Introduction
Breast cancer is one of the most commonly diagnosed
cancers among female around the world (1). Clinically defined, patients do not die
from the primary tumor, but from metastasis, which is resistant to
systemic therapy. It is acknowledged that a series of genetic
alterations promote normal epithelial cells to transform into
malignant cancer cells, resulting in dysregulated cell growth and
metastasis to distant sites. Therefore, a better understanding of
the detailed mechanisms of breast cancer progression is urgently
needed.
microRNAs (miRNAs), are a class of single-stranded
small non-coding RNA molecules of ~22 nucleotides, and function as
negative gene regulators to downregulate the expression of target
genes. miRNAs play a vital role as both oncogenes and tumor
suppressors, and are implicated in the hallmarks of breast cancer
(2–5). Increased evidence indicates that
aberrant expression of miRNAs plays important roles in diverse
biological processes, including development, differentiation,
growth and metabolism (6–8). miRNAs can function either as oncogenes
or tumor suppressors during cancer development and progression.
Deregulation of miRNAs is observed in many types of cancers,
including gastric cancer, liver cancer, bladder cancer, prostate
cancer, lung cancer, glioma and breast cancer (9–11).
Aberrant expression of miR-410-3p has been observed
in various types of cancers, suggesting that miR-410-3p plays a
significant role in cancer development and progression (12–19).
Previous research has shown that miR-410-3p functions as a tumor
suppressor by targeting MDM2 in gastric cancer or the angiotensin
II type 1 receptor in pancreatic cancer, respectively (16,17,19).
Furthermore, miR-410-3p regulates MET to influence the
proliferation and invasion of glioma (14). However, several studies indicate
that miR-410-3p functions as an oncogene to promote cancer
proliferation (13,15,19),
indicating that miR-410-3p plays dual roles in different types of
cancers. The role of miR-410-3p in breast cancer development and
progression remains unclear.
In the present study, we found that expression of
miR-410-3p was downregulated in breast cancer tissues compared with
that noted in paired normal breast tissues. Moreover,
overexpression of miR-410-3p promoted cell proliferation and
invasion in breast cancer. Furthermore, Snail, an
epithelial-mesenchymal transition (EMT)-related factor, was
identified as a target of miR-410-3p. These results suggest that
high expression of miR-410-3p may be involved in breast cancer
progression.
Materials and methods
Cell culture and tissue specimens
The MCF7, T47D, BT474, BT549, MDA-MB-468 and
MDA-MB-231 cell lines were obtained from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China) and were cultured as
previously described (20).
Breast cancer specimens were obtained from Tianjin
Medical University Cancer Institute and Hospital. A total of 30
primary breast cancer tissues and paired adjacent normal breast
tissue specimens were included in this study. All tumors were from
patients with a newly diagnosed breast cancer who had received no
therapy before sample collection. After mastectomy surgery, the
primary breast cancer tissues and the adjacent normal tissues were
flash-frozen in liquid nitrogen and stored at −80°C. This study was
approved by the Institutional Review Board of the Tianjin Medical
University Cancer Institute and Hospital, and written consent was
obtained from all participants.
To further validate the expression of miR-410-3p in
breast cancer, we analyzed miR-410-3p expression profiling data set
from The Cancer Genome Atlas (TCGA) including 683 cases of breast
cancer tissues and 87 cases of normal breast tissues.
Plasmid, miRNA and antibodies
In pcDNA3.1-HA, annealed oligonucleotides encoding
the HA tag were ligated into the HindIII and BamHI
sites of pcDNA3.1 (Invitrogen). The ORF of human Snail was
generated from MDA-MB-231 cells, the resultant PCR product of which
was connected together with pcDNA3.1 tagged HA (Snail-HA), the
Snail 3′-UTR containing the miR-410-3p binding site or the
miR-410-3p binding site mutated fragments were cloned into the
pGL3-Control vector (Snail-3′UTR-wt and Snail-3′UTR-mu; Promega,
Madison, WI, USA) and the resulting constructs were confirmed by
DNA sequencing. miRNAs were purchased from RiboBio (Guangzhou,
China). Antibodies against Snail (Abcam, Cambridge, MA, USA) and
β-actin (Cell Signaling Technology, Beverly, MA, USA) were used.
Recombinant human TGFβ1 was purchased from R&D Systems
(Redmond, WA, USA).
RNA extraction and reverse transcription
quantitative-PCR
Total RNA from the cultured cells and surgically
resected fresh breast tissues was extracted using mirVana PARIS kit
(Life Technologies) according to the manufacturer's instructions.
For miRNA detection, miRNA was reverse transcribed using the TaqMan
MicroRNA Reverse Transcription kit and real-time quantitative PCR
was performed using TaqMan miR-410-3p and U6 RNA (used as a
normalizer) assays (Life Technologies) following the manufacturer's
instructions.
Western blot analysis
Cells were lysed in RIPA buffer protease inhibitor
cocktail (Roche Molecular Biochemicals, Indianapolis, IN, USA).
Samples were denatured for 5 min at 95°C and subjected to 10%
SDS/PAGE. The separated proteins were transferred to a PVDF
membrane (Millipore, Bedford, MA, USA). The membrane was blocked in
5% (w/v) skim milk-TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween-20,
pH 8.3) solution, followed by incubation with the primary
antibodies diluted in skim milk-TBST solution overnight at 4°C.
Then the membrane was incubated with the corresponding horseradish
peroxidase-conjugated secondary antibody (Cell Signaling
Technology) for 1 h at room temperature, and the immunoreactive
protein bands were visualized by enhanced chemiluminescence
reagents (Millipore).
Transfection and luciferase assay
For transfection, the cells were plated at a density
of 2×105 cells/well in 6-well plates. When the cells
reached 60% confluency, 50 nmol/l miRNA or 4 µg Snail-HA was
transfected into the cells using Lipofectamine 3000 (Invitrogen)
for 48 h, according to the manufacturer's recommendations. After
transfection, the RNA and protein were extracted after 24 and 48 h,
respectively.
Luciferase assay was carried out on extracts from
the different breast cancer cells co-transfected for 24/48 h with
the corresponding plasmids or miRNAs using a dual-luciferase assay
kit (Promega) according to the manufacturer's recommendations. The
results were normalized against Renilla luciferase activity.
All transfections were performed in triplicate.
Cell proliferation assay
MTT, plate colony formation and EdU assays were used
to evaluate the ability of cell proliferation.
For the MTT assay, the cells were seeded in 96-well
plates (5×103/well). Cell viability was examined during
the following 5 days. After incubation for the indicated time, the
cells were incubated with 20 µl MTT (5 mg/ml in PBS;
Sigma-Aldrich) at 37°C for 4 h. Then, the medium was removed and
the formazan was dissolved in 150 µl of dimethyl sulfoxide
(DMSO; Sigma-Aldrich). The absorbance was measured at 570 nm using
a microplate auto-reader (Bio-Rad).
For the colony formation assay, 24 h after
transfection, the cells were seeded into 6-well plates at a density
of 500 cells/well. After ~15 days, the cells grew to visible
colonies and were stained with crystal violet. The colonies were
counted and compared with the control cells.
The EdU assay was performed using the EdU
labeling/detection kit (RiboBio) according to the manufacturer's
protocol. Briefly, after transfection for 48 h, the cells were
incubated with 25 µM EdU for 12 h. The cells were fixed with
4% formaldehyde for 30 min at room temperature and treated with
0.5% Triton X-100 for 15 min at room temperature for
permeabilization. After washing with PBS, the cells were reacted
with Apollo reaction cocktail for 30 min. Times before fixation,
permeabilization, and EdU staining. Subsequently, cell nuclei were
stained with Hoechst 33342 at a concentration of 5 µg/ml for
30 min. Then the cells were observed under a fluorescence
microscope. The pecentage of EdU-positive cells was examined by
fluorescence microscopy.
Invasion assay
The invasive ability of the breast cancer cells
in vitro was evaluated using Matrigel-coated Transwell
inserts (BD Biosciences, San Diego, CA, USA), respectively.
Briefly, 5×104 cells in 500 µl serum-free medium
were added to the upper chamber, and medium containing 20% FBS was
added into the lower chamber. Twenty-four hours later, the
migrating cells that had attached to the lower surface were fixed
with 20% methanol and stained for 20 min with crystal violet. The
membranes were then carved and embedded under coverslips with the
cells on the top. The number of migrating cells was counted under a
microscope in five predetermined fields.
Statistical analysis
All the experiments were performed at least twice
independently, and data are presented as mean ± standard error
mean. All statistical analyses were performed using SPSS18.0
software system for Windows (SPSS Inc.). Statistical significance
of difference was calculated using the Student's t-test with
significant differences defined as at least a P-value of
<0.05.
Results
miR-410-3p is downregulated in breast
cancer
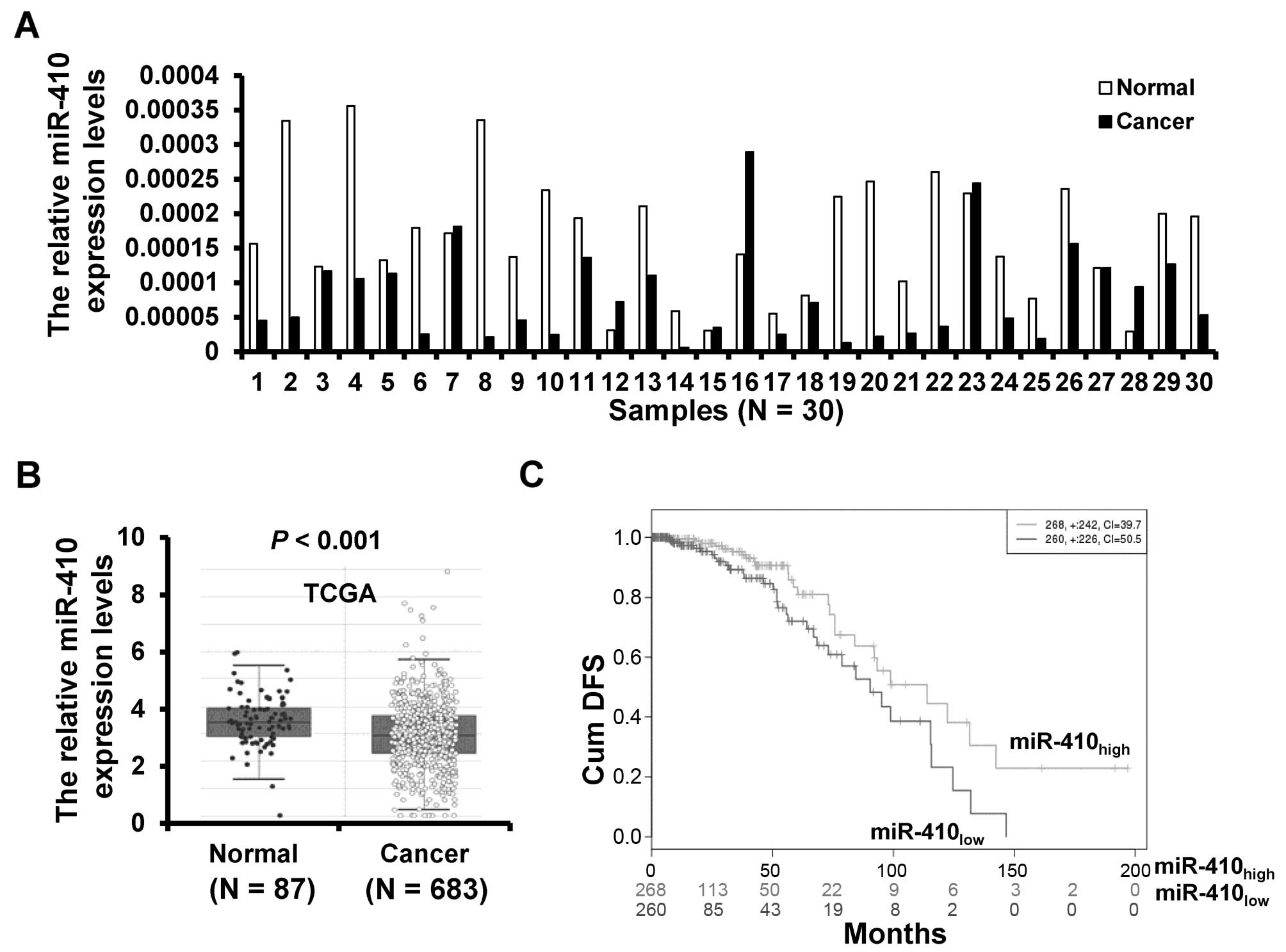
The expression of miR-410-3p in 30 breast cancer
tissues and paired adjacent normal tissues was detected by RT-qPCR.
The expression of miR-410-3p was downregulated in 23 (76.7%) of the
30 breast cancer samples (Fig. 1A).
To further validate the expression of miR-410-3p in breast cancer,
we analyzed miR-410-3p expression profiling data set from The
Cancer Genome Atlas (TCGA) including 683 cases of breast cancer
tissues and 87 cases of normal breast tissues. The validation data
confirmed that the miR-410-3p expression was downregulated in
breast cancer tissues (Fig. 1B).
Moreover, we compared the cumulative disease-free survival (cum
DFS) between patients with high miR-410-3p expression and low
miR-410-3p expression and found that the cum DFS of patients with
high miR-410-3p expression was higher (n=268) than that of patients
with low miR-410-3p expression (n=260) according to the TCGA data
(Fig. 1C). Taken together, these
results indicate that miR-410-3p is downregulated in breast
cancer.
miR-410-3p inhibits breast cancer cell
proliferation and invasion
Next, we assessed the miR-410-3p expression levels
in various breast cancer cell lines by RT-qPCR. We observed that
the miR-410-3p expression was highly expressed in the MCF7 cells
and lowly expressed in the MDA-MB-231 cells by RT-qPCR (Fig. 2A). Next, we investigated the
influence of miR-410-3p on cell proliferation by transiently
transfecting the miR-410-3p mimic or inhibitor, as well as their
corresponding controls in the MDA-MB-231 and MCF7 cell lines
(Fig. 2B). miR-410-3p
overexpression reduced cell growth, colony formation and the number
of EdU-positive cells in the MDA-MB-231 cells (Fig. 2C–E; left panels). In contrast,
inhibition of miR-410-3p in the MCF7 cells resulted in a higher
proliferation rate as assessed by MTT assay, plate colony formation
and EdU assays (Fig. 2C–E; right
panels). To investigate the role of miR-410-3p in cell invasion we
used a Transwell assay. miR-410-3p overexpression reduced cell
invasion in the MDA-MB-231 cells, while its inhibition enhanced
cell invasion in the MCF7 cells as compared to the control cells
(Fig. 2F). These results indicated
that miR-410-3p inhibits breast cancer cell proliferation and
invasion, suggesting that miR-410-3p is a tumor suppressor in
breast cancer.
Snail is a target of miR-410-3p
To elucidate the biological mechanisms underlying
the role of miR-410-3p in the inhibition of breast cancer
progression, we investigated the potential targets of miR-410-3p.
Target prediction programs, miRanda and TargetScan, were applied to
identify Snail as a putative miR-410-3p target (Fig. 3A). To further confirm this
regulation, Snail 3′-UTR and its mutant containing the putative
miR-410-3p binding sites were cloned into the downstream of the
luciferase ORF (Fig. 3A). These
reporter constructs were co-transfected into MDA-MB-231 cells with
the miR-410-3p mimic. Overexpression of miR-410-3p significantly
suppressed luciferase activity with inhibition rates of 40%
compared to that of the control MDA-MB-231 cells (Fig. 3B; left panel). These effects were
abolished when mutated Snail 3′-UTR, in which the binding sites for
miR-410-3p were inactivated by site-directed mutagenesis (Fig. 3B; right panel). Functional
regulation of Snail expression by miR-410-3p was analyzed by
modulating miR-410-3p levels via overexpression in MDA-MB-231 and
depletion in MCF7 cells. The Snail mRNA level was decreased in the
miR-410-3p-overexpressing MDA-MB-231 cells compared with that in
the control cells (Fig. 3C; left
panel). Meanwhile, the protein level of Snail was also reduced in
the miR-410-3p-overexpressing MDA-MB-231 cells (Fig. 3D; left panel). On the other hand,
depletion of miR-410-3p in MCF7 cells resulted in elevated mRNA and
protein levels of Snail (Fig. 3C and
D; right panels). Collectively, these data support the
bioinformatic prediction of Snail as a direct target of
miR-410-3p.
miR-410-3p regulates EMT by targeting
Snail in breast cancer cells
To ascertain whether miR-410-3p regulates breast
cancer progression through its interaction with Snail, we performed
a rescue experiment. We overexpressed Snail in the
miR-410-3p-overexpressing MDA-MB-231 cells (Fig. 4A and B) and observed that Snail
overexpression greatly impaired the anti-proliferative properties
of miR-410-3p, as documented by MTT, colony formation and EdU
assays (Fig. 4C–E). Similarly, the
ability of invasion was rescued by Snail overexpression (Fig. 4F). To address whether the expression
of miR-410-3p is associated with its target, Snail mRNA expression
was examined in 30 cases of primary breast cancer tissues by
RT-qPCR. There was a significant inverse correlation between
miR-410-3p and Snail expression in the breast cancer tissues
(Fig. 4G).
miR-410-3p-overexpressing MDA-MB-231 cells exhibited a significant
upregulation of β-catenin and E-cadherin, while mesenchymal marker
vimentin was dramatically downregulated as determined by RT-PCR
(Fig. 4H) and western blot analysis
(Fig. 4I). Furthermore,
overexpression of Snail rescued the effects of miR-410-3p
overxpression on the expresssion of β-catenin and vimentin
(Fig. 4H and I). Together, these
data indicate that miR-410-3p regulates the EMT phenotype by
targeting Snail in breast cancer cells.
Discussion
Aberrantly expressed miRNAs play a crucial role in
tumor development and progression (21–25).
In recent years, increasing studies have indicated that the
deregulation of miRNAs is involved in many processes of
carcinogenesis, functioning as either an oncogene or tumor
suppressor (26,27). In the present study, we found that
the expression of miR-410-3p was lower in breast cancer tissues
compared with that noted in the paired normal breast tissues.
Moreover, overexpression of miR-410-3p suppressed cell
proliferation and invasion in breast cancer cells. Furthermore, we
identified Snail as a direct target of miR-410-3p. In addition,
re-expression of Snail reversed the miR-410-3p-induced inhibition
of the EMT phenotype and breast cancer progression. Clinically, the
expression of miR-410-3p was downregulated and was inversely
correlated with expression of Snail in breast cancer tissues. These
findings suggest that miR-410-3p may be involved in breast
tumorigenesis and progression.
Aberrant expression of miR-410-3p is common in a
variety of cancers, suggesting that miR-410-3p may play an
important role in cancer development and progression (12–19).
Previous research showed that high expression of miR-410-3p is
associated with favorable disease-free survival in patients with
non-MYCN-amplified localized neuroblastoma (28). Furthermore, miR-410-3p was reported
to suppress the migration and invasion of gastric cancer and glioma
cells (14,17). Other studies showed that miR-410-3p
functions as an oncogene in non-small cell lung cancer, liver
cancer and colorectal cancer (13,15).
These data indicate that dyregulation of miR-410-3p may occur in a
tissue-specific manner in different types of cancer. However, the
roles of miR-410-3p in breast cancer are still unknown. In the
present study, the expression of miR-410-3p was upregulated in
breast cancer tissues compared with that in paired adjacent normal
breast. Furthermore, miR-410-3p suppressed breast cancer cell
proliferation and invasion. These results suggest that miR-410-3p
may act as a tumor suppressor in breast cancer progression.
We also demonstrated that miR-410-3p bound to the
3′-UTR of Snail. miR-410-3p downregulated Snail mRNA and protein
expression. We also observed that Snail could mediate the function
of miR-410-3p in breast cancer progression. EMT is a crucial
process in cancer progression that causes epithelial cells to
acquire fibroblast-like properties and show reduced intercellular
adhesion and increased motility (29,30).
Snail is overexpressed in various malignancies and is one of the
master regulators that promotes EMT and mediates invasiveness as
well as metastasis in many different types of malignant tumors
(31–34). Our data demonstrated that miR-410-3p
inhibits the EMT phenotype in breast cancer cells and the effect of
miR-410-3p on EMT was rescued by overexpression of Snail.
Furthermore, the expression of Snail was inversely correlated with
expression of miR-410-3p in breast cancer tissues. We first
demonstrated that miR-410-3p is a novel regulator of Snail in
breast cancer cells, which provided one possible mechanism for the
role of miR-410-3p in breast cancer progression.
In conclusion, we demonstrated for the first time
that miR-410-3p acts as a tumor suppressor in breast cancer through
inhibition of the expression of Snail. These data suggest a
potential therapeutic application of miR-410-3p in breast
cancer.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (nos. 81372843, 81472472 and
81502518).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cortés-Sempere M and Ibáñez de Cáceres I:
microRNAs as novel epigenetic biomarkers for human cancer. Clin
Transl Oncol. 13:357–362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sreekumar R, Sayan BS, Mirnezami AH and
Sayan AE: MicroRNA control of invasion and metastasis pathways.
Front Genet. 2:582011. View Article : Google Scholar
|
|
4
|
Shi KQ, Lin Z, Chen XJ, Song M, Wang YQ,
Cai YJ, Yang NB, Zheng MH, Dong JZ, Zhang L, et al: Hepatocellular
carcinoma associated microRNA expression signature: Integrated
bioinformatics analysis, experimental validation and clinical
significance. Oncotarget. 6:25093–25108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Z, Yu X, Shen J, Wu WK and Chan MT:
MicroRNA expression and its clinical implications in Ewing's
sarcoma. Cell Prolif. 48:1–6. 2015. View Article : Google Scholar
|
|
6
|
Lee HK, Finniss S, Cazacu S, Bucris E,
Ziv-Av A, Xiang C, Bobbitt K, Rempel SA, Hasselbach L, Mikkelsen T,
et al: Mesenchymal stem cells deliver synthetic microRNA mimics to
glioma cells and glioma stem cells and inhibit their cell migration
and self-renewal. Oncotarget. 4:346–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pichler M and Calin GA: MicroRNAs in
cancer: From developmental genes in worms to their clinical
application in patients. Br J Cancer. 113:569–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Zhang E and Lin C: MicroRNAs in
tumor angiogenesis. Life Sci. 136:28–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao D, Jia P, Wang W and Zhang G:
VEGF-mediated suppression of cell proliferation and invasion by
miR-410 in osteosarcoma. Mol Cell Biochem. 400:87–95. 2015.
View Article : Google Scholar
|
|
13
|
Wang Y, Fu J, Jiang M, Zhang X, Cheng L,
Xu X, Fan Z, Zhang J, Ye Q and Song H: miR-410 is overexpressed in
liver and colorectal tumors and enhances tumor cell growth by
silencing FHL1 via a direct/indirect mechanism. PLoS One.
9:e1087082014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen L, Zhang J, Feng Y, Li R, Sun X, Du
W, Piao X, Wang H, Yang D, Sun Y, et al: miR-410 regulates MET to
influence the proliferation and invasion of glioma. Int J Biochem
Cell Biol. 44:1711–1717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li D, Yang Y, Zhu G, Liu X, Zhao M, Li X
and Yang Q: MicroRNA-410 promotes cell proliferation by targeting
BRD7 in non-small cell lung cancer. FEBS Lett. 589:2218–2223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo R, Gu J, Zhang Z, Wang Y and Gu C:
MicroRNA-410 functions as a tumor suppressor by targeting
angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life.
67:42–53. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen J, Niu W, Zhou M and Zhang H, Ma J,
Wang L and Zhang H: MicroRNA-410 suppresses migration and invasion
by targeting MDM2 in gastric cancer. PLoS One. 9:e1045102014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen N, Wang J, Hu Y, Cui B, Li W, Xu G,
Liu L and Liu S: MicroRNA-410 reduces the expression of vascular
endothelial growth factor and inhibits oxygen-induced retinal
neovascularization. PLoS One. 9:e956652014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Müssnich P, Raverot G, Jaffrain-Rea ML,
Fraggetta F, Wierinckx A, Trouillas J, Fusco A and D'Angelo D:
Downregulation of miR-410 targeting the cyclin B1 gene plays a role
in pituitary gonadotroph tumors. Cell Cycle. 14:2590–2597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu Y, Zhao Y, Sun XH, Ge J, Zhang B, Wang
X and Cao XC: Down-regulation of miR-129-5p via the Twist1-Snail
feedback loop stimulates the epithelial-mesenchymal transition and
is associated with poor prognosis in breast cancer. Oncotarget.
6:34423–34436. 2015.PubMed/NCBI
|
|
21
|
Yu X and Li Z: MicroRNA expression and its
implications for diagnosis and therapy of tongue squamous cell
carcinoma. J Cell Mol Med. 20:10–16. 2016. View Article : Google Scholar
|
|
22
|
Li Z, Yu X, Shen J, Law PT, Chan MT and Wu
WK: MicroRNA expression and its implications for diagnosis and
therapy of gallbladder cancer. Oncotarget. 6:13914–13921. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu X and Li Z: The role of microRNAs
expression in laryngeal cancer. Oncotarget. 6:23297–23305. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takahashi RU, Miyazaki H and Ochiya T: The
roles of microRNAs in breast cancer. Cancers (Basel). 7:598–616.
2015. View Article : Google Scholar
|
|
25
|
Goh JN, Loo SY, Datta A, Siveen KS, Yap
WN, Cai W, Shin EM, Wang C, Kim JE, Chan M, et al: microRNAs in
breast cancer: Regulatory roles governing the hallmarks of cancer.
Biol Rev Camb Philos Soc. Jan 28–2015.Epub ahead of print.
PubMed/NCBI
|
|
26
|
Bracken CP, Khew-Goodall Y and Goodall GJ:
Network-based approaches to understand the roles of miR-200 and
other microRNAs in cancer. Cancer Res. 75:2594–2599. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang J, Zhang SY, Gao YM, Liu YF, Liu YB,
Zhao ZG and Yang K: MicroRNAs as oncogenes or tumour suppressors in
oesophageal cancer: Potential biomarkers and therapeutic targets.
Cell Prolif. 47:277–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gattolliat CH, Thomas L, Ciafrè SA,
Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P,
Valteau-Couanet D, et al: Expression of miR-487b and miR-410
encoded by 14q32.31 locus is a prognostic marker in neuroblastoma.
Br J Cancer. 105:1352–1361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kotiyal S and Bhattacharya S: Breast
cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res
Commun. 453:112–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saitoh M: Epithelial-mesenchymal
transition is regulated at post-transcriptional levels by
transforming growth factor-β signaling during tumor progression.
Cancer Sci. 106:481–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK,
Jo HJ, Kim HS, Oh N, Song GA and Park Y: Snail and serpinA1 promote
tumor progression and predict prognosis in colorectal cancer.
Oncotarget. 6:20312–20326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cho HJ, Park SM, Kim IK, Nam IK, Baek KE,
Im MJ, Yoo JM, Park SH, Ryu KJ, Han HT, et al: RhoGDI2 promotes
epithelial-mesenchymal transition via induction of Snail in gastric
cancer cells. Oncotarget. 5:1554–1564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cichon MA and Radisky DC: ROS-induced
epithelial-mesenchymal transition in mammary epithelial cells is
mediated by NF-κB-dependent activation of Snail. Oncotarget.
5:2827–2838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pilli VS, Gupta K, Kotha BP and Aradhyam
GK: Snail-mediated Cripto-1 repression regulates the cell cycle and
epithelial-mesenchymal transition-related gene expression. FEBS
Lett. 589:1249–1256. 2015. View Article : Google Scholar : PubMed/NCBI
|