Introduction
Lung cancers with epidermal growth factor receptor
(EGFR) gene mutations are a well-understood subgroup of lung
adenocarcinomas (LACs). This subgroup is characterized by
sensitivity to EGFR tyrosine kinase inhibitors (TKIs; e.g.
gefitinib or erlotinib), and has high occurrence rates in females,
non-smokers and Asians (1).
However, a median therapy (10–14 months) with EGFR TKI may
aggravate the disease progression in most patients in this
subgroup. The mechanisms of EGFR-TKI resistance are multi-factorial
and several of them have been reported, such as T790M mutation in
exon 20 of EGFR, MET amplification and depletion of phosphatase and
tensin homolog (PTEN) (2–4). However, nearly 30% of resistance
mechanisms are unknown. Thus, elucidating the molecular mechanisms
of EGFR-TKI resistance is essential for the identification of key
biomarkers.
In the last two decades, non-coding RNAs (ncRNAs)
have been gradually accepted as key factors in the process of
epigenetic regulation rather than 'transcription noise' (5). As recently reported, ncRNAs take part
in the pathogenesis of non-small cell lung cancer (NSCLC),
providing new biological insights into this disease (6,7). Long
non-coding RNAs (lncRNAs), which are non-protein coding transcripts
with length >200 nucleotides (nt), are involved in regulating
the occurrence, invasion, metastasis and chemotherapeutic
resistance of lung cancer (6,8,9).
lncRNAs also take part in the regulation of EGFR-TKI resistance.
GAS5, one lncRNA, was found to be overexpressed in
EGFR-TKI-sensitive cells compared with its expression in resistant
cells, and was found to enhance gefitinib-induced cell death in
innate EGFR-TKI-resistant LAC cells with wild-type EGFR via
downregulation of IGF-1R expression (10). These findings indicate that lncRNAs
may be promising biomarkers as diagnostic and therapeutic targets
in the resistance of EGFR-TKIs.
The present study presents the lncRNA expression
profiles in 3 replicate gefitinib-sensitive HCC827 and
gefitinib-resistant HCC827-8-1 cells in pairs by microarray. Then,
7 of the differentially expressed lncRNAs were validated by
real-time quantitative PCR (RT-qPCR) in HCC827 and HCC827-8-1
cells. We also predicted the functions of differentially expressed
lncRNAs through their co-expressed protein-coding genes.
Materials and methods
Cell culture
The human NSCLC H827 cell line harboring the EGFR
exon 19 deletion (Del E746-A750) was obtained from the Shanghai
Institutes for Biological Sciences, Chinese Academy of Cell
Resource Center. We generated a gefitinib-resistant cell line by
exposing HCC827 cells to increasing concentrations of gefitinib as
described in our previous study (11). One individual clone HCC827-8-1 was
isolated and independently confirmed to be resistant to gefitinib.
HCC827-8-1 cells were 348-fold more resistant to gefitinib than the
parental HCC827 cells. The cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad,
CA, USA) at 37°C, in a 5% CO2 and humidified
atmosphere.
RNA extraction
HCC827 and HCC827-8-1 cells were seeded in 6-well
plates (1×105 cells/well) for 72 h in 3 replicate wells,
and then resuspended in 500 μl lysis buffer. Total RNA was
extracted from lysis buffer using the mirVana miRNA Isolation kit
procedure (Applied Biosystem, Foster City, CA, USA), according to
the manufacturer's specifications, and eluted with 100 ml of
nuclease-free water. The yield of RNA was quantified by the
NanoDrop ND 2000 (Thermo Scientific, Waltham, MA, USA) and the RNA
integrity was assessed using Agilent Bioanalyzer 2100 (Agilent
Technologies, Santa Clara, CA, USA).
lncRNA and mRNA microarray expression
profiling
Total RNA was transcribed to double-stranded cDNA,
and then synthesized into cRNA and labeled with cyanine 3-CTP. The
labeled cRNAs were hybridized onto the microarray. The Agilent
human lncRNA array (4×180K) contains 32,776 human mRNAs and 78,243
human lncRNAs, which are derived from authoritative databases,
including RefSeq, Ensemble, GenBank and the Broad Institute. After
washing, the arrays were scanned by the Agilent Scanner G2505C
(Agilent Technologies). Raw datum was extracted using Feature
Extraction (version 10.7.1.1; Agilent Technologies). The microarray
profiling was conducted in the laboratory of the OE Biotechnology
Co. (Shanghai, China).
RT-qPCR validation of 7 differentially
expressed lncRNAs
Total RNA was extracted from HCC827 and HCC827-8-1
cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. One microgram of
total RNA was reverse transcribed in a final volume of 20 μl
using PrimerScript RT Master Mix (Takara, Dalian, China). The
reverse transcription reaction was carried out under the following
conditions: 37°C for 15 min; 85°C for 5 sec, and then hold on 4°C.
The RT-qPCR was performed using SYBR-Green PCR Mix (Roche,
Mannheim, Germany) on the ABI 7900 system (Applied Biosystems)
according to the manufacturer's instructions. The primer sequences
are listed in Table I. β-actin was
used as an internal control to normalize the amount of total RNA in
each sample. Each sample was run in triplicate for analysis. At the
end of the PCR cycles, melting curve analysis was performed to
validate the specific generation of the expected PCR product. The
expression levels of lncRNAs were normalized to internal control
gene β-actin and were calculated using the 2−ΔΔCt method
(12).
 | Table IThe primer sequences used in the
present study. |
Table I
The primer sequences used in the
present study.
| Target ID | Forward primer | Reverse primer |
|---|
| FR165245 |
GAGGGTTTGGCTGTTTGCTG |
ACCCCCACTTAGAGACCAGAA |
|
ENST00000464359 |
GCAACAACCACTTGGCTCAG |
GCAGAGGACACGAACTCACA |
|
ENST00000602301 |
GTTACCTCCTCATGCCGGAC |
AAAAGGGTCAGTAAGCACCCG |
| NONHSAT107900 |
GGCTGCATTTGTTTCTCGCA |
CCCGCCCAGCTATAGTCAAG |
| NONHSAT082241 |
TGCCAAAACTCACCAGCTACA |
GGAGCGGTATGTGCTAGACC |
|
ENST00000434951 |
TGGGAGTGAATGTTCCGGTG |
CAAGAGGAGCTGTTGTTTGTCC |
| NONHSAG031748 |
GGATGTGCACGCATGAACTG |
ACTCCAGCCAAGGTGGTTTT |
| β-actin |
GATGAGATTGGCATGGCTTT |
CACCTTCACCGTTCCAGTTT |
Differential expression level of mRNAs
and lncRNAs from the microarray
After quantile normalization, raw signals from the
microarray were log2 transformed. Differential expression of an
mRNA or lncRNA was defined by the absolute value of fold-change
(FC) >2 (gefitinib-sensitive HCC827 cells=1) and P-value
<0.05 (Student's t-test), with the 3 parallel samples in the
HCC827 or HCC827-8-1 group having detectable signals compared with
the background. The differentially expressed mRNAs were submitted
to the NCBI (gene_go_information) and KEGG database analyzed by
Python program to be classified into different Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation
groups.
Co-expression of lncRNAs with mRNAs and
functional prediction
Most of the lncRNAs in the current databases have
not yet been functionally annotated. Thus, the prediction of their
functions is based on the functional annotations of their
co-expressed mRNAs. This method was originally described by Guttman
et al (13). In brief,
first, for every dysregulated lncRNA, Pearson correlation
coefficient (PCC) of its expression with that of each dysregulated
mRNA was calculated to find its co-expressed mRNAs, with PCC
>0.7 or ≤0.7 and P-value of PCC <0.05 being statistically
significant. Then, a functional enrichment analysis of the
co-expressed mRNAs was conducted using the hypergeometric
cumulative distribution function, and the enriched GO/KEGG pathway
annotations were assigned to the lncRNA as its predicted functions.
The threshold of statistical significance is set as a P-value
<0.05 and false discovery rate (FDR) <0.05 [under the control
of the Benjamini and Hochberg procedure (14)].
In addition, on the basis of co-expression, we
further explored how these dysregulated lncRNAs may exert their
functions through cis- and/or trans-regulating
protein-coding genes. We defined cis-regulated genes as
protein-coding genes co-expressed with one dysregulated lncRNA and
within 300 kb in genomic distance in the same allele.
We potentially defined trans-regulated
protein-coding genes as co-expressed and beyond 300 kb in genomic
distance from, or, on the other allele of, differentially expressed
lncRNAs. In other words, for any protein-coding gene co-expressed
with an lncRNA, if it did not fit the criteria of
cis-regulated, it was categorized as potentially
trans-regulated.
According to Guttman et al (13,15),
specific lncRNAs participating in certain biological pathways are
transcriptionally regulated by key transcription factors (TFs) that
regulate these pathways. Thus, to categorize lncRNAs that possibly
have trans-regulating functions, we compared the mRNAs that
co-expressed with these lncRNAs with the mRNAs that are regulatory
targets of certain TFs. If the intersection of these 2 groups is
large enough (P<0.05; calculated by hypergeometric cumulative
distribution function and FDR <0.05, under the control of the
Benjamini and Hochberg procedure), then, we predict that these
lncRNAs possibly participate in pathways regulated by these TFs.
The lncRNA-TF network was constructed using hypergeometric
cumulative distribution function with the help of Perl. The graph
of the lncRNA-TF network was drawn with the help of Cytoscape
3.01.
Results
General expression profiles of the
differentially expressed lncRNAs and mRNAs
We found that 1,476 lncRNA transcripts were
differentially expressed between the HCC827-8-1 and paired HCC827
cells, with 703 being upregulated and 773 being downregulated.
Among the dysregulated lncRNA transcripts, ENST00000464359 (probe
CUST_89973_PI429545380) was the most upregulated, with an FC of
136.11, whereas ENST00000602301 (probe CUST_1064_PI429545384) was
the most downregulated, FC being 81.46. According to the absolute
value of FC, the dysregulated lncRNA transcripts were stratified
into 4 groups: 2 transcripts with FCs >100, 38 transcripts
between 10 and 100, 117 transcripts between 5 and 10 and 1,319
transcripts between 2 and 5.
Using the same criteria of lncRNAs, we found that
1,026 mRNA transcripts were dysregulated, with 516 being
upregulated and 510 being downregulated. The most upregulated and
downregulated mRNA transcripts were FOXP2 (A_24_P16559) and MCOLN2
(A_23_P23639), with FCs of 74.25 and 31.03, respectively. Table II lists the top 20 upregulated and
downregulated lncRNAs and mRNAs from our microarray.
 | Table IITop 20 upregulated and downregulated
lncRNAs and mRNAs in the HCC827 cells compared with the
gefitinib-resistant HCC827-8-1 cells. |
Table II
Top 20 upregulated and downregulated
lncRNAs and mRNAs in the HCC827 cells compared with the
gefitinib-resistant HCC827-8-1 cells.
Upregulated lncRNAs
| Downregulated
lncRNAs
| Upregulated mRNAs
| Downregulated mRNAs
|
|---|
| lncRNAs | FC | lncRNAs | FC | mRNAs | FC | mRNAs | FC |
|---|
|
ENST00000464359 | 136.11 |
ENST00000602301 | 81.46 | FOXP2 | 74.25 | MCOLN2 | 31.03 |
| NONHSAT130274 | 107.33 | NONHSAG002294 | 68.92 | PYDC2 | 29.44 | RNF183 | 27.43 |
| NONHSAT122833 | 47.29 |
ENST00000542980 | 33.87 | CHI3L1 | 26.90 | THNSL2 | 27.18 |
| NONHSAT122828 | 38.98 | NONHSAT122257 | 23.86 | SFTPB | 19.99 | PGBD5 | 17.46 |
| NONHSAG048587 | 30.11 | NONHSAG032896 | 22.43 | GJA1 | 16.79 | FAM26F | 15.87 |
| uc.225+ | 27.11 | NONHSAT082241 | 22.04 | CTGF | 14.14 | ZNF655 | 15.23 |
| NONHSAT122829 | 22.16 | NONHSAT060965 | 20.40 | CALB1 | 13.04 | IGFBP3 | 15.12 |
| NONHSAT122825 | 19.67 | NONHSAT101176 | 19.36 | ATP8A1 | 12.95 | ZNF560 | 13.81 |
| NONHSAT127642 | 14.70 | NONHSAT118619 | 15.69 | WNT9A | 10.87 | RPS6KA6 | 13.41 |
| NONHSAT093968 | 14.13 |
ENST00000423737 | 15.53 | SPTLC3 | 10.79 | FRMD3 | 13.17 |
| NONHSAT060927 | 13.25 |
ENST00000424690 | 15.04 | SPOCK1 | 9.81 | BACE2 | 13.16 |
| NONHSAT127638 | 12.60 | NONHSAT120476 | 14.96 | SPTLC3 | 9.48 | GPR133 | 12.70 |
| FR173955 | 12.50 | NONHSAG046899 | 14.92 | SFTPB | 9.16 | CYP4F12 | 12.26 |
| NONHSAG050711 | 12.28 | NONHSAT118621 | 14.67 | BMP4 | 8.77 | PTGER2 | 12.23 |
| XR_254376.1 | 11.98 | NONHSAT101177 | 13.85 | AQP3 | 8.62 | CKMT1A | 11.58 |
| NONHSAT107900 | 11.85 | NONHSAT060785 | 12.80 | EGR1 | 8.27 | FAM129A | 11.11 |
| NONHSAT103972 | 11.56 | NONHSAT082244 | 12.59 | TOX2 | 8.20 | C17orf104 | 10.12 |
| uc.222+ | 10.73 | NONHSAT053086 | 12.33 | GMPR | 7.92 | ICAM2 | 9.63 |
| TCONS_00012260 | 10.37 | NONHSAT122256 | 11.48 | ADORA1 | 7.90 | MUC22 | 9.61 |
| FR165245 | 9.81 | NONHSAT004232 | 11.24 | CEACAM6 | 7.81 | LOC643988 | 9.53 |
In the GO pathway analysis, the most common pathways
that the dysregulated mRNAs were involved in included chondroitin
sulfate metabolic process (GO, biological processes), extracellular
space (GO, cellular components) and receptor binding (GO, molecular
functions). The most common KEGG pathways involved were
graft-versus-host disease, type I diabetes mellitus and viral
myocarditis.
In unsupervised hierarchical clustering analysis,
the differentially expressed lncRNAs were used to generate a heat
map; and they clearly self-segregated into HCC827 and HCC827-8-1
clusters, as shown in Fig. 1.
RT-qPCR validation
To validate the results of the microarray, we chose
a total of 7 differentially expressed lncRNA transcripts for
RT-qPCR. They could be divided into 2 groups: the first group,
including FR165245, which was randomly chosen. In the other group,
NONHSAT107900 and NONHSAT082241 were chosen since they were
predicted to have cis-regulating potential. ENST00000434951
and ONHSAG031748 were chosen since they were predicted to have
trans-regulating potential and involved in constituting the
top 100 lncRNA-TF pairs with the most credentiality.
ENST00000464359 and ENST00000602301 were chosen for being the most
upregulated and downregulated lncRNAs (Table II).
The RT-qPCR results were consistent with that of the
microarray, in that all 7 lncRNA transcripts were differentially
expressed with the same trend (upregulated or downregulated) and
reached statistical significance (P<0.05 for each lncRNA;
Student's t-test) as shown in Fig.
2.
lncRNA and mRNA co-expression profiles
and lncRNA function prediction
Hundreds of lncRNAs were co-expressed with thousands
of mRNAs. For example, ENST00000412387 (CUST_86592_PI429545380) was
co-expressed with 3,487 mRNA transcripts and NONHSAT060786
(CUST_61584_PI429545406) with 4,642 mRNA transcripts.
The functions of the differentially expressed
lncRNAs were predicted by the GO and KEGG pathway annotations of
their co-expressed mRNAs. The lncRNAs were clustered into hundreds
of GO and KEGG pathway annotations. Certain pathways are known to
be involved in the mechanism of gefitinib resistance, including
focal adhesion, cell cycle, cell proliferation and apoptosis
(16–18). For example, 186 lncRNAs were
clustered into apoptosis and 79 in the focal adhesion. As expected,
one lncRNA can participate in more than one GO/KEGG pathway
involved in the mechanism of gefitinib resistance. For example,
ENST00000412387 (CUST_86592_PI429545380) was predicted to have
functions in apoptosis, cell cycle, focal adhesion and pathways in
cancer.
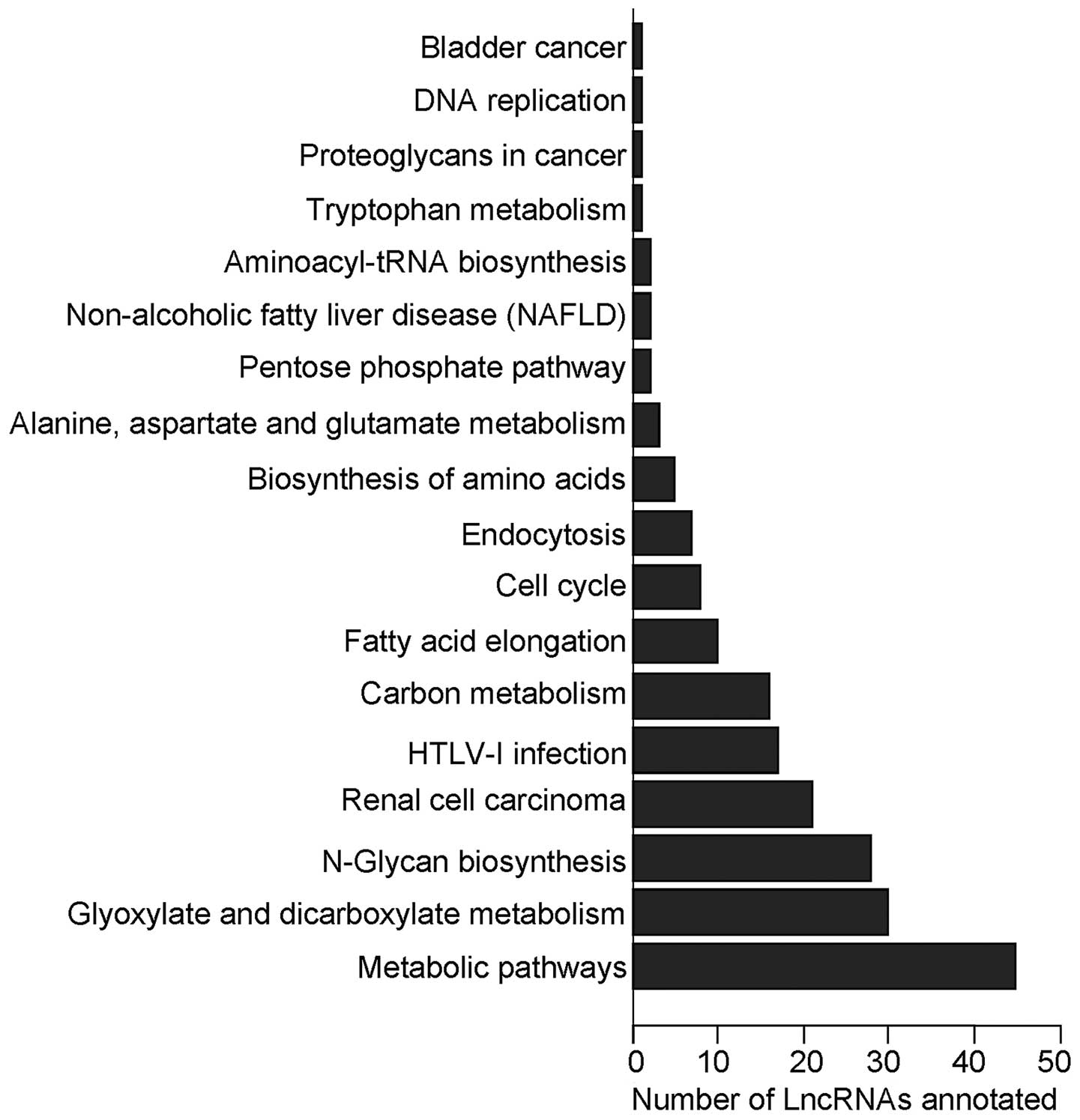
From the matrix of the lncRNAs and their
corresponding KEGG pathway annotations of co-expressed
protein-coding genes, we counted and summarized the top 200
annotations with the most credentiality (the lowest P-values). The
most frequently predicted functions of the differentially expressed
lncRNAs were metabolic pathways, glyoxylate and dicarboxylate
metabolism and N-glycan biosynthesis as shown in Fig. 3.
cis-regulation of lncRNAs
A total of 149 lncRNA transcripts with their
predicted cis-regulated protein-coding genes were found
through accurate genomic mapping, using the above mentioned
criteria. Table III lists all the
lncRNA transcripts and their potentially cis-regulated mRNA
transcripts. For instance, lncRNA NONHSAG010348 was predicted to
cis-regulate five mRNAs (corresponding to five mRNA
transcripts): COPS7A, TPI1, EMG1, USP5 and LRRC23.
 | Table IIIDysregulated lncRNA transcripts and
their potentially cis-regulated mRNA transcripts. |
Table III
Dysregulated lncRNA transcripts and
their potentially cis-regulated mRNA transcripts.
| lncRNA | mRNA | PCC | P-valuea | Chrom |
|---|
|
ENST00000423737 | LOC284930 | 0.990763962 | 0.000128 | 22 |
|
ENST00000425104 | SLC35F3 | −0.871875729 | 0.023572 | 1 |
|
ENST00000431813 | DAPK1 | 0.944254301 | 0.004575 | 9 |
|
ENST00000438810 | LOC284930 | 0.988689739 | 0.000191 | 22 |
|
ENST00000499202 | CD27 | −0.927815863 | 0.007628 | 12 |
|
ENST00000499202 | COPS7A | −0.915741752 | 0.010350 | 12 |
|
ENST00000499202 | GAPDH | −0.820431489 | 0.045472 | 12 |
|
ENST00000499202 | LTBR | −0.958751121 | 0.002517 | 12 |
|
ENST00000499202 | NCAPD2 | −0.926349955 | 0.007937 | 12 |
|
ENST00000499202 | PLEKHG6 | 0.886477026 | 0.018600 | 12 |
|
ENST00000508616 | OR5H15 | −0.914857473 | 0.010565 | 3 |
|
ENST00000510682 | COMMD10 | 0.931176055 | 0.006942 | 5 |
|
ENST00000558536 | C15orf48 | 0.948581359 | 0.003898 | 15 |
|
ENST00000563635 | LMO7 | 0.962112937 | 0.002126 | 13 |
|
ENST00000567305 | RHBDD1 | −0.860850737 | 0.027697 | 2 |
|
ENST00000605692 | ATP6V1F | 0.869839077 | 0.024310 | 7 |
|
ENST00000605692 | CCDC136 | 0.829361383 | 0.041192 | 7 |
|
ENST00000605692 | FAM71F1 | 0.889362332 | 0.017684 | 7 |
|
ENST00000605692 | FAM71F2 | 0.919989247 | 0.009346 | 7 |
|
ENST00000605692 | HILPDA | −0.983130671 | 0.000424 | 7 |
|
ENST00000605692 | IRF5 | 0.95923013 | 0.002459 | 7 |
|
ENST00000605692 | LOC100130705 | 0.954248076 | 0.003092 | 7 |
|
ENST00000606008 | ZNF35 | 0.890496793 | 0.017330 | 3 |
|
ENST00000606008 | ZNF501 | 0.847741084 | 0.033009 | 3 |
| FR009982 | LOC100131490 | −0.86144213 | 0.027467 | 12 |
| FR070335 | ADAMTS15 | 0.915157825 | 0.010492 | 11 |
| FR070335 | ST14 | −0.911123761 | 0.011497 | 11 |
| FR078172 | SENP3 | 0.891854464 | 0.016911 | 17 |
| FR078172 | WRAP53 | 0.851342123 | 0.031506 | 17 |
| FR085258 | PIM3 | 0.89707096 | 0.015346 | 22 |
| FR148984 | COQ10A | −0.918928534 | 0.009592 | 12 |
| FR148984 | ESYT1 | −0.859157792 | 0.028358 | 12 |
| FR148984 | GDF11 | −0.861243067 | 0.027544 | 12 |
| FR148984 | MYL6 | −0.939508853 | 0.005378 | 12 |
| FR148984 | MYL6B | −0.818664102 | 0.046343 | 12 |
| FR148984 | OBFC2B | 0.949121375 | 0.003817 | 12 |
| FR148984 | ORMDL2 | −0.938671908 | 0.005526 | 12 |
| FR148984 | RAB5B | −0.92713364 | 0.007771 | 12 |
| FR171954 | SEMA4B | −0.901277827 | 0.014138 | 15 |
| FR230846 | SERF1B | 0.949144988 | 0.003814 | 5 |
| FR314507 | KCNK6 | −0.914052441 | 0.010763 | 19 |
| FR314507 | SIPA1L3 | 0.900771256 | 0.014281 | 19 |
| FR314507 | SPRED3 | 0.885802839 | 0.018817 | 19 |
| FR331033 | EFNB1 | 0.967665213 | 0.001551 | X |
| NONHSAG002294 | NTNG1 | 0.835837077 | 0.038212 | 1 |
| NONHSAG004747 | LGALS8 | 0.941744836 | 0.004992 | 1 |
| NONHSAG006325 | PPIF | −0.832471447 | 0.039748 | 10 |
| NONHSAG006325 | ZMIZ1 | 0.944693182 | 0.004504 | 10 |
| NONHSAG010348 | COPS7A | 0.984517242 | 0.000358 | 12 |
| NONHSAG010348 | EMG1 | 0.931371288 | 0.006903 | 12 |
| NONHSAG010348 | LRRC23 | 0.900711998 | 0.014298 | 12 |
| NONHSAG010348 | TPI1 | 0.953578734 | 0.003182 | 12 |
| NONHSAG010348 | USP5 | 0.918972659 | 0.009582 | 12 |
| NONHSAG010749 | ARNTL2 | 0.917332369 | 0.009968 | 12 |
| NONHSAG025545 | FXYD5 | 0.924498207 | 0.008336 | 19 |
| NONHSAG025545 | FXYD7 | −0.927092642 | 0.007779 | 19 |
| NONHSAG025545 | ZNF181 | 0.893862023 | 0.016300 | 19 |
| NONHSAG025545 | ZNF30 | 0.93136471 | 0.006905 | 19 |
| NONHSAG025545 | ZNF302 | 0.830684654 | 0.040575 | 19 |
| NONHSAG028996 | IL1RN | 0.956644173 | 0.002779 | 2 |
| NONHSAG034063 | SERHL2 | 0.905613504 | 0.012943 | 22 |
| NONHSAG035134 | HEMK1 | −0.964351703 | 0.001884 | 3 |
| NONHSAG035134 | MAPKAPK3 | 0.817490481 | 0.046925 | 3 |
| NONHSAG037825 | DCAF4L1 | −0.886889957 | 0.018467 | 4 |
| NONHSAG039533 | CYP4V2 | 0.967426574 | 0.001574 | 4 |
| NONHSAG039533 | FAM149A | 0.986343279 | 0.000278 | 4 |
| NONHSAG041216 | MAN2A1 | −0.863109335 | 0.026826 | 5 |
| NONHSAG042317 | C5orf25 | 0.898935246 | 0.014805 | 5 |
| NONHSAG042611 | FOXQ1 | −0.908075468 | 0.012287 | 6 |
| NONHSAG047857 | C7orf42 | 0.950970866 | 0.003547 | 7 |
| NONHSAG048587 | MDFIC | 0.942137396 | 0.004925 | 7 |
| NONHSAG049128 | GIMAP4 | −0.859295231 | 0.028304 | 7 |
| NONHSAG049128 | GIMAP8 | −0.900385381 | 0.014390 | 7 |
| NONHSAG049128 | REPIN1 | −0.912314796 | 0.011196 | 7 |
| NONHSAG052187 | DCAF10 | −0.886559031 | 0.018573 | 9 |
| NONHSAG052187 | RG9MTD3 | 0.974819506 | 0.000943 | 9 |
| NONHSAG052737 | DAPK1 | 0.991651051 | 0.000104 | 9 |
| NONHSAG053520 | COQ4 | 0.816256551 | 0.047541 | 9 |
| NONHSAT002986 | C1orf190 | 0.944107317 | 0.004599 | 1 |
| NONHSAT002986 | LOC100133124 | −0.923578795 | 0.008537 | 1 |
| NONHSAT002986 | NSUN4 | −0.856846302 | 0.029273 | 1 |
| NONHSAT002986 | RAD54L | −0.864794791 | 0.026185 | 1 |
| NONHSAT004254 | CYR61 | 0.990548945 | 0.000134 | 1 |
| NONHSAT004989 | GPSM2 | 0.887936322 | 0.018134 | 1 |
| NONHSAT005081 | CSF1 | 0.870913412 | 0.023920 | 1 |
| NONHSAT005942 | ANKRD34A | 0.91139166 | 0.011429 | 1 |
| NONHSAT005942 | LIX1L | 0.969652404 | 0.001367 | 1 |
| NONHSAT005942 | PDZK1 | 0.982557048 | 0.000454 | 1 |
| NONHSAT005942 | RBM8A | 0.86427635 | 0.026381 | 1 |
| NONHSAT005942 | RNF115 | −0.849741206 | 0.032170 | 1 |
| NONHSAT006288 | C1orf51 | 0.888664251 | 0.017903 | 1 |
| NONHSAT006288 | CA14 | 0.895183669 | 0.015904 | 1 |
| NONHSAT006288 | PRPF3 | 0.812873365 | 0.049248 | 1 |
| NONHSAT006799 | LAMTOR2 | 0.877579069 | 0.021563 | 1 |
| NONHSAT006799 | LMNA | −0.940089303 | 0.005276 | 1 |
| NONHSAT006799 | PMF1-BGLAP | −0.865484674 | 0.025925 | 1 |
| NONHSAT006799 | RXFP4 | −0.816428591 | 0.047455 | 1 |
| NONHSAT006799 | SEMA4A | −0.865518945 | 0.025912 | 1 |
| NONHSAT006799 | SLC25A44 | −0.92529896 | 0.008162 | 1 |
| NONHSAT006799 | TMEM79 | 0.864331726 | 0.026360 | 1 |
| NONHSAT007139 | NDUFS2 | −0.811560982 | 0.049918 | 1 |
| NONHSAT007139 | NIT1 | 0.850528627 | 0.031843 | 1 |
| NONHSAT007139 | TOMM40L | 0.981849759 | 0.000491 | 1 |
| NONHSAT007139 | UFC1 | 0.91161181 | 0.011373 | 1 |
| NONHSAT007139 | USP21 | −0.908563094 | 0.012159 | 1 |
| NONHSAT010406 | LGALS8 | 0.920135293 | 0.009313 | 1 |
| NONHSAT010413 | LGALS8 | 0.934268515 | 0.006339 | 1 |
| NONHSAT010414 | LGALS8 | 0.94625791 | 0.004255 | 1 |
| NONHSAT011117 | AKR1E2 | 0.887463422 | 0.018284 | 10 |
| NONHSAT016347 | DUSP5 | 0.923683122 | 0.008514 | 10 |
| NONHSAT016347 | MXI1 | −0.921167433 | 0.009077 | 10 |
| NONHSAT016347 | RBM20 | 0.816015258 | 0.047662 | 10 |
| NONHSAT018220 | NUCB2 | 0.968114991 | 0.001509 | 11 |
| NONHSAT018821 | LDLRAD3 | 0.830563596 | 0.040631 | 11 |
| NONHSAT021952 | C11orf20 | −0.939345932 | 0.005407 | 11 |
| NONHSAT021952 | COX8A | −0.849229323 | 0.032384 | 11 |
| NONHSAT021952 | FERMT3 | 0.908490858 | 0.012178 | 11 |
| NONHSAT021952 | FKBP2 | 0.880614632 | 0.020529 | 11 |
| NONHSAT021952 | NAA40 | 0.950492833 | 0.003616 | 11 |
| NONHSAT021952 | OTUB1 | 0.96848133 | 0.001474 | 11 |
| NONHSAT021952 | PLCB3 | −0.927522493 | 0.007689 | 11 |
| NONHSAT021952 | PRDX5 | 0.873645855 | 0.022939 | 11 |
| NONHSAT021952 | STIP1 | 0.97759167 | 0.000748 | 11 |
| NONHSAT021952 | VEGFB | −0.98828861 | 0.000205 | 11 |
| NONHSAT023878 | BIRC2 | 0.879025568 | 0.021067 | 11 |
| NONHSAT023878 | C11orf70 | 0.932341202 | 0.006712 | 11 |
| NONHSAT026185 | COPS7A | 0.988490936 | 0.000198 | 12 |
| NONHSAT026185 | EMG1 | 0.942136719 | 0.004925 | 12 |
| NONHSAT026185 | LRRC23 | 0.928925218 | 0.007398 | 12 |
| NONHSAT026185 | PTPN6 | 0.831260781 | 0.040307 | 12 |
| NONHSAT026185 | TPI1 | 0.964345232 | 0.001884 | 12 |
| NONHSAT026185 | USP5 | 0.935201329 | 0.006162 | 12 |
| NONHSAT028274 | SCN8A | −0.926130113 | 0.007984 | 12 |
| NONHSAT028274 | SLC4A8 | 0.88116699 | 0.020343 | 12 |
| NONHSAT028356 | C12orf44 | 0.989758866 | 0.000157 | 12 |
| NONHSAT030840 | LOC100129447 | 0.824754125 | 0.043376 | 12 |
| NONHSAT031072 | HSPB8 | 0.955579328 | 0.002916 | 12 |
| NONHSAT034980 | ITGBL1 | 0.960755647 | 0.002280 | 13 |
| NONHSAT035015 | ERCC5 | 0.814310544 | 0.048520 | 13 |
| NONHSAT037446 | ARG2 | 0.928999759 | 0.007383 | 14 |
| NONHSAT037446 | PLEKHH1 | 0.983562296 | 0.000403 | 14 |
| NONHSAT037452 | ARG2 | 0.937491109 | 0.005739 | 14 |
| NONHSAT037452 | PLEKHH1 | 0.974920148 | 0.000936 | 14 |
| NONHSAT051795 | NME4 | 0.885330103 | 0.018970 | 16 |
| NONHSAT051795 | RAB40C | 0.889230112 | 0.017725 | 16 |
| NONHSAT051795 | WFIKKN1 | 0.837341388 | 0.037535 | 16 |
| NONHSAT054629 | LOC100130580 | 0.875832347 | 0.022169 | 17 |
| NONHSAT054629 | PDK2 | 0.895328542 | 0.015861 | 17 |
| NONHSAT054629 | RSAD1 | −0.87520844 | 0.022388 | 17 |
| NONHSAT054629 | SPATA20 | 0.868769216 | 0.024702 | 17 |
| NONHSAT054629 | TMEM92 | 0.988506098 | 0.000197 | 17 |
| NONHSAT056331 | CCDC40 | 0.938792121 | 0.005505 | 17 |
| NONHSAT056331 | SLC26A11 | 0.868405134 | 0.024836 | 17 |
| NONHSAT060927 | ANGPTL4 | 0.946117654 | 0.004277 | 19 |
| NONHSAT060927 | HNRNPM | 0.913205652 | 0.010973 | 19 |
| NONHSAT060927 | 2-Mar | 0.8868303 | 0.018486 | 19 |
| NONHSAT060927 | OR2Z1 | 0.917317857 | 0.009972 | 19 |
| NONHSAT060927 | RAB11B | 0.95392537 | 0.003135 | 19 |
| NONHSAT066483 | CEACAM3 | 0.990570871 | 0.000133 | 19 |
| NONHSAT066483 | CEACAM6 | 0.974843312 | 0.000941 | 19 |
| NONHSAT074144 | WBP11 | −0.924683038 | 0.008295 | 2 |
| NONHSAT075349 | PHOSPHO2 | 0.918978696 | 0.009581 | 2 |
| NONHSAT077950 | FARP2 | 0.842995975 | 0.035040 | 2 |
| NONHSAT077950 | ING5 | 0.919706694 | 0.009412 | 2 |
| NONHSAT077950 | LOC100129675 | 0.945435006 | 0.004385 | 2 |
| NONHSAT082241 | BACE2 | 0.97176878 | 0.001184 | 21 |
| NONHSAT083016 | COL6A2 | 0.84242078 | 0.035290 | 21 |
| NONHSAT086623 | LOC100652769 | 0.947578927 | 0.004050 | 22 |
| NONHSAT086623 | NOL12 | 0.831885348 | 0.040018 | 22 |
| NONHSAT086623 | SH3BP1 | 0.964024253 | 0.001918 | 22 |
| NONHSAT086623 | TRIOBP | 0.956519314 | 0.002795 | 22 |
| NONHSAT086830 | FAM83F | 0.935060728 | 0.006189 | 22 |
| NONHSAT089680 | KLHDC8B | 0.968550457 | 0.001468 | 3 |
| NONHSAT089680 | TCTA | 0.820457886 | 0.045459 | 3 |
| NONHSAT090112 | PXK | 0.98306533 | 0.000428 | 3 |
| NONHSAT092654 | TM4SF4 | −0.900748794 | 0.014287 | 3 |
| NONHSAT092847 | GMPS | 0.890300599 | 0.017391 | 3 |
| NONHSAT093875 | HRG | −0.937122594 | 0.005806 | 3 |
| NONHSAT093875 | ST6GAL1 | 0.987402781 | 0.000237 | 3 |
| NONHSAT093968 | CCDC50 | 0.884575249 | 0.019215 | 3 |
| NONHSAT093968 | PYDC2 | 0.963323686 | 0.001993 | 3 |
| NONHSAT094657 | FGFRL1 | −0.957980604 | 0.002611 | 4 |
| NONHSAT094657 | SPON2 | 0.925043387 | 0.008217 | 4 |
| NONHSAT095682 | FLJ39653 | 0.924889987 | 0.008250 | 4 |
| NONHSAT096163 | LIMCH1 | 0.972549746 | 0.001120 | 4 |
| NONHSAT096163 | UCHL1 | 0.918539886 | 0.009683 | 4 |
| NONHSAT096168 | DCAF4L1 | −0.868194518 | 0.024914 | 4 |
| NONHSAT096168 | LIMCH1 | 0.991159228 | 0.000117 | 4 |
| NONHSAT099638 | CYP4V2 | 0.942816522 | 0.004811 | 4 |
| NONHSAT099638 | FAM149A | 0.986547438 | 0.000270 | 4 |
| NONHSAT099643 | CYP4V2 | 0.969996619 | 0.001337 | 4 |
| NONHSAT099643 | FAM149A | 0.920009458 | 0.009342 | 4 |
| NONHSAT101145 | PTGER4 | 0.876795864 | 0.021834 | 5 |
| NONHSAT105337 | C5orf25 | 0.946965914 | 0.004144 | 5 |
| NONHSAT107900 | GMPR | 0.924577154 | 0.008318 | 6 |
| NONHSAT107917 | CAP2 | 0.969062042 | 0.001421 | 6 |
| NONHSAT107973 | KDM1B | 0.946171234 | 0.004268 | 6 |
| NONHSAT113449 | C6orf57 | 0.849248329 | 0.032376 | 6 |
| NONHSAT114226 | PRDM1 | 0.938342631 | 0.005585 | 6 |
| NONHSAT119003 | C7orf26 | 0.987782187 | 0.000223 | 7 |
| NONHSAT119003 | RAC1 | 0.935823609 | 0.006046 | 7 |
| NONHSAT119451 | DNAH11 | 0.985235314 | 0.000325 | 7 |
| NONHSAT119452 | DNAH11 | 0.980786037 | 0.000550 | 7 |
| NONHSAT119495 | KLHL7 | −0.917044909 | 0.010037 | 7 |
| NONHSAT120737 | CCT6A | 0.898610052 | 0.014899 | 7 |
| NONHSAT120737 | GBAS | 0.948686683 | 0.003882 | 7 |
| NONHSAT120737 | MRPS17 | 0.811824769 | 0.049783 | 7 |
| NONHSAT120737 | SUMF2 | 0.948263714 | 0.003946 | 7 |
| NONHSAT121617 | ZP3 | 0.861523702 | 0.027436 | 7 |
| NONHSAT122253 | CPSF4 | 0.908475946 | 0.012182 | 7 |
| NONHSAT122253 | ZNF655 | 0.949103779 | 0.003820 | 7 |
| NONHSAT122253 | ZNF789 | −0.94433753 | 0.004561 | 7 |
| NONHSAT122254 | CPSF4 | 0.937348472 | 0.005765 | 7 |
| NONHSAT122254 | ZNF655 | 0.949540597 | 0.003755 | 7 |
| NONHSAT122254 | ZNF789 | −0.841096553 | 0.035869 | 7 |
| NONHSAT122256 | CPSF4 | 0.94869328 | 0.003881 | 7 |
| NONHSAT122256 | ZNF655 | 0.982379785 | 0.000463 | 7 |
| NONHSAT122256 | ZNF789 | −0.891613102 | 0.016985 | 7 |
| NONHSAT122257 | CPSF4 | 0.979295885 | 0.000639 | 7 |
| NONHSAT122257 | ZNF789 | −0.90462309 | 0.013211 | 7 |
| NONHSAT122826 | FOXP2 | 0.984524991 | 0.000357 | 7 |
| NONHSAT122828 | FOXP2 | 0.963307259 | 0.001995 | 7 |
| NONHSAT122828 | MDFIC | 0.957629177 | 0.002655 | 7 |
| NONHSAT122829 | FOXP2 | 0.972231129 | 0.001146 | 7 |
| NONHSAT122829 | MDFIC | 0.890381753 | 0.017366 | 7 |
| NONHSAT122833 | MDFIC | 0.925397662 | 0.008141 | 7 |
| NONHSAT122928 | CAPZA2 | 0.959462687 | 0.002432 | 7 |
| NONHSAT122928 | ST7 | 0.98816268 | 0.000209 | 7 |
| NONHSAT122929 | CAPZA2 | 0.953017879 | 0.003259 | 7 |
| NONHSAT123242 | ATP6V1F | 0.936690382 | 0.005885 | 7 |
| NONHSAT123242 | CCDC136 | 0.869648072 | 0.024380 | 7 |
| NONHSAT123242 | HILPDA | −0.843372365 | 0.034877 | 7 |
| NONHSAT123242 | LOC100130705 | 0.843594245 | 0.034781 | 7 |
| NONHSAT125539 | LOC254896 | 0.873918742 | 0.022843 | 8 |
| NONHSAT125539 | TNFRSF10C | 0.976171332 | 0.000845 | 8 |
| NONHSAT127008 | ADHFE1 | 0.832589152 | 0.039694 | 8 |
| NONHSAT127008 | RRS1 | 0.827125523 | 0.042245 | 8 |
| NONHSAT129523 | LY6E | 0.868444037 | 0.024822 | 8 |
| NONHSAT129523 | ZFP41 | 0.875838462 | 0.022167 | 8 |
| NONHSAT129523 | ZNF696 | −0.885112169 | 0.019041 | 8 |
| NONHSAT134920 | LCN2 | 0.960849658 | 0.002269 | 9 |
| NONHSAT134920 | ODF2 | 0.86450331 | 0.026295 | 9 |
| NONHSAT136475 | SMS | −0.976071345 | 0.000852 | X |
| NONHSAT136568 | IL1RAPL1 | 0.976788703 | 0.000802 | X |
| NONHSAT140499 | ATF7IP2 | 0.881735051 | 0.020153 | 16 |
| NONHSAT140500 | ATF7IP2 | 0.913532878 | 0.010892 | 16 |
| NONHSAT140506 | ATF7IP2 | 0.839342422 | 0.036643 | 16 |
| NONHSAT140507 | ATF7IP2 | 0.835899063 | 0.038184 | 16 |
| NONHSAT142843 | CCDC113 | −0.952964178 | 0.003267 | 16 |
| NONHSAT142843 | NDRG4 | 0.983697804 | 0.000396 | 16 |
| NONHSAT142849 | CCDC113 | −0.959233693 | 0.002459 | 16 |
| NONHSAT145291 | TXNDC17 | 0.914040222 | 0.010766 | 17 |
| NONHSAT145291 | XAF1 | 0.990386145 | 0.000138 | 17 |
| NR_002834.1 | OBSCN | 0.855142567 | 0.029956 | 1 |
| NR_002834.1 | RNF187 | −0.865681416 | 0.025851 | 1 |
| NR_027459.2 | SYT14 | 0.871422894 | 0.023735 | 1 |
| NR_027621.1 | GPR112 | −0.818615818 | 0.046367 | X |
| NR_040072.1 | CCDC113 | −0.975161687 | 0.000918 | 16 |
| NR_040072.1 | NDRG4 | 0.989689573 | 0.000159 | 16 |
| NR_046396.1 | LOC100652883 | −0.816083992 | 0.047627 | 17 |
| NR_046396.1 | SLC16A13 | −0.852523846 | 0.031020 | 17 |
| NR_046396.1 | TXNDC17 | 0.93099277 | 0.006979 | 17 |
| NR_046396.1 | XAF1 | 0.97916361 | 0.000647 | 17 |
| NR_073032.1 | KCNK6 | −0.945478809 | 0.004378 | 19 |
| NR_073032.1 | SIPA1L3 | 0.942774847 | 0.004818 | 19 |
| NR_073032.1 | SPRED3 | 0.955022355 | 0.002989 | 19 |
| NR_102308.1 | MYADM | 0.856205852 | 0.029529 | 19 |
| NR_102308.1 | PRKCG | 0.950132007 | 0.003668 | 19 |
| NR_102308.1 | TSEN34 | 0.863372316 | 0.026725 | 19 |
| NR_103533.1 | RUNX2 | −0.823610352 | 0.043926 | 6 |
| TCONS_00001307 | SRP9 | 0.881615264 | 0.020193 | 1 |
| TCONS_00009873 | CCT5 | −0.874191191 | 0.022746 | 5 |
| TCONS_00009873 | 6-Mar | 0.886701263 | 0.018528 | 5 |
| TCONS_00012995 | ANLN | −0.859883705 | 0.028073 | 7 |
| TCONS_00013626 | GIMAP4 | −0.815740282 | 0.047800 | 7 |
| TCONS_00013626 | GIMAP8 | −0.857174368 | 0.029142 | 7 |
| TCONS_00013626 | REPIN1 | −0.931532021 | 0.006871 | 7 |
| TCONS_00026596 | CCDC165 | 0.877457033 | 0.021605 | 18 |
| TCONS_00026596 | LOC284219 | 0.870249439 | 0.024161 | 18 |
| TCONS_00027489 | CYP4F12 | 0.968533169 | 0.001470 | 19 |
| TCONS_00027489 | CYP4F3 | 0.979154806 | 0.000647 | 19 |
| TCONS_00027489 | CYP4F8 | 0.831544704 | 0.040176 | 19 |
| TCONS_00027489 | TPM4 | 0.830964632 | 0.040445 | 19 |
|
TCONS_l2_00015621 | PSMD1 | 0.869747246 | 0.024344 | 2 |
|
TCONS_l2_00021700 | C4orf29 | 0.873207805 | 0.023095 | 4 |
Trans-regulation of the lncRNAs
Using the threshold of P<0.05 and FDR <0.01,
3,429 lncRNA-TF pairs were found, corresponding to 48 TFs. Then, we
generated a core network using the top 100 lncRNA-TF pairs with the
most credentiality (lowest P-values and FDRs), as shown by Fig. 4 and are listed in Table IV. We can see that most of these
potential trans-regulatory lncRNAs participate in pathways
regulated by four TFs: E2F transcription factor 1 (E2F1), E2F
transcription factor 4 (E2F4), upstream transcription factor 1
(USF1) and transcription factor AP-2γ (TFAP2C). In the core network
of lncRNA-TF pairs, E2F1 participates in 31 of the 100 pairs, E2F4
in 10 pairs, USF1 in 17 pairs and TFAP2C in 30 pairs.
 | Table IVThe top 100 lncRNA-TF pairs with the
most credentiality. |
Table IV
The top 100 lncRNA-TF pairs with the
most credentiality.
| lncRNA | TF | P-value | FDR |
|---|
| NONHSAG029883 | E2F1 | 0.000100199 | 0.011522898 |
| NONHSAT098423 | E2F1 | 0.00010995 | 0.012644217 |
| FR165245 | E2F1 | 0.000114954 | 0.013219731 |
| NONHSAT001522 | E2F4 | 0.00011977 | 0.005932082 |
|
TCONS_l2_00015621 | YY1 | 0.000120282 | 0.013591822 |
| NONHSAG035406 | E2F1 | 0.000120559 | 0.013623208 |
| NONHSAT005942 | E2F1 | 0.000133523 | 0.015355119 |
| NONHSAT008438 | USF1 | 0.000134804 | 0.007751239 |
| NONHSAT030840 | E2F4 | 0.000136262 | 0.010899038 |
| NONHSAG050806 | TFAP2C | 0.000140853 | 0.008099028 |
| NONHSAT122253 | TFAP2C | 0.000141107 | 0.008113627 |
| NONHSAT011117 | USF1 | 0.00014186 | 0.008156953 |
| NONHSAT093783 | E2F1 | 0.000148833 | 0.016966919 |
|
ENST00000508616 | E2F1 | 0.000150861 | 0.017047245 |
| TCONS_00001679 | ATF3 | 0.000153448 | 0.005882192 |
| NONHSAT001522 | E2F1 | 0.00015475 | 0.005932082 |
| NONHSAT081186 | USF1 | 0.000163155 | 0.009218242 |
| NR_027459.2 | E2F1 | 0.000165025 | 0.018977858 |
|
ENST00000420044 | TFAP2C | 0.000165234 | 0.009500979 |
| NONHSAT092847 | E2F1 | 0.000170233 | 0.00970326 |
| FR009982 | USF1 | 0.000174482 | 0.02006547 |
|
ENST00000598393 | E2F1 | 0.000175642 | 0.010099417 |
| NONHSAT028356 | USF1 | 0.000182684 | 0.010504343 |
|
ENST00000438810 | TFAP2C | 0.000184836 | 0.010628064 |
| NONHSAT030840 | E2F1 | 0.000189548 | 0.010899038 |
| NONHSAT095682 | TFAP2C | 0.000191811 | 0.01102914 |
| NONHSAT129523 | ELK4 | 0.000192126 | 0.022094508 |
| NONHSAT010620 | E2F1 | 0.000194595 | 0.02198926 |
|
ENST00000533220 | E2F4 | 0.000195146 | 0.011123299 |
| NONHSAT017390 | E2F1 | 0.000201551 | 0.022372179 |
| NONHSAG001064 | E2F1 | 0.000201777 | 0.022800789 |
| NONHSAT140499 | TFAP2C | 0.00020195 | 0.011612109 |
| FR042098 | USF1 | 0.000202339 | 0.011634494 |
|
TCONS_l2_00021700 | PPARGC1A | 0.000216816 | 0.024283375 |
| NONHSAT120476 | TFAP2C | 0.00022388 | 0.012761147 |
| TCONS_00016368 | E2F4 | 0.000243819 | 0.027795322 |
| NONHSAT037766 | ELK4 | 0.000256704 | 0.017378335 |
| NONHSAG031748 | USF1 | 0.000264772 | 0.023070091 |
| FR224199 | E2F1 | 0.000266261 | 0.030619965 |
| NONHSAG003987 | TFAP2C | 0.000275049 | 0.015815294 |
|
TCONS_l2_00020217 | E2F1 | 0.000276553 | 0.031250531 |
| NONHSAT090951 | E2F4 | 0.000283269 | 0.032575881 |
| NONHSAT138382 | E2F1 | 0.000291224 | 0.032617038 |
| NONHSAT031072 | TFAP2C | 0.00029954 | 0.017223562 |
| NONHSAT118621 | TFAP2C | 0.00029985 | 0.017091435 |
| NONHSAT037766 | E2F1 | 0.000302232 | 0.017378335 |
| NONHSAT122928 | TFAP2C | 0.000307854 | 0.017701585 |
| NONHSAT026185 | TFAP2C | 0.000308128 | 0.017717359 |
|
ENST00000412387 | E2F4 | 0.000310984 | 0.035763147 |
|
ENST00000602721 | TFAP2C | 0.000311238 | 0.017896208 |
|
ENST00000434951 | TFAP2C | 0.000318988 | 0.020166596 |
| NR_002834.1 | USF1 | 0.000319364 | 0.018363452 |
| NONHSAT030171 | USF1 | 0.00032151 | 0.036652133 |
| NONHSAT035083 | E2F1 | 0.000325007 | 0.03672582 |
| NONHSAT060965 | TFAP2C | 0.00032951 | 0.018946816 |
| NONHSAT093783 | E2F4 | 0.000334903 | 0.019089467 |
| TCONS_00013626 | E2F1 | 0.00034006 | 0.039106915 |
| NONHSAG039347 | E2F1 | 0.000343688 | 0.039524106 |
| FR314507 | ZBTB7A | 0.000348142 | 0.020018175 |
| NONHSAT022221 | PPARGC1A | 0.000348948 | 0.019715575 |
|
ENST00000434951 | TFAP2A | 0.0003538 | 0.020166596 |
| NR_073125.1 | TFAP2C | 0.000355186 | 0.020068009 |
| FR070335 | E2F1 | 0.000360267 | 0.041430707 |
| NONHSAT026990 | USF1 | 0.000375935 | 0.021616259 |
| NONHSAT136810 | TFAP2C | 0.00037671 | 0.021660808 |
| NONHSAT010513 | E2F1 | 0.000396964 | 0.045650912 |
| TCONS_00029195 | TFAP2C | 0.000398087 | 0.015818714 |
| NONHSAT119003 | TFAP2C | 0.000403253 | 0.022985416 |
| FR055729 | E2F1 | 0.000403717 | 0.0464274 |
|
ENST00000417120 | E2F1 | 0.000406492 | 0.045527124 |
| NONHSAG031748 | E2F4 | 0.00040832 | 0.023070091 |
| NONHSAT130274 | TFAP2C | 0.00040865 | 0.023497401 |
| TCONS_00029195 | TFAP2A | 0.000412662 | 0.015818714 |
| NONHSAT096163 | E2F4 | 0.000424883 | 0.024218343 |
| NONHSAT056714 | TFAP2C | 0.000425979 | 0.024493803 |
| NONHSAG037825 | USF1 | 0.000427744 | 0.024595276 |
| NR_104426.1 | TFAP2C | 0.000431261 | 0.024581897 |
| NONHSAT122833 | TFAP2C | 0.000436645 | 0.025107059 |
| TCONS_00027489 | TFAP2C | 0.000442076 | 0.025419365 |
| NONHSAT093875 | USF1 | 0.000443636 | 0.02550908 |
| NONHSAT140507 | E2F1 | 0.00045296 | 0.030884797 |
| NONHSAT023895 | USF1 | 0.000454218 | 0.051326598 |
| NONHSAT077173 | ATF3 | 0.000457842 | 0.026325887 |
| NONHSAT093946 | TFAP2C | 0.000459005 | 0.026392802 |
| NONHSAT119495 | E2F1 | 0.000466442 | 0.053640826 |
| NONHSAT136475 | E2F4 | 0.000466481 | 0.026589441 |
| NONHSAT096168 | TFAP2C | 0.000466948 | 0.026616051 |
| NONHSAG046899 | USF1 | 0.000471352 | 0.026867058 |
| NONHSAG050557 | E2F1 | 0.000472068 | 0.05428779 |
| NONHSAT032403 | E2F1 | 0.000478104 | 0.054025738 |
|
ENST00000561486 | TFAP2C | 0.000479431 | 0.027567279 |
| TCONS_l2_00015621
U | SF1 | 0.000487468 | 0.02320976 |
| NONHSAT006799 | TFAP2C | 0.000492218 | 0.028302546 |
| NONHSAT034980 | E2F1 | 0.000493572 | 0.056760748 |
| NONHSAT004232 | TFAP2C | 0.000505112 | 0.028791388 |
| NONHSAT079625 | USF1 | 0.000516961 | 0.058933596 |
| NONHSAT095682 | TFAP2A | 0.000525617 | 0.020148659 |
| NONHSAT077174 | TFAP2C | 0.000525879 | 0.024031165 |
| FR280917 | USF1 | 0.00052622 | 0.029994527 |
| TCONS_00005559 | TFAP2A | 0.000534675 | 0.030743841 |
Discussion
We assessed genome-wide lncRNA expression patterns
in gefitinib-resistant human NSCLC HCC827-8-1 cells, compared with
their parental HCC827 cells by microarray and explored their
possible functions by analyzing their co-expressed protein-coding
mRNAs. HCC827-8-1 is an individual clone isolated from resistant
HCC827 cells, which is 348-fold more resistant to gefitinib than
HCC827 cells with high stability (11). We used HCC827-8-1 cells to study the
EGFR-TKI resistance mechanisms and to obtain more accurate
results.
The microarray showed that 1,476 lncRNA and 1,026
mRNA transcripts were dysregulated. Then we chose and validated 7
of the differentially expressed lncRNAs by RT-qPCR. Hundreds of
lncRNAs were co-expressed with thousands of mRNAs, and some of the
lncRNAs may be active in the mechanism of gefitinib resistance
through cis-regulation and/or in trans-regulation of
these mRNAs.
We predicted the roles of lncRNAs through the
co-expressed mRNAs. The most widely used technique for function
prediction is the 'guilt by association' method, which aims to
interrogate the co-expressed protein-coding genes and relevant
bio-pathways (13). Among the
hundreds of pathways as-predicted, some are key pathways in the
mechanisms of gefitinib resistance, such as focal adhesion, cell
proliferation, cell cycle and apoptosis (16–18).
We compared the differentially expressed mRNAs with other studies,
and the high consistency strongly supported our predictions. For
example, in the mRNA expression analysis using Agilent SurePrint G3
Human Gene Expression 8×60K array of a gefitinib-resistant lung
cancer cell line (19), IGFBP3 was
found to be expressed differentially, which is in accordance with
our results. Additionally, the constitutive activation of EGFR
tyrosine kinase receptor caused by mutation and/or amplification is
closely correlated with the development, progression and poor
prognosis of NSCLC, through the activation of various downstream
signaling pathways. These pathways include the RAS/RAF/MAPK pathway
that induces signals associated with proliferative activity, and
the PI3K/AKT pathway that results in an anti-apoptotic effect
(16). In the present study, the
expression levels of AKT and RAS were higher in the HCC827-8-1
cells than that in the HCC827 cells.
cis-regulation is the intrinsic property of
ncRNAs, since mRNAs are only effective in dissociation,
transportation and translation (5).
Moreover, the cis-regulation of nearby protein-coding genes
is a common mechanism of lncRNAs (20). In the present study, we identified
149 lncRNA transcripts that were able to cis-regulate the
nearby protein-coding genes. Although most of the 149 lncRNAs were
uncharacterized, several of the lncRNA-mRNAs attracted our
interest. lncRNAs that are related to dysregulated mRNAs known to
be involved in EGFR-TKI resistance are valuable for further study.
For example, one lncRNA is NONHSAT021952 (CUST_7952_PI429545402),
which probably cis-regulates VEGF-B mRNA. As reported,
VEGF-B plays important roles in promoting cancer metastasis through
the remodeling of microvasculature of NSCLC, which is related to
EGFR-TKI resistance (21).
NONHSAT021952 (chr11: 63988306-63990592+) has no annotated
functions and lies near VEGF-B (chr11: 64006111-64006170+) in the
same chromosome. Thus, there may be a new way that nearby lncRNAs
regulate the expression of VEGF-B.
Alhough some lncRNAs are proven as
cis-regulators, the vast majority of lncRNAs with known
functions are actually trans-regulators (22). In contrast to cis-regulation,
the trans-regulatory lncRNAs, after isolation from the
synthesis site, affect the genes at a long genomic distance away,
accumulate and take effect in large regulatory networks (5). We predicted the functions of
trans-regulatory lncRNAs through the relevant
expression-regulating TFs. In the core network of lncRNA-TF pairs,
the lncRNAs were divided into four groups of pathways that are
controlled by E2F4, E2F1, USF1 and TFAP2C, respectively.
E2F4 and E2F1 which belong to the E2F family are
proposed to be primary transcription repressors. The E2F family is
pivotal in the regulation of cell cycle and the action of tumor
suppressor proteins. The E2F family is also a target of the
transforming proteins of small DNA tumor viruses (23).
E2F1 regulates the transcription of DNA
synthesis-related genes and thus, the binding of hypophosphorylated
pRB to E2F1 will arrest G1 phase cells (24). Gefitinib treatment downregulates the
expression of E2F1 mRNA and protein. The antiproliferative effect
of gefitinib is attributed, or at least partially, to the
inhibition of E2F1 expression and increases the proportion of G1
phase cells in human LACs (A431 and A549) (25). E2F1 is also downregulated by
gefitinib treatment in gefitinib-resistant NSCLC cells with MET
amplification, but not in resistant NSCLC cells harboring the T790M
mutation of EGFR (26). The above
studies indicate that E2F1 is closely associated with the
resistance to EGFR-TKIs. The lncRNAs predicted as regulatory
targets of E2F1 may take part in the acquisition of gifitinib
resistance, which should be validated by further functional
tests.
E2F4 takes part in diverse functions, such as cell
cycle control, DNA damage repair, apoptosis, mRNA processing and
ubiquitination (27,28). The upregulation of E2F4 nuclear
expression in breast cancer is related to poor prognosis in
patients with larger tumors, recurrence, distant metastasis and
poorer outcome (29). E2F4 protein
expression in human colorectal tumors is also upregulated by
affecting the G1/S phase transition and the proliferation of colon
cancer cells (30). In comparison,
E2F4 protein expression is down-regulated in sporadic Burkitt
lymphoma (sBL) cells but not in immortal B-cells. Besides the
ability to reduce E2F1 levels, E2F4 expression in BL cells also
selectively weakens the tumorigenic properties and BL cell
proliferation and increases the proportions of G2/M
cells (31). Since E2F4 has diverse
effects in the regulation of tumor progression, it may also play a
role in lung cancer.
USF1 is a basic helix-loop-helix (bHLH) TF encoded
by distinct genes that are heterodimerized and functionally
overlapped (32). USF1 also
mediates the transcription of many neuropeptides (33–36),
surfactant protein A in the lung (37), genes in lung tumors (38) which include hTERT as the
immortalizing telomerase subunit (39,40)
and CDK4 as the cell cycle regulator (41). USF1 can also regulate neuropeptide
genes in lung cancer, particularly arginine vasopressin (AVP)
(42,43). USF1 mRNA expression is downregulated
in LACs compared with that in normal tissues (44). In lung cancer, USF1 and USF2
dimerize to mediate the transcription via E-box motifs in target
genes (45). Moreover, with the
E-box motif, USF1 activates GATA5 gene expression (46), which is pivotal in cell
differentiation and tissue development in lung cancer (47,48).
Since USF1 is closely related to lung cancer occurrence,
USF1-regulated lncRNAs may be involved in EGFR-TKI resistance.
TFAP2C, also known as AP2-γ, belongs to the AP-2 TF
family, which is composed of five closely related 50-kDa isoforms
(49). By regulating the expression
of many downstream genes, TFAP2C mediates various cellular
processes, including induction, differentiation, survival,
proliferation and apoptosis under diverse developmental contexts
(50–54). The internalization of activated EGFR
involves the AP-2 complex (55).
TFAP2C is pivotal in human cancer development. For example, through
transcriptional regulation of EGFR, TFAP2C mediates the tumor
development, cell growth and survival of HER2-amplified breast
cancer (56). Moreover, after 10
years of diagnosis, higher TFAP2C scores are correlated with poor
overall survival in ERα-positive and endocrine therapy-treated
patients (57). Since TFAP2C
affects tumorigenesis by mediating EGFR expression, TFAP2C may be
an important cause of EGFR-TKI-resistant lung cancer. Thus, the
lncRNAs that take part in the TFAP2C-regulated pathways, as
predicted, are candidate participants in the mechanisms of EGFR-TKI
resistance.
The present study has also a few limitations.
Firstly, we only used one cell line to study the mechanisms of
EGFR-TKI resistance in vitro. Our results are not integrated
but typical, and thus further studies with larger sample size in
vivo are needed. Second, we only predicted lncRNA functions
indirectly through network and pathway analyses of co-expressed
protein-coding genes. The reason is that the majority of the
lncRNAs identified to date are not functionally characterized
(58). The 'guilt by association''
method for hypothesis generation is a key primary step for
functional studies in the future, such as loss/acquisition of
functions (59).
In conclusion, nearly 1,500 lncRNAs were differently
expressed between the gefitinib-sensitive HCC827 cells and the
gefitinib-resistant HCC827-8-1 cells. Many of these lncRNAs play
important roles in regulating EGFR-TKI resistance through the
cis- and/or trans-regulation of target protein-coding
genes. The present study underlies future functional studies on
lncRNAs related to EGFR resistance and provides a candidate
reservoir for their application as diagnostic and therapeutic
targets.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (81372396).
References
|
1
|
Jackman DM, Miller VA, Cioffredi LA, Yeap
BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV and Johnson
BE: Impact of epidermal growth factor receptor and KRAS mutations
on clinical outcomes in previously untreated non-small cell lung
cancer patients: Results of an online tumor registry of clinical
trials. Clin Cancer Res. 15:5267–5273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Godin-Heymann N, Bryant I, Rivera MN,
Ulkus L, Bell DW, Riese DJ II, Settleman J and Haber DA: Oncogenic
activity of epidermal growth factor receptor kinase mutant alleles
is enhanced by the T790M drug resistance mutation. Cancer Res.
67:7319–7326. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McDermott U, Pusapati RV, Christensen JG,
Gray NS and Settleman J: Acquired resistance of non-small cell lung
cancer cells to MET kinase inhibition is mediated by a switch to
epidermal growth factor receptor dependency. Cancer Res.
70:1625–1634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamamoto C, Basaki Y, Kawahara A,
Nakashima K, Kage M, Izumi H, Kohno K, Uramoto H, Yasumoto K,
Kuwano M, et al: Loss of PTEN expression by blocking nuclear
translocation of EGR1 in gefitinib-resistant lung cancer cells
harboring epidermal growth factor receptor-activating mutations.
Cancer Res. 70:8715–8725. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar :
|
|
8
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Z1, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21WAF1/CIP1 expression. PLoS One.
8:e772932013. View Article : Google Scholar
|
|
10
|
Dong S, Qu X, Li W, Zhong X, Li P, Yang S,
Chen X, Shao M and Zhang L: The long non-coding RNA, GAS5, enhances
gefitinib-induced cell death in innate EGFR tyrosine kinase
inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR
via downregulation of the IGF-1R expression. J Hematol Oncol.
8:432015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y, Yu DD, Hu Y, Cao HX, Yu SR, Liu SW
and Feng JF: LXR ligands sensitize EGFR-TKI-resistant human lung
cancer cells in vitro by inhibiting Akt activation. Biochem Biophys
Res Commun. 467:900–905. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
13
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Efron B and Tibshirani R: Empirical bayes
methods and false discovery rates for microarrays. Genet Epidemiol.
23:70–86. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Remon J, Morán T, Majem M, Reguart N,
Dalmau E, Márquez-Medina D and Lianes P: Acquired resistance to
epidermal growth factor receptor tyrosine kinase inhibitors in
EGFR-mutant non-small cell lung cancer: A new era begins. Cancer
Treat Rev. 40:93–101. 2014. View Article : Google Scholar
|
|
17
|
Li LH, Wu P, Lee JY, Li PR, Hsieh WY, Ho
CC, Ho CL, Chen WJ, Wang CC, Yen MY, et al: Hinokitiol induces DNA
damage and autophagy followed by cell cycle arrest and senescence
in gefitinib-resistant lung adenocarcinoma cells. PLoS One.
9:e1042032014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ju L and Zhou C: Association of integrin
beta1 and c-MET in mediating EGFR TKI gefitinib resistance in
non-small cell lung cancer. Cancer Cell Int. 13:152013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Terai H, Soejima K, Yasuda H, Sato T,
Naoki K, Ikemura S, Arai D, Ohgino K, Ishioka K, Hamamoto J, et al:
Long-term exposure to gefitinib induces acquired resistance through
DNA methylation changes in the EGFR-mutant PC9 lung cancer cell
line. Int J Oncol. 46:430–436. 2015.
|
|
20
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang X, Zhang Y, Hosaka K, Andersson P,
Wang J, Tholander F, Cao Z, Morikawa H, Tegnér J, Yang Y, et al:
VEGF-B promotes cancer metastasis through a VEGF-A-independent
mechanism and serves as a marker of poor prognosis for cancer
patients. Proc Natl Acad Sci USA. 112:E2900–E2909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vaishnav YNV, Vaishnav MY and Pant V: The
molecular and functional characterization of E2F-5 transcription
factor. Biochem Biophys Res Commun. 242:586–592. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stewart ZA, Westfall MD and Pietenpol JA:
Cell-cycle dysregulation and anticancer therapy. Trends Pharmacol
Sci. 24:139–145. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suenaga M, Yamaguchi A, Soda H, Orihara K,
Tokito Y, Sakaki Y, Umehara M, Terashi K, Kawamata N, Oka M, et al:
Antiproliferative effects of gefitinib are associated with
suppression of E2F-1 expression and telomerase activity. Anticancer
Res. 26:3387–3391. 2006.PubMed/NCBI
|
|
26
|
Okabe T, Okamoto I, Tsukioka S, Uchida J,
Hatashita E, Yamada Y, Yoshida T, Nishio K, Fukuoka M, Jänne PA, et
al: Addition of S-1 to the epidermal growth factor receptor
inhibitor gefitinib overcomes gefitinib resistance in non-small
cell lung cancer cell lines with MET amplification. Clin Cancer
Res. 15:907–913. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lindeman GJ, Gaubatz S, Livingston DM and
Ginsberg D: The subcellular localization of E2F-4 is cell-cycle
dependent. Proc Natl Acad Sci USA. 94:5095–5100. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rakha EA, Pinder SE, Paish EC, Robertson
JF and Ellis IO: Expression of E2F-4 in invasive breast carcinomas
is associated with poor prognosis. J Pathol. 203:754–761. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garneau H, Paquin MC, Carrier JC and
Rivard N: E2F4 expression is required for cell cycle progression of
normal intestinal crypt cells and colorectal cancer cells. J Cell
Physiol. 221:350–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Molina-Privado I, Jiménez-P R,
Montes-Moreno S, Chiodo Y, Rodríguez-Martínez M, Sánchez-Verde L,
Iglesias T, Piris MA and Campanero MR: E2F4 plays a key role in
Burkitt lymphoma tumorigenesis. Leukemia. 26:2277–2285. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sirito M, Lin Q, Deng JM, Behringer RR and
Sawadogo M: Overlapping roles and asymmetrical cross-regulation of
the USF proteins in mice. Proc Natl Acad Sci USA. 95:3758–3763.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nielsen FC, Pedersen K, Hansen TV, Rourke
IJ and Rehfeld JF: Transcriptional regulation of the human
cholecystokinin gene: Composite action of upstream stimulatory
factor, Sp1, and members of the CREB/ATF-AP-1 family of
transcription factors. DNA Cell Biol. 15:53–63. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Viney TJ, Schmidt TW, Gierasch W, Sattar
AW, Yaggie RE, Kuburas A, Quinn JP, Coulson JM and Russo AF:
Regulation of the cell-specific calcitonin/calcitonin gene-related
peptide enhancer by USF and the Foxa2 forkhead protein. J Biol
Chem. 279:49948–49955. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paterson JM, Morrison CF, Mendelson SC,
McAllister J and Quinn JP: An upstream stimulatory factor (USF)
binding motif is critical for rat preprotachykinin-A promoter
activity in PC12 cells. Biochem J. 310:401–406. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hadsell DL, Bonnette S, George J, Torres
D, Klimentidis Y, Gao S, Haney PM, Summy-Long J, Soloff MS, Parlow
AF, et al: Diminished milk synthesis in upstream stimulatory factor
2 null mice is associated with decreased circulating oxytocin and
decreased mammary gland expression of eukaryotic initiation factors
4E and 4G. Mol Endocrinol. 17:2251–2267. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao E, Wang Y, Alcorn JL and Mendelson CR:
Transcription factor USF2 is developmentally regulated in fetal
lung and acts together with USF1 to induce SP-A gene expression. Am
J Physiol Lung Cell Mol Physiol. 284:L1027–L1036. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Coulson JM, Edgson JL, Marshall-Jones ZV,
Mulgrew R, Quinn JP and Woll PJ: Upstream stimulatory factor
activates the vasopressin promoter via multiple motifs, including a
non-canonical E-box. Biochem J. 369:549–561. 2003. View Article : Google Scholar
|
|
39
|
McMurray HR and McCance DJ: Human
papillomavirus type 16 E6 activates TERT gene transcription through
induction of c-Myc and release of USF-mediated repression. J Virol.
77:9852–9861. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goueli BS and Janknecht R: Regulation of
telomerase reverse transcriptase gene activity by upstream
stimulatory factor. Oncogene. 22:8042–8047. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pawar SA, Szentirmay MN, Hermeking H and
Sawadogo M: Evidence for a cancer-specific switch at the CDK4
promoter with loss of control by both USF and c-Myc. Oncogene.
23:6125–6135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Coulson JM, Fiskerstrand CE, Woll PJ and
Quinn JP: E-box motifs within the human vasopressin gene promoter
contribute to a major enhancer in small-cell lung cancer. Biochem
J. 344:961–970. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Grace CO, Fink G and Quinn JP:
Characterization of potential regulatory elements within the rat
arginine vasopressin proximal promoter. Neuropeptides. 33:81–90.
1999. View Article : Google Scholar
|
|
44
|
Khattar NH, Lele SM and Kaetzel CS:
Down-regulation of the polymeric immunoglobulin receptor in
non-small cell lung carcinoma: Correlation with dysregulated
expression of the transcription factors USF and AP2. J Biomed Sci.
12:65–77. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ocejo-Garcia M, Baokbah TA, Ashurst HL,
Cowlishaw D, Soomro I, Coulson JM and Woll PJ: Roles for USF-2 in
lung cancer proliferation and bronchial carcinogenesis. J Pathol.
206:151–159. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen B, Hsu R, Li Z, Kogut PC, Du Q,
Rouser K, Camoretti-Mercado B and Solway J: Upstream stimulatory
factor 1 activates GATA5 expression through an E-box motif. Biochem
J. 446:89–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kakita T, Hasegawa K, Morimoto T, Kaburagi
S, Wada H and Sasayama S: p300 protein as a coactivator of GATA-5
in the transcription of cardiac-restricted atrial natriuretic
factor gene. J Biol Chem. 274:34096–34102. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu
MM, Epstein JA, Morrisey EE and Gruber PJ: Gata4 and Gata5
cooperatively regulate cardiac myocyte proliferation in mice. J
Biol Chem. 285:1765–1772. 2010. View Article : Google Scholar :
|
|
49
|
Tummala R, Romano RA, Fuchs E and Sinha S:
Molecular cloning and characterization of AP-2 epsilon, a fifth
member of the AP-2 family. Gene. 321:93–102. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hoffman TL, Javier AL, Campeau SA, Knight
RD and Schilling TF: Tfap2 transcription factors in zebrafish
neural crest development and ectodermal evolution. J Exp Zoolog B
Mol Dev Evol. 308:679–691. 2007. View Article : Google Scholar
|
|
51
|
Kuckenberg P, Kubaczka C and Schorle H:
The role of transcription factor Tcfap2c/TFAP2C in trophectoderm
development. Reprod Biomed Online. 25:12–20. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li W and Cornell RA: Redundant activities
of Tfap2a and Tfap2c are required for neural crest induction and
development of other non-neural ectoderm derivatives in zebrafish
embryos. Dev Biol. 304:338–354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li X, Glubrecht DD and Godbout R: AP2
transcription factor induces apoptosis in retinoblastoma cells.
Genes Chromosomes Cancer. 49:819–830. 2010.PubMed/NCBI
|
|
54
|
Van Otterloo E, Li W, Garnett A, Cattell
M, Medeiros DM and Cornell RA: Novel Tfap2-mediated control of soxE
expression facilitated the evolutionary emergence of the neural
crest. Development. 139:720–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Rappoport JZ and Simon SM: Endocytic
trafficking of activated EGFR is AP-2 dependent and occurs through
preformed clathrin spots. J Cell Sci. 122:1301–1305. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Park JM, Wu T, Cyr AR, Woodfield GW, De
Andrade JP, Spanheimer PM, Li T, Sugg SL, Lal G, Domann FE, et al:
The role of Tcfap2c in tumorigenesis and cancer growth in an
activated Neu model of mammary carcinogenesis. Oncogene.
34:6105–6104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Perkins SM, Bales C, Vladislav T, Althouse
S, Miller KD, Sandusky G, Badve S and Nakshatri H: TFAP2C
expression in breast cancer: Correlation with overall survival
beyond 10 years of initial diagnosis. Breast Cancer Res Treat.
152:519–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|