Introduction
Prostate cancer (PCa) is the most commonly diagnosed
solid organ malignancy in the United States and remains the second
leading cause of cancer-related death among American men.
Approximately 29,480 deaths due to prostate cancer were recorded in
the US in 2014 (1). Prostate cancer
deaths are typically the result of metastatic castration-resistant
prostate cancer (mCRPC), and historically the median survival for
men with mCRPC is less than 2 years. Similar to other human
diseases, PCa is associated with a wide spectrum of genetic
aberrations. However, the mechanisms implicated in the initiation
and progression of PCa remain unclear (2).
MicroRNAs (miRNAs) are small (18 to 24 nucleotides
in length), single-stranded, endogenous non-coding RNAs that
regulate gene expression post-transcriptionally. The aberrant
expression of miRNAs is closely associated with the proliferation,
cell cycle, apoptosis, differentiation, migration, metabolism and
prognosis of various types of cancers, including PCa (3–6).
miR-195 is a member of the miR-15/16 family, which consists of a
group of miRNAs (miR-195, miR-15a, miR-15b, miR-16-1 and miR-16-2)
that share a similar seed sequence (7). The sequence of mature miR-195 is
conserved across mammalian species (8). miR-195 has been reported to be
deregulated in certain types of cancer, including upregulation in
chronic and acute lymphocytic leukemia (9) and metastatic melanoma (10) but is downregulated in
adrenocortical, hepatocellular carcinoma and squamous cell
carcinoma (11–13). However, the exact role of miR-195 in
PCa remains elusive.
In this study, we found that miR-195 expression
levels were decreased in human PCa samples and positively
correlated with prognosis. Next, we used RNA sequence and
bioinformation to screen the biological effect of miR-195 in PCa
cells and found miR-195 may influence cell cycle. Further study
showed that overexpression of miR-195 inhibited cell proliferation,
cell cycle progression, and tumorigenesis in vitro and in
vivo. In addition, HMGA1 was identified as the target gene of
miR-195 in PCa and its high expression was inversely correlated
with the prognosis of PCa patients. Downregulation of the
expression of HMGA1 had an effect similar to that of miR-195 in the
PCa cells. All of these results indicate that miR-195 and its
downstream target gene HMGA1 can be used to predict the prognosis
or even suppress the development of PCa.
Materials and methods
Data mining and bioinformation
analysis
Gene expression data were downloaded from the
GSE35988 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35988)
(14) and GSE21034 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21034)
(15). Detailed information on
patients can be obtained from the Oncomine database (https://www.oncomine.org/resource/main.html). Gene set
enrichment analysis (GSEA) was used to identify pathway gene sets
that were correlated with the miR-195 expression profile
(http://www.broadinstitute.org/gsea/index.jsp). The
gene sets were derived from the Molecular Signatures Database. The
normalized ES was used to compare the analysis results across the
various gene sets.
Cell culture
Human prostate cancer cells, DU-145 and PC-3, were
purchased from Shanghai Cell Bank, Chinese Academy of Sciences.
DU-145 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Hyclone, Beijing, China) plus 10% fetal bovine serum (FBS;
Hyclone, Gaithersburg, MD, USA), 100 mg/ml streptomycin and 100
U/ml penicillin (Hyclone, Beijing, China). PC-3 cells were cultured
in DMEM/F12 (Hyclone, Beijing, China) medium supplemented with 10%
FBS and antibiotics. Cells were incubated at 37°C in a humidified
atmosphere of 5% CO2 in air.
miRNA/siRNA transfections
Cells were plated in 6-well plates in their normal
growth medium without antibiotics for 24 h before transfections.
Transient transfections of miRNA mimics/siRNA (both from
GenePharma) were carried out using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's instructions,
when cells reached 50–70% confluency. miR-195 mimics, the negative
control (miR-NC), HMGA1 siRNA or the universal scrambled negative
control (NC siRNA) were purchased from GenePharma (Shanghai,
China).
MTT assay
The transfected cells were plated at 3,000/well in
96-well plates. Every 24 h after plating, respectively, 20
µl of 5 mg/ml MTT in PBS was added per well; cells were
lysed after 4 h by addition of 200 µl dimethyl sulfoxide
(DMSO; Sigma). Absorbance was measured at 570 nm.
Colony formation assay
After transfection, the cells were seeded in 6-well
plates at a density of 3,000 cells/well and cultured for 9–14 days
until visible colonies appeared. Then the cells were stained with
crystal violet. The number of colonies was counted only if they
contained more than 50 cells.
Cell cycle assays
Cell cycle distribution was detected by flow
cytometry (FCM). After 48 h of transient transfections, the cells
were harvested by trypsinisation and fixed in 70% ice-cold ethanol
the overnight. Then cells were washed with cold PBS and resuspended
in propidium iodide (PI) nuclear staining for cell cycle
analysis.
Western blotting
Whole cell extracts were prepared in RIPA buffer
containing protease inhibitor PMSF (both from Beyotime, Shanghai,
China). Protein concentration was measured using the BCA assay
(Beyotime) according to the manufacturer's instructions. Total
protein was electrophoresed by SDS-PAGE. Then the proteins were
transferred to a polyvinylidene fluoride membrane (Millipore,
Billerica, MA, USA) and blocked for 1 h with 5% skim milk at room
temperature. Incubation with primary antibodies was conducted
overnight at 4°C. The blots were incubated with horseradish
peroxidase labelled secondary antibodies and the signal was
detected using ECL (Beyotime). The following antibodies were used
for western blotting: rabbit anti-GAPDH (1:500; Xianzhi
Biotechnology, Hangzhou, China), rabbit anti-HMGA1 (1:500; Cell
Signaling Technology, USA), HRP-labelled goat anti-rabbit secondary
antibody (1:3,000; Zhongshan Golden Bridge Biotechnology, Beijing,
China).
Construction of the plasmid vectors
HMGA1 3′UTR-luciferase reporter vectors were cloned
into psiCHECK-2™ vectors. In brief, wide and mutant 3′UTR regions
were chemically synthesised and cloned into the XhoI and
NotI sites of the psiCHECK-2™ vector (Promega, Madison, WI,
USA).
Luciferase assay
For the luciferase reporter assay, the cells were
cultured in 24-well plates and transiently co-transfected with 50
nM miRNA (miR-195 mimics or scrambled miR-195 negative control) and
250 ng reporter vectors (wild-type reporter vectors or mutant-type
reporter vectors), using Lipofectamine™ 2000. Luciferase activities
were measured using a Dual-Luciferase assay kit (Promega) according
to the manufacturer's instructions at 48 h post-transfection
Lentiviral packaging and establishment of
stable cell lines
The lentiviral packaging kit was purchased from Open
Biosystems (Huntsville, AL, USA). The lentivirus carrying
hsa-miR-195 or hsa-miR-negative control (NC) was packaged following
the manufacturer's manual. Stable cell lines were established by
infecting the lentivirus into PC-3 cells and selection by
puromycin.
Tumor growth assay in mice and
immunohistochemical staining
Nude mice (female BALB/c-nu, 4-weeks old) were
purchased from the Shanghai Experimental Animal Center (Chinese
Academy of Sciences, Shanghai, China), and maintained under special
pathogen-free (SPF) conditions. Six mice were randomly divided into
two groups. PC-3 cells stably expressing miR-195 were injected
subcutaneously into the flank of nude mice (4×106 cells
in 100 µl). PC-3 cells stably expressing miR-NC were used as
the negative control. Tumor size was measured using a vernier
caliper every week, and tumor volume was calculated according to
the formula: Volume = 0.5 × length × width2.
Immunohistochemical staining assay was performed as previously
described (2).
Results
High level of miR-195 is associated with
a good prognosis of PCa
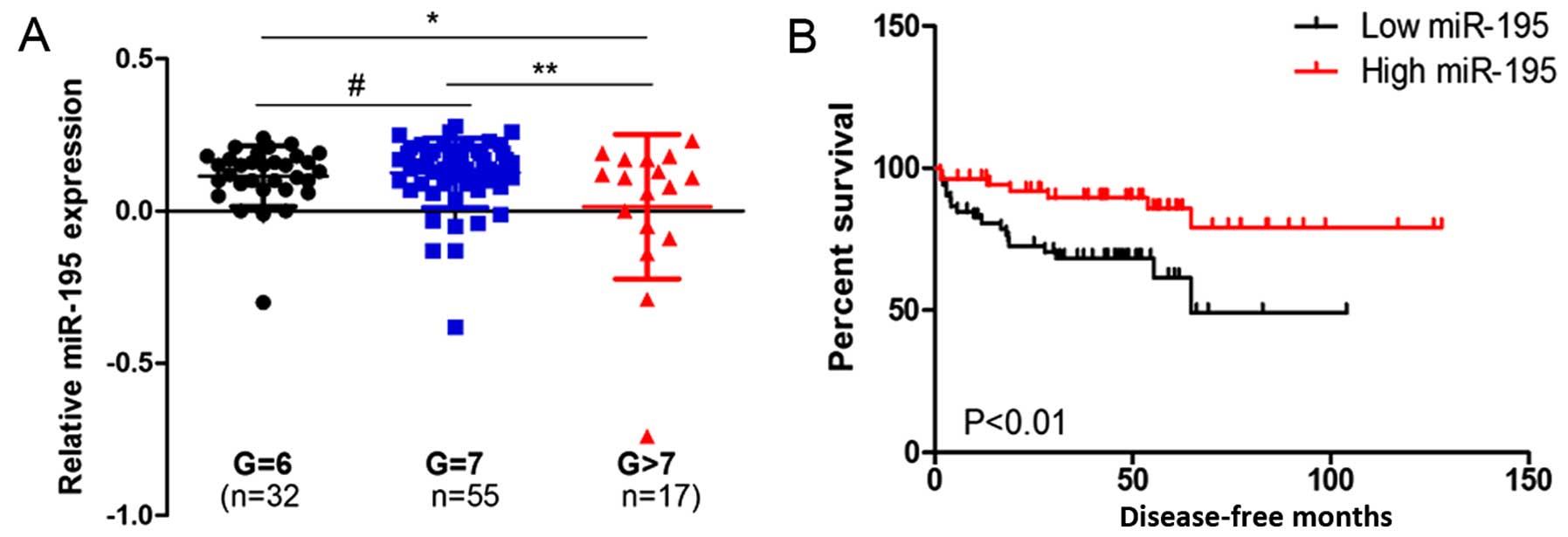
In the GSE21304 data, we analyzed the miR-195
expression pattern in different pathological grade PCa tissues and
found that miR-195 expression was downregulated in high grade PCa
tissues compared to low grade tissues (Fig. 1A). Next, the relationship between
miR-195 expression and patient prognosis was investigated. The
result indicated that a high level of miR-195 was associated with
longer disease-free survival (Fig.
1B).
miR-195 overexpression inhibits
proliferation and induces cell cycle arrest in prostate cancer
cells
To assess the role of miR-195, we overexpressed
miR-195 in prostate cancer cell lines (DU145 and PC3) followed by
RNA sequence and bioinformational analysis. GSEA analysis showed
that there was negatively enriched expression of gene sets involved
in cell cycle progression in the miR-195-transfected cells
(Fig. 2A). Consistent with the GSEA
analysis, miR-195 overexpression inhibited DU145 and PC3 cell
proliferation in the MTT assay (Fig.
2B). miR-195 overexpression also decreased clonogenicity of the
DU145 and PC3 cells compared to that noted in the miR-NC (Fig. 2C). FCM analysis showed that
overexpression of miR-195 led to a significant increase in the
percentage of G0/G1 phase DU145 and PC3 cells (Fig. 2D). This indicated that miR-195
overexpression suppressed the growth of prostate cancer cells.
miR-195 targets HMGA1 in prostate cancer
cells
To find the target of miR-195, bioinformatic
analysis was performed using TargetScan Human 6.2 and miRanda.
HMGA1 was found to possess potential miR-195 target sites within
its 3′UTR (Fig. 3A). Guided by this
finding, we performed western blot analysis in the DU145 and PC3
cells that were transfected with miR-195 or miR-NC. The results
showed that overexpression of miR-195 inhibited HMGA1 expression
(Fig. 3B). In order to investigate
HMGA1 as a direct target of miR-195, transient transfection of the
HMGA1 3′UTR plasmid along with miR-195/miR-NC was performed in the
DU145 and PC3 cells. A significant decrease was observed in
luciferase activities when compared with the mutant plasmid
(Fig. 3C). These results showed
that HMGA1 is a direct target of miR-195.
The role of HMGA1 in PCa
In the GSE35988 dataset, we initially analyzed the
HMGA1 expression pattern in benign prostate hyperplasia (BPH)
tissues and different PCa tissues and found that HMGA1 expression
was elevated in the CRPC tissues compared to the level noted in the
BPH tissues. Furthermore, HMGA1 expression was significantly
increased in the CRPC tissues when compared with that noted in the
androgen-dependent prostate cancer (ADPC) tissues (Fig. 4A). GSEA was used to evaluate the
pathways that were differentially expressed between patients with
high levels of HMGA1 expression and those with low levels of HMGA1
expression. The data revealed that HMGA1 regulates genes primarily
associated with PCa cell cycle progression (Fig. 4B). Next, we investigated the
correlation between HMGA1 expression and survival using
Kaplan-Meier survival curve analysis with a log-rank comparison.
PCa samples expressing higher than median levels of HMGA1 were
associated with decreased biochemical relapse-free survival and
overall survival relative to those with HMGA1 levels lower than the
median (P<0.05) in the Oncomine data (Fig. 4C and D).
Then, HMGA1 was knocked down in the DU145 and PC3
cells by introducing siRNA. The knockdown efficiency was verified
through western blot analysis and reduced expression of HMGA1 was
observed (Fig. 5B). MTT assay
showed that low expression of HMGA1 inhibited the proliferation of
the PCa cells (Fig. 5A). FCM
analysis also indicated that low expression of HMGA1 induced G0/G1
phase arrest (Fig. 5C).
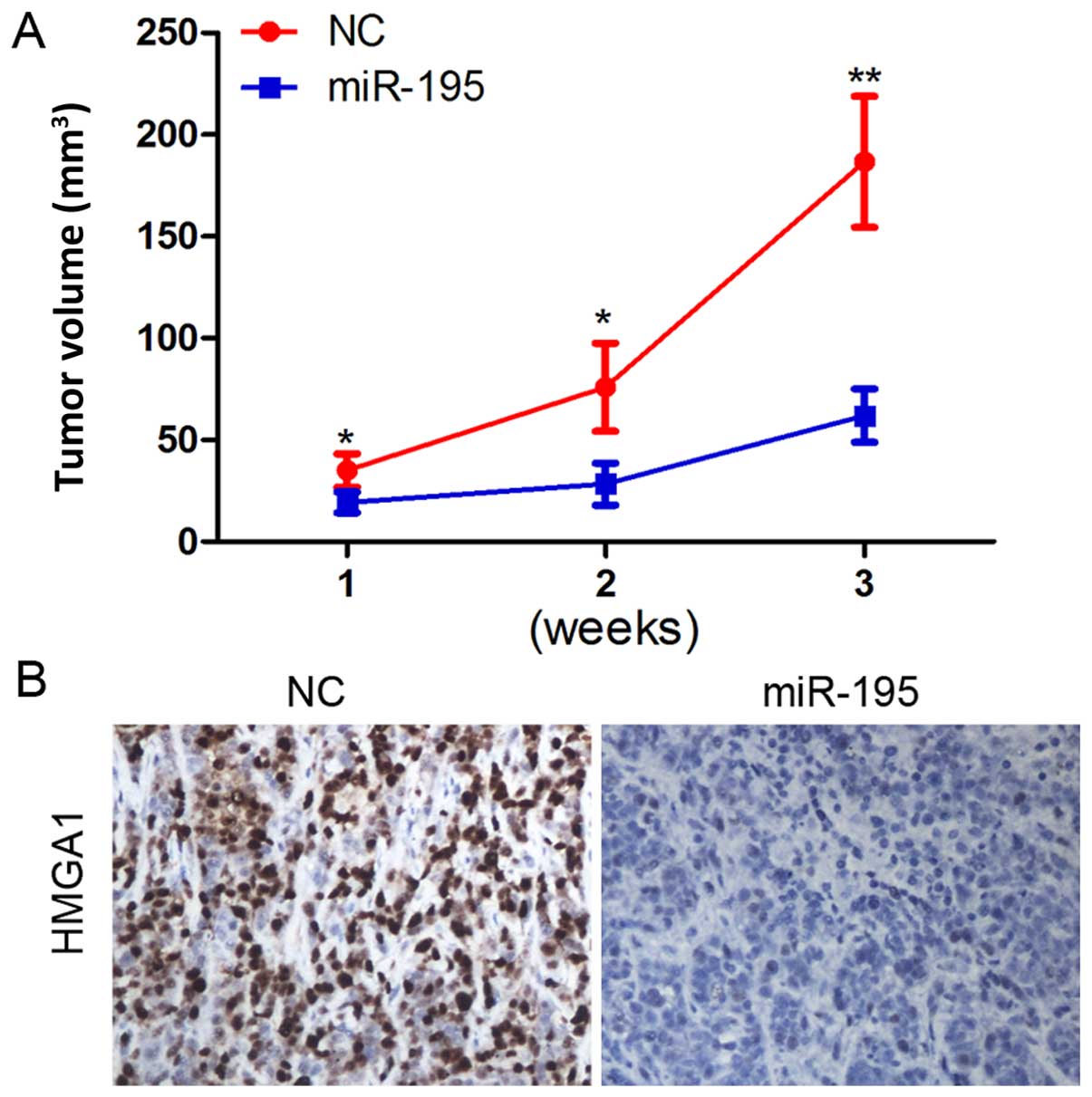
MiR-195 inhibits PCa tumorigenesis and
downregulates HMGA1 in vivo
In the previous experiment, miR-195 was found to be
downregulated in human PCa tissues and to play an important role in
proliferation and cell cycle distribution. We further examined the
effect of miR-195 in a nude mouse PCa xenograft model.
Overexpression of miR-195 inhibited PCa xenograft growth in
vivo (Fig. 6A).
Immunohistochemical analysis revealed that HMGA1 was markedly
decreased following miR-195 treatment (Fig. 6B).
Discussion
miR-195, derived from the miR-497/miR-195 locus at
human chromosome 17p13.1, has been found to be aberrantly
deregulated in tumorigenesis (11).
For example, it is upregulated in metastatic melanoma (10) and some cases of lung cancer
(16), and hepatocellular carcinoma
(17). By contrast, it is
preferentially downregulated in breast cancer (18), gastric cancer (17,19),
colorectal cancer (20–22), and bladder cancer (23,24).
However, inconsistent and inconclusive results have been reported
concerning the expression and function of miR-195 in PCa (17,25).
These controversial observations indicate the complexity of miR-195
during tumorigenesis and development and also call for further
investigation of the role in PCa. In the present study, we found
that miR-195 expression levels were decreased in human PCa samples
and were positively correlated with prognosis. Overexpression of
miR-195 significantly suppressed the cell cycle and proliferation
of PCa cells. In addition, our results in PCa cells indicated that
miR-195 targets HMGA1 and negatively regulates its expression at
the translational level which indicates that miR-195 may suppress
PCa malignant progression by downregulating the HMGA1 oncogene.
HMGA proteins are encoded by two genes, HMGA1 and
HMGA2, located at chromosome 6p21 and 12q13-15, respectively.
Numerous studies have confirmed the association of HMGA, in
particular HMGA1 overexpression, with a high malignant phenotype as
outlined by chemoresistance, the spread of metastases, and a global
poor survival (26). For example,
overexpression of HMGA1 in colon carcinoma was found to be strongly
associated with invasive ability, staining being more intense in
invasion-positive cases in comparison to invasion-negative ones
(27), in advanced stage (T3 and T4
tumors) and with the presence of distant, but not regional,
metastases. Several reports have indicated that HMGA1 is abundantly
expressed in pancreas adenocarcinomas, where overexpression of
HMGA1 correlates with advanced grade and, though less frequently,
in pancreas intraepithelial neoplasias (28). Moreover, HMGA1 is not expressed in
the normal epithelium surface where adenocarcinomas originate, but
it was found to be highly expressed in invasive ovarian carcinomas,
and weakly expressed in ovarian carcinomas with low invasive
potential (29). To evaluate the
expression and role of HMGA1 in PCa, we downloaded mRNA expression
and clinical follow-up data from the Oncomine and GSE database. Our
analysis showed upregulation of HMGA1 in CRPC samples in comparison
to ADPC and BPH tissues. In addition, high HMGA1 expression was
associated with the poor prognosis of PCa patients. Moreover, HMGA1
knockdown significantly decreased the potential of cell
proliferation and cell cycle progression in the PCa cells in
vitro, which had effects similar to those for miR-195
overexpression.
In conclusion, the results presented here
demonstrate that miR-195 has significant biological effects on PCa
development. Overexpression of miR-195 downregulated the expression
of HMGA1 protein, suggesting that miR-195 functions as a tumor
suppressor probably through downregulation of HMGA1 in PCa.
Furthermore, there are other putative miR-195 target genes which
could potentially be key players in the malignant progression of
PCa cells. miR-195 may prove to be a promising gene therapeutic
agent.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (81370849, 81300472, and 81572517), the
Natural Science Foundation of Jiangsu Province (BL2013032 and
BK2012336) and Nanjing City (201201053) and Southeast University
(3290002402), Science Foundation of Ministry of Education of China
(20120092120071).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tao T, Liu D, Liu C, Xu B, Chen S, Yin Y,
Ang L, Huang Y, Zhang X and Chen M: Autoregulatory feedback loop of
EZH2/miR-200c/E2F3 as a driving force for prostate cancer
development. Biochim Biophys Acta. 1839:858–865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao
L, Cai H, Li P, Cao Q, Ju X, et al: miR-152 controls migration and
invasive potential by targeting TGFα in prostate cancer cell lines.
Prostate. 73:1082–1089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu D, Tao T, Xu B, Chen S, Liu C, Zhang
L, Lu K, Huang Y, Jiang L, Zhang X, et al: miR-361-5p acts as a
tumor suppressor in prostate cancer by targeting signal transducer
and activator of transcription-6 (STAT6). Biochem Biophys Res
Commun. 445:151–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu B, Wang N, Wang X, Tong N, Shao N, Tao
J, Li P, Niu X, Feng N, Zhang L, et al: miR-146a suppresses tumor
growth and progression by targeting EGFR pathway and in a
p-ERK-dependent manner in castration-resistant prostate cancer.
Prostate. 72:1171–1178. 2012. View Article : Google Scholar
|
|
6
|
Tao T, Li G, Dong Q, Liu D, Liu C, Han D,
Huang Y, Chen S, Xu B and Chen M: Loss of SNAIL inhibits cellular
growth and metabolism through the miR-128-mediated
RPS6KB1/HIF-1α/PKM2 signaling pathway in prostate cancer cells.
Tumour Biol. 35:8543–8550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36(Database): D154–D158. 2008. View Article : Google Scholar :
|
|
8
|
Finnerty JR, Wang WX, Hébert SS, Wilfred
BR, Mao G and Nelson PT: The miR-15/107 group of microRNA genes:
Evolutionary biology, cellular functions, and roles in human
diseases. J Mol Biol. 402:491–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zanette DL, Rivadavia F, Molfetta GA,
Barbuzano FG, Proto-Siqueira R, Silva-Jr WA, Falcão RP and Zago MA:
miRNA expression profiles in chronic lymphocytic and acute
lymphocytic leukemia. Braz J Med Biol Res. 40:1435–1440. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhattacharya A, Schmitz U, Wolkenhauer O,
Schönherr M, Raatz Y and Kunz M: Regulation of cell cycle
checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene.
32:3175–3183. 2013. View Article : Google Scholar
|
|
11
|
Wang X, Wang J, Ma H, Zhang J and Zhou X:
Downregulation of miR-195 correlates with lymph node metastasis and
poor prognosis in colorectal cancer. Med Oncol. 29:919–927. 2012.
View Article : Google Scholar
|
|
12
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soon PS, Tacon LJ, Gill AJ, Bambach CP,
Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson
BG, et al: miR-195 and miR-483-5p identified as predictors of poor
prognosis in adrenocortical cancer. Clin Cancer Res. 15:7684–7692.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
et al: The mutational landscape of lethal castration-resistant
prostate cancer. Nature. 487:239–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al: Genome-wide screening
reveals that miR-195 targets the TNF-α/NF-κB pathway by
down-regulating IκB kinase alpha and TAB3 in hepatocellular
carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of miR-195 and
miR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wang K, Wang G, Ba Y, et al: Role of miR-143
targeting KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo ST, Jiang CC, Wang GP, Li YP, Wang CY,
Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, et al: MicroRNA-497
targets insulin-like growth factor 1 receptor and has a tumour
suppressive role in human colorectal cancer. Oncogene.
32:1910–1920. 2013. View Article : Google Scholar :
|
|
22
|
Liu L, Chen L, Xu Y, Li R and Du X:
microRNA-195 promotes apoptosis and suppresses tumorigenicity of
human colorectal cancer cells. Biochem Biophys Res Commun.
400:236–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Y, Wu J, Chen H, Mao Y, Liu Y, Mao Q,
Yang K, Zheng X and Xie L: Cyclin-dependent kinase 4 is a novel
target in micoRNA-195-mediated cell cycle arrest in bladder cancer
cells. FEBS Lett. 586:442–447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Itesako T, Seki N, Yoshino H, Chiyomaru T,
Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa
M, et al: The microRNA expression signature of bladder cancer by
deep sequencing: The functional significance of the miR-195/497
cluster. PLoS One. 9:e843112014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katoh M: Cardio-miRNAs and onco-miRNAs:
Circulating miRNA-based diagnostics for non-cancerous and cancerous
diseases. Front Cell Dev Biol. 2:612014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Kim M, Choi YH, Goemans B, Yeung
C, Hu Z, Zhan S, Seth P and Helman LJ: Diminished G1 checkpoint
after gamma-irradiation and altered cell cycle regulation by
insulin-like growth factor II overexpression. J Biol Chem.
274:13118–13126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piscuoglio S, Zlobec I, Pallante P, Sepe
R, Esposito F, Zimmermann A, Diamantis I, Terracciano L, Fusco A
and Karamitopoulou E: HMGA1 and HMGA2 protein expression correlates
with advanced tumour grade and lymph node metastasis in pancreatic
adenocarcinoma. Histopathology. 60:397–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Masciullo V, Baldassarre G, Pentimalli F,
Berlingieri MT, Boccia A, Chiappetta G, Palazzo J, Manfioletti G,
Giancotti V, Viglietto G, et al: HMGA1 protein over-expression is a
frequent feature of epithelial ovarian carcinomas. Carcinogenesis.
24:1191–1198. 2003. View Article : Google Scholar : PubMed/NCBI
|