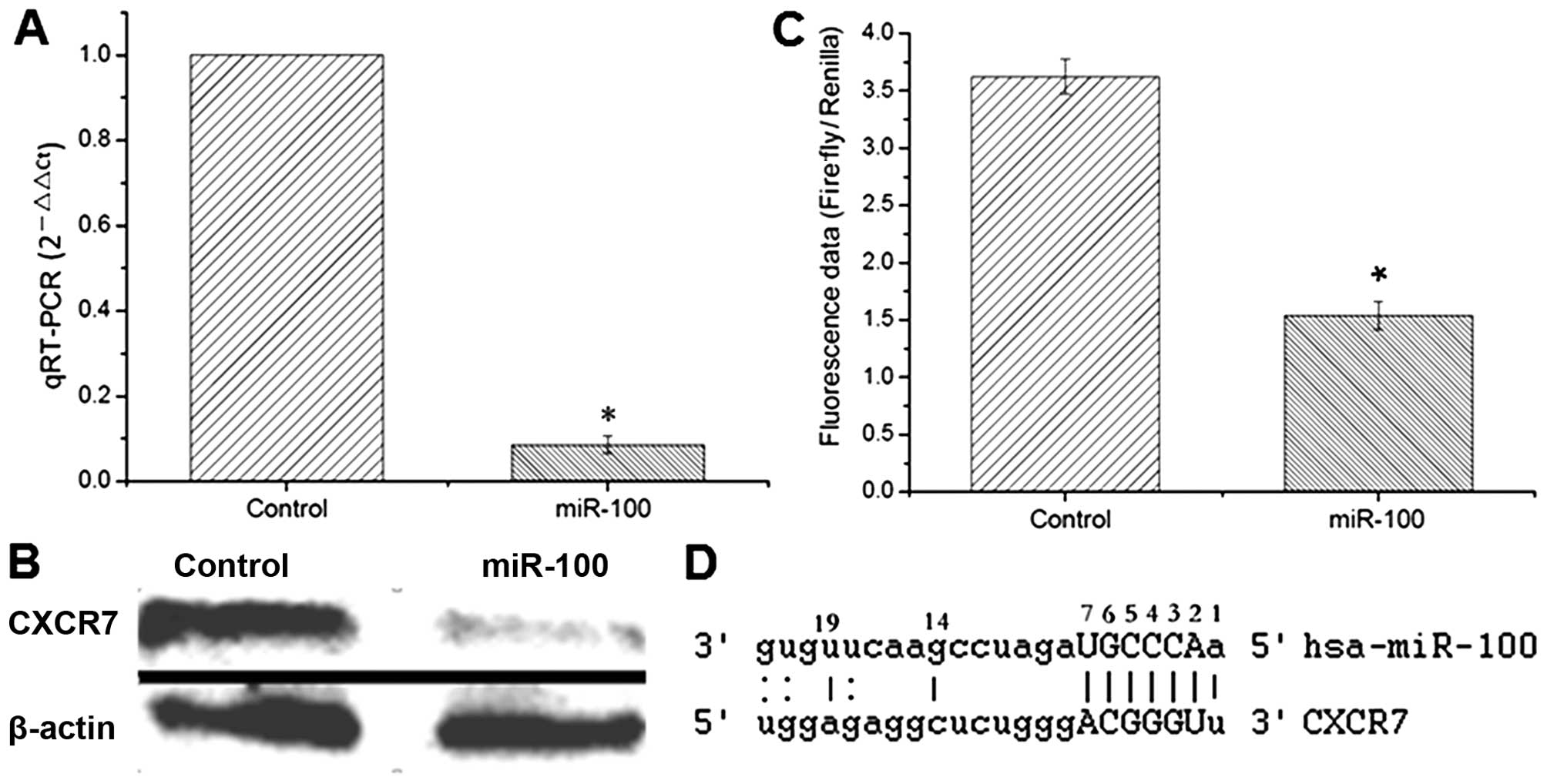
CXCR7 is a target gene of miR-100
Several computational prediction websites such as
TargetScan, PicTar and MiRanda and the target prediction methods as
previously reported (12,13) were used to identify functionally
relevant targets of miR-100. The expression of CXCR7 which contains
a putative target sequence of miR-100 in 3′-UTR was quantitatively
examined by both RT-PCR and western blotting. The data showed that
the expression of CXCR7 was significantly decreased in the
miR-100-expressing cells compared to that noted in the control
cells (Fig. 5A and B). Luciferase
assay was thereafter used to investigate whether CXCR7 is a direct
target of miR-100. The 3′-UTR region of the CXCR7-encoding gene
containing the putative binding site of miR-100 was subcloned into
the luciferase reporter vector and co-transfected with miR-100 into
esophageal cancer cells. Cells co-transfected with the empty vector
and miR-100 were used as a control. The results showed that
luciferase activity was markedly reduced in the reporter construct
containing 3′-UTR of CXCR7 compared to that in the empty vector
control (Fig. 5C), indicating that
CXCR7 is a direct downstream target gene of miR-100 and miR-100
downregulates the expression of CXCR7 through direct binding to its
3′-UTR (Fig. 5D).
Discussion
Downregulation of miR-100 has been reported in a
variety of different types of cancers including ESCC (14–18).
In the present study, miR-100 was overexpressed in ESCC cells to
elucidate the biological functions of miR-100 in ESCC. The obtained
data showed that exogenous expression of miR-100 in ESCC cells was
able to significantly suppress cell proliferation, migration and
invasion and inhibit tumor growth in vivo, suggesting a
tumor-suppressive role of miR-100 in esophageal cancer cells. A
number of tumor-associated genes such as FGFR3, HOXA1, mTOR,
IGF1-R, AGO2 and Rac1 have been reported to be modulated by miR-100
in different types of tumors and are involved in the
miR-100-mediated inhibitory effects on cell proliferation,
migration and invasion (11,19–23).
in the present study, we showed that CXCR7 is a direct target gene
of miR-100 as validated by luciferase assay and it is significantly
downregulated in miR-100-overexpressing cells.
CXCR7 was initially named receptor dog cDNA 1
(RDC1), which was cloned from a canine thyroid cDNA library and
identified as an atypical chemokine receptor (24). The human RDC1 gene is located at
chromosome region 2q37.3. Subsequent to showing that RDC1 acts as a
receptor of both chemokines of CXCL12 and CXCL11, RDC1 was
officially renamed CXCR7 as a seventh receptor of the CXC class of
the chemokine receptor family (25,26).
Chemokine receptors are seven-transmembrane receptors coupled to
G-proteins and are responsible to initiate a cascade of signal
transduction events (27).
increasing evidence suggests that chemokines and their receptors
play pivotal roles in tumor growth, angiogenesis and distant
metastases to lymph nodes and bone marrow. Chemokine CXCL12 is a
broadly expressed cytokine (28–30).
CXCR7 is highly expressed in the majority of tumor cells and
activation of the CXCR7 signaling pathway by CXCL12 has been found
to be coupled with the enhanced proliferation and increased
metastasis and invasive activities of tumor cells (29,30).
Accordingly, it has been shown that CXCR7 knockdown by gene
silencing significantly inhibited the proliferation, migration and
invasion of hepatocellular carcinoma cells and suppressed tumor
growth, angiogenesis and lung metastasis in a xenograft model of
hepatocellular carcinoma (31,32).
However, CXCR7 is overexpressed in esophageal cancer, particularly
in ESCC (33), suggesting the
critical regulatory effects of CXCR7 on ESCC progression.
In conclusion, CXCR7, as a chemokine receptor, is
tightly implicated in the initiation, adhesion, angiogenesis and
metastasis of various cancers. Previous evidence suggests that
CXCR7 is a potential therapeutic target against cancer (34,35).
in the present study, we showed that CXCR7 is a direct downstream
target of miR-100, and overexpression of miR-100 efficiently
suppresses CXCR7 expression, indicating a great potential for
miR-100 as an anticancer therapeutic candidate.
Acknowledgments
The present study was supported by grants from the
NSFC (31440059), the Project of Construction of Innovative Teams
and Teacher Career Development for Universities and Colleges Under
Beijing Municipality (IDHT20140504), the Beijing Natural Science
Foundation (5153024), and the Beijing City Board of Education
Science and Technology Program (KM201510005027).
References
|
1
|
Demeester SR: Epidemiology and biology of
esophageal cancer. Gastrointest Cancer Res. 3(Suppl): S2–S5.
2009.PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Medina PP and Slack FJ: microRNAs and
cancer: An overview. Cell Cycle. 7:2485–2492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo Y, Chen Z, Zhang L, Zhou F, Shi S,
Feng X, Li B, Meng X, Ma X, Luo M, et al: Distinctive microRNA
profiles relating to patient survival in esophageal squamous cell
carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu Y, Correa AM, Hoque A, Guan B, Ye F,
Huang J, Swisher SG, Wu TT, Ajani JA and Xu XC: Prognostic
significance of differentially expressed miRNAs in esophageal
cancer. Int J Cancer. 128:132–143. 2011. View Article : Google Scholar
|
|
8
|
Li X, Wainscott C and Xi Y: MicroRNA
provides insight into understanding esophageal cancer. Thorac
Cancer. 2:134–142. 2011. View Article : Google Scholar
|
|
9
|
Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen
X, Gao Z, Xin J, Zhou S, Zhou Z, et al: MiR-203 suppresses tumor
growth and invasion and down-regulates miR-21 expression through
repressing Ran in esophageal cancer. Cancer Lett. 342:121–129.
2014. View Article : Google Scholar
|
|
10
|
Qin C, Huang RY and Wang ZX: Potential
role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumour
Biol. 36:1403–1409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao
Y, Sun N, Xu X, Shao K and He J: MicroRNA-99a/100 promotes
apoptosis by targeting mTOR in human esophageal squamous cell
carcinoma. Med Oncol. 30:4112013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Laganà A, Forte S, Russo F, Giugno R,
Pulvirenti A and Ferro A: Prediction of human targets for
viral-encoded microRNAs by thermodynamics and empirical
constraints. J RNAi Gene Silencing. 6:379–385. 2010.PubMed/NCBI
|
|
13
|
Veksler-Lublinsky I, Shemer-Avni Y, Kedem
K and Ziv-Ukelson M: Gene bi-targeting by viral and human miRNAs.
BMC Bioinformatics. 11:2492010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi W, Alajez NM, Bastianutto C, Hui AB,
Mocanu JD, Ito E, Busson P, Lo KW, Ng R, Waldron J, et al:
Significance of Plk1 regulation by miR-100 in human nasopharyngeal
cancer. Int J Cancer. 126:2036–2048. 2010.
|
|
15
|
Fu HL, Wu P, Wang XF, Wang JG, Jiao F,
Song LL, Xie H, Wen XY, Shan HS, Du YX, et al: Altered miRNA
expression is associated with differentiation, invasion, and
metastasis of esophageal squamous cell carcinoma (ESCC) in patients
from Huaian, China. Cell Biochem Biophys. 67:657–668. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang N, Fu H, Song L, Ding Y, Wang X,
Zhao C and Zhao Y, Jiao F and Zhao Y: MicroRNA-100 promotes
migration and invasion through mammalian target of rapamycin in
esophageal squamous cell carcinoma. Oncol Rep. 32:1409–1418.
2014.PubMed/NCBI
|
|
18
|
Gebeshuber CA and Martinez J: miR-100
suppresses IGF2 and inhibits breast tumorigenesis by interfering
with proliferation and survival signaling. Oncogene. 32:3306–3310.
2013. View Article : Google Scholar
|
|
19
|
Bi Y, Jing Y and Cao Y: Overexpression of
miR-100 inhibits growth of osteosarcoma through FGFR3. Tumour Biol.
36:8405–8411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang
J, Yang J, Liao H and Guo L: Downregulation of HOXA1 gene affects
small cell lung cancer cell survival and chemoresistance under the
regulation of miR-100. Eur J Cancer. 50:1541–1554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou HC, Fang JH, Luo X, Zhang L, Yang J,
Zhang C and Zhuang SM: Downregulation of microRNA-100 enhances the
ICMT-Rac1 signaling and promotes metastasis of hepatocellular
carcinoma cells. Oncotarget. 5:12177–12188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang M, Ren D, Guo W, Wang Z, Huang S, Du
H, Song L and Peng X: Loss of miR-100 enhances migration, invasion,
epithelial-mesenchymal transition and stemness properties in
prostate cancer cells through targeting Argonaute 2. Int J Oncol.
45:362–372. 2014.PubMed/NCBI
|
|
23
|
Huang JS, Egger ME, Grizzle WE and McNally
LR: MicroRNA-100 regulates IGF1-receptor expression in metastatic
pancreatic cancer cells. Biotech Histochem. 88:397–402. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Libert F, Parmentier M, Lefort A, Dumont
JE and Vassart G: Complete nucleotide sequence of a putative G
protein coupled receptor: RDC1. Nucleic Acids Res. 18:19171990.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tarnowski M, Liu R, Wysoczynski M,
Ratajczak J, Kucia M and Ratajczak MZ: CXCR7: A new SDF-1-binding
receptor in contrast to normal CD34+ progenitors is
functional and is expressed at higher level in human malignant
hematopoietic cells. Eur J Haematol. 85:472–483. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Naumann U, Cameroni E, Pruenster M,
Mahabaleshwar H, Raz E, Zerwes HG, Rot A and Thelen M: CXCR7
functions as a scavenger for CXCL12 and CXCL11. PLoS One.
5:e91752010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eva C and Sprengel R: A novel putative G
protein-coupled receptor highly expressed in lung and testis. DNA
Cell Biol. 12:393–399. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miao Z, Luker KE, Summers BC, Berahovich
R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A,
Luker GD, et al: CXCR7 (RDC1) promotes breast and lung tumor growth
in vivo and is expressed on tumor-associated vasculature. Proc Natl
Acad Sci USA. 104:15735–15740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun X, Cheng G, Hao M, Zheng J, Zhou X,
Zhang J, Taichman RS, Pienta KJ and Wang J: CXCL12 / CXCR4 / CXCR7
chemokine axis and cancer progression. Cancer Metastasis Rev.
29:709–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Keeley EC, Mehrad B and Strieter RM: CXC
chemokines in cancer angiogenesis and metastases. Adv Cancer Res.
106:91–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xue TC, Chen RX, Han D, Chen J, Xue Q, Gao
DM, Sun RX, Tang ZY and Ye SL: Down-regulation of CXCR7 inhibits
the growth and lung metastasis of human hepatocellular carcinoma
cells with highly metastatic potential. Exp Ther Med. 3:117–123.
2012.PubMed/NCBI
|
|
32
|
Zheng K, Li HY, Su XL, Wang XY, Tian T, Li
F and Ren GS: Chemokine receptor CXCR7 regulates the invasion,
angiogenesis and tumor growth of human hepatocellular carcinoma
cells. J Exp Clin Cancer Res. 29:312010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tachezy M, Zander H, Gebauer F, von Loga
K, Pantel K, Izbicki JR and Bockhorn M: CXCR7 expression in
esophageal cancer. J Transl Med. 11:2382013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Heckmann D, Maier P, Laufs S, Li L,
Sleeman JP, Trunk MJ, Leupold JH, Wenz F, Zeller WJ, Fruehauf S, et
al: The disparate twins: A comparative study of CXCR4 and CXCR7 in
SDF-1α-induced gene expression, invasion and chemosensitivity of
colon cancer. Clin Cancer Res. 20:604–616. 2014. View Article : Google Scholar
|
|
35
|
Guillemot E, Karimdjee-Soilihi B, Pradelli
E, Benchetrit M, Goguet-Surmenian E, Millet MA, Larbret F, Michiels
JF, Birnbaum D, Alemanno P, et al: CXCR7 receptors facilitate the
progression of colon carcinoma within lung not within liver. Br J
Cancer. 107:1944–1949. 2012. View Article : Google Scholar : PubMed/NCBI
|