Introduction
MicroRNAs (miRNAs) are a class of non-coding RNAs
that are ~22 nucleotides in length. Since their discovery in 1993,
miRNAs have been shown to play a profound role in the regulation of
many aspects of cell function (1,2).
miRNAs are often upregulated or downregulated in human types of
cancers, and can affect different stages of tumor development to
promote or inhibit tumor growth by post-transcriptionally
downregulating or suppressing mRNA expression (3,4). In
particular, many miRNAs that are expressed at low levels have been
demonstrated to play a role in tumor suppression in cancers
including laryngeal squamous cell carcinomas (LSCCs).
LSCC, is one of the most common head and neck
cancers worldwide and, accounts for nearly 90% of all malignant
laryngeal cancers (5). Despite
recent advances in chemoradiotherapy and surgery, there have been
no clear improvements in the prognosis of this disease (6). Although the pathology of LSCC has been
attributed to many factors, the accumulation of genetic and
epigenetic changes remains the most favored basic mechanism of
tumorigenesis (7). Recent studies
have identified many miRNAs that are abnormally expressed in
multiple solid tumors, such as gastric (8), liver (9) and ovarian cancer (10), including LSCC (11). We previously investigated the
functions of a series of miRNAs in LSCC. It was found that the
expression levels of miR-19a, miR-21, miR-129 and miR-206 were
decreased, whereas the expression levels of miR-203 and miR-205
were increased in LSCC (11–17).
Functional analyses indicated that these miRNAs, which function as
oncogenes or tumor suppressors, have indeed been implicated in the
carcinogenesis of LSCC. Recently, studies have shown that
synergistic expression of certain miRNAs could jointly effect tumor
behavior (18,19). To validate the function of the
coordination among miRNAs, the present study aimed to investigate
whether the combined expression of miRNAs have superior
tumor-suppression effects to those of a single miRNA.
miR-375 is located on human chromosome 2q35
(20). To date, low levels of
miR-375 expression have been detected in prostate (21) and esophageal cancer (22), and other types of cancers. miR-206
is considered to promote muscle differentiation by downregulating
the DNA polymerase P180 subunit and transcription factors (23). We previously found that miR-206 was
downregulated in LSCC. Transfection of miR-206 in Hep-2 cells
suppressed tumor proliferation, invasion and induced apoptosis,
indicating that miR-206 functions as a novel tumor-suppressor gene
in this cancer (13–17).
Krüppel-like factor 4 (KLF4), also known as
gut-enriched Krüppel-like factor, was recently identified as a
Krüppel-type transcription factor with three
C2H2 zinc fingers (24). KLF4 regulates the phenotype of tumor
and stem cells (25), inhibits the
formation of tumors in sites such as the gastrointestinal tract
(26), and can promote malignant
characteristics in other tissues, including breast and skin tissues
(27–29). Metacore pathway analysis predicted
that KLF4 is a target of miR-206, which was further supported by
sequence complementarity alignment (30), and miR-375 directly inhibits the
expression of KLF4 by targeting its 3′ untranslated region
(7).
In the present study, it was found that the
expression of miR-375 was downregulated in LSCC tissues. miR-375
was found to inhibit the tumor cell proliferation, migration and
invasion, and induce the apoptosis of LSCC cells, by regulating
KLF4 as a target gene. These effects were superior to that of the
combined expression of miR-375 and miR-206. miR-375 may act as a
tumor suppressor and serve as a potential therapeutic target in
LSCC.
Materials and methods
Patients and tissue collection
A retrospective review of 60 adult patients with
pathologically confirmed primary LSCC was performed. The use of
clinical materials was approved by the local Ethics Committee of
the Second Affiliated Hospital of Harbin Medical University.
Between 2012 and 2015, these patients underwent a partial or total
laryngectomy at the Department of Otorhinolaryngology and Head and
Neck Surgery at The Second Affiliated Hospital of Harbin Medical
University. The matched specimens of LSCCs and the corresponding
adjacent non-neoplastic tissues obtained from 60 patients were
preserved in liquid nitrogen within 5 min after tumor resection,
and were then stored at −80°C. The study protocol used was in
accordance with the Institutional Guidelines for Human Research and
was approved by the ethics committee.
Lentiviral vectors for miR-206 and
miR-375
Human miR-375 and miR-206 lentivirus gene transfer
vectors harboring green fluorescent protein (GFP) sequence were
constructed by GeneChem (Shanghai, China): miR-206,
5′-TCCCAGTGATCTTCTCGCTAAGAGTTTCCTGCCTGGGCAAGGAGGAAAGATGCTACAAGTGGCCCACTTCTGAGATGCGGGCTGCTTCTGGATGACACTGCTTCCCGAGGCCACATGCTTCTTTATATCCCCATATGGATTACTTTGCTATGGAATGTAAGGAAGTGTGTGGTTTCGGCAAGTGCCTCCTCGCTGGCCCCAGGGTACCACCCGGAGCACAGGTTTGGTGACCTTCTTCCTCATCAGGGCTTTGTGCCAGCAAATGACTCCCTCACCAAGGAAGTTTTTT-3′
and miR-375,
5′-AGGCTAGCGGGGCGCTGTGCAGCACTGAGCTCGCGGAAGACCAGGACCAGGAGATCACCGAGGGCGACCGCCAGGCCCCGGGCCCTCCGCTCCCGCCCCGCGACGAGCCCCTCGCACAAACCGGACCTGAGCGTTTTGTTCGTTCGGCTCGCGTGAGGCAGGGGCGGCCTCTCAGCACCAGCCCGGGGGCCGGCCTGATCGCCACGCAGGCACCTGCCGCCGCCA-3′.
The recombinant lentiviruses of miR-206 and miR-375, and the
control lentivirus (GFP-lentivirus) were prepared and titered to
108 TU (transfection units)/ml, according to the
manufacturer's guidelines (GeneChem).
Cell culture and transfections
The human LSCC cell line, Hep-2, was purchased from
the Cell Bank of the Chinese Academy of Science (Shanghai, China).
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
HyClone, Logan City, UT, USA) with high glucose, supplemented with
10% fetal bovine serum (FBS; Shanghai Shenggong Co., Ltd.,
Shanghai, China) and 1% penicillin/streptomycin (Beyotime
Biotechnology, Shanghai, China) and were maintained at 37°C under a
humidified atmosphere containing 5% CO2. Six-well plates
were maintained at a concentration of 1×105 cells/well
for transfection. After 24 h, when the cells reached ~70–80%
confluency, 1 ml of complete medium containing lentivirus
(108 TU/ml) preparations and Polybrene (5 μg/ml)
were added to each well. The cells were incubated at 37°C for 12 h.
The supernatant from the cells was then removed, and DMEM
containing 10% FBS and 1% penicillin-streptomycin was added. After
24 h, the culture medium was replaced with fresh DMEM. At 72 h
post-transfection, the mean percentage of GFP-positive cells
observed in each well was calculated from three random fields of
view at a magnification of ×200 using a fluorescence microscope
(IX70; Olympus, Tokyo, Japan).
Extraction of total RNA and quantitative
real-time polymerase chain reaction
Total RNA was extracted using TRIzol (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol. RNA
concentrations were determined using absorbance readings at 260 nm,
while RNA purity was evaluated using the optical density
(OD)260/OD280 absorption ratios. cDNA was
reverse transcribed using an All-in-One™ miRNA quantitative
polymerase chain reaction (qPCR) detection kit (GeneCopoeia,
Rockville, MD, USA). The reverse transcription (RT) reactions were
incubated at 37°C for 60 min and then at 70°C for 5 min. Real-time
qPCR was performed using a SYBR-Green Master Mix and a 7500 Fast
Real-Time PCR system (both from Applied Biosystems, Carlsbad, CA,
USA). Reactions were incubated at 95°C for 10 min, followed by 40
cycles at 95°C for 10 sec, 60°C for 20 sec and 72°C for 30 sec. For
the measurement of the KLF4 transcript from total RNA, total cDNA
was synthesized using ReverTra Ace qPCR RT kit (Toyobo Co., Ltd.,
Osaka, Japan). Real-time PCR was performed using SYBR Real-Time PCR
Master Mix (Toyobo Co., Ltd.). The ubiquitin 6 small nuclear RNA
(snRNA) and β-actin were used as endogenous controls for miRNA and
mRNA, respectively. The ΔΔCt method was used to determine relative
quantitation of miRNA and expression of mRNA in tissue samples, and
fold-change was determined as 2−ΔΔCt. The primer
sequences of miR-375 (product code, CD201-0173) and endogenous
control U6 (CD201-01730145) were applied by Tiangen Biotech
(Beijing, China). KLF4, 5′-CTTCCTGCCCGATCAGATGC-3′ and
5′-TCGCAGGTGTGCCTTGAGT-3′; β-actin, 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′
and 5′-CTGTCACCTTCACCGTTCCAGTTT-3′ were from Applied Bioneer
(Daejeon, Korea). Each reaction was performed in triplicate.
Western blot analysis
At 72 h post-transfection with miR-375 and miR-206,
the cells were subjected to western blot analysis. Cells were
incubated in cell lysis buffer for 30 min on ice. Cell lysates were
separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and were then transferred to polyvinylidene
fluoride membranes. After incubating the membranes in 5% skim milk
in Tris-buffered saline containing 0.05% Tween-20 (TBST), the
membranes were incubated with primary antibodies overnight at 4°C.
The primary antibodies used for western blotting included anti-KLF4
(1:1,000; Cell Signaling Technology, Boston, MA, USA). The
membranes were then washed with TBST and incubated with
species-appropriate horseradish peroxidase-conjugated secondary
antibodies for 1 h at 37°C. β-actin (1:5,000; Bioworld Technology,
Inc., St. Louis Park, MN, USA) served as a loading control, and
bands were quantified using ImageJ software (National Institutes of
Health, Bethesda, MD, USA). Three independent experiments were
performed.
Cell proliferation assay
After transfection of Hep-2 cells by lentivirus for
varying durations of time: 12, 24, 48 and 72 h, 100 μl of
sterile Cell Counting Kit-8
[2-(2-methoxy-4-nitro-phenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium
monosodium salt] (Dojindo Co., Ltd., Shanghai, China) was added and
incubated for another 4 h at 37°C. Later, spectrometric absorbance
at a wavelength of 450 nm was measured on an enzyme immunoassay
analyzer (model 680; Bio-Rad Laboratories, Hercules, CA, USA). The
rate of cell growth was calculated using the following formula:
Cell growth rate (%) = (mean absorbance in 96-wells of the
treatment group/mean absorbance in 6-wells of the cells in the
control group) × 100.
Transwell chamber invasion assay
Cells were washed, resuspended in complete RPMI-1640
medium (1×105 cells/ml), and added to the upper chamber
of Boyden chambers (24-well, 8-μm pores) coated with
Matrigel (Becton-Dickinson Labware, Franklin Lakes, NJ, USA). The
lower chamber contained 600 μl conditioned medium. After 24
h of incubation, the inserts and cells were mechanically removed
from the upper side of the filters. Filters were fixed in 4%
paraformaldehyde and stained with hematoxylin and eosin. Cells were
observed on a microscope, and five randomly selected fields were
observed at a magnification of ×400. Tests were repeated in
triplicate.
Cell cycle assay
Cells were fixed with cold ethanol at 4°C for 1 h,
and then stored at −20°C until analysis. Cells were washed with
phosphate-buffered saline (PBS), treated with RNase A (50
μg/ml) and stained with ethidium bromide for 20 min at 37°C.
Cells were analyzed for DNA content using flow cytometry
(FACSCalibur; Becton-Dickinson Immunocytometry Systems, San Jose,
CA, USA) and the distribution of cell cycle phases was determined
using ModFit LT for Mac V 3.0.
Apoptosis assay
The Annexin V-fluorescein isothiocyanate (FITC) and
propidium iodide (PI) double staining detection kit was used to
measure apoptosis according to the manufacturer's protocol
(Wanleibio, Shenyang, China). The cells were resuspended in binding
buffer (Beyotime Biotechnology) at a concentration of
1×106 cells/ml. The cells were incubated with 5
μl Annexin V-FITC, stained with 5 μl PI, and then
incubated in the dark at room temperature for 15 min. Cells were
analyzed using flow cytometry within 1 h.
Statistical analyses
Data are expressed as the means ± SEM of three
independent experiments, each performed in triplicate. Statistical
significance was tested using SPSS 19.0 software. An independent
t-test was used to analyze differences in KLF4 mRNA levels after
transfection, KLF4 protein scoring, cell viability, invasive
phenotype, apoptosis induction and cell cycle distribution between
groups. For analysis of differences in miR-375 mRNA and KLF4 mRNA
between tumor samples and adjacent normal tissues, the data were
checked by paired t-test. P-value <0.05 was considered to
indicate a statistically significant result.
Results
miR-375 is downregulated in human
LSCCs
We previously reported that the expression of
miR-206 is decreased in LSCCs (13). In the present study, the expression
of miR-375 was assessed in 60 patients. The clinicopathological
findings of the patients are shown in Table I. To determine the expression of
miR-375 in LSCC, qRT-PCR was used to assess the expression levels
of RNA in the LSCC tissue samples and adjacent normal tissues
collected from the same 60 patients. Similar to miR-206, the
expression of miR-375 was also decreased in the tumor samples
(0.524±0.009), when compared with that in the adjacent
non-cancerous tissues (2.179±0.019) (P<0.01). The
tumor/non-cancerous tissue (T/N) ratios for the expression of
miR-375 were found to be significantly correlated with T stage,
tumor differentiation, neck nodal metastasis and clinical stage.
Decreased expression of miR-375 was found in the tumors from
patients with advanced clinical stage, T3-4 grade or lymph node
metastasis.
 | Table ICorrelation between the miR-375
expression level and the clinicopathological parameters of the LSCC
patients. |
Table I
Correlation between the miR-375
expression level and the clinicopathological parameters of the LSCC
patients.
|
Characteristics | n | miR-375 (T/N
expression ratio) | P-value |
|---|
| Gender | | | 0.255 |
| Male | 52 | 0.224±0.006 | |
| Female | 8 | 0.228±0.003 | |
| Age (years) | | | 0.520 |
| ≥60 | 27 | 0.245±0.010 | |
| <60 | 33 | 0.239±0.004 | |
| T
classification | | | 0.016 |
| T1–2 | 38 | 0.251±0.007 | |
| T3–4 | 22 | 0.227±0.003 | |
| Lymph node
metastasis | | | 0.032 |
| Negative | 35 | 0.251±0.008 | |
| Positive | 25 | 0.230±0.003 | |
| Primary
location | | | 0.853 |
| Supraglottic | 31 | 0.241±0.009 | |
| Glottic | 29 | 0.243±0.003 | |
| Clinical stage | | | 0.003 |
| I–II | 34 | 0.254±0.008 | |
| III–IV | 26 | 0.226±0.002 | |
Transfection of miR-375 and miR-206
inhibits cell viability, respectively
To investigate the biological function of miR-375
and miR-206 in LSCC, recombinant lentiviruses containing miR-375
and miR-206 mimics as well as a GFP cassette, were created. At 72 h
after Hep-2 cells were transfected with miRNAs and GFP control
lentiviruses, >80% of the Hep-2 cells were found to express GFP,
indicating the efficiency and stability of the transductions
(Fig. 1). The expression of miRNAs
in the different experimental groups was then confirmed using
qRT-PCR (data not shown).
All the experimental cells in the present study were
categorized into five groups: blank control group that contained
untreated Hep-2 cells; GFP control group that contained Hep-2 cells
transfected with lentiviruses without miRNAs; miR-375 group that
contained Hep-2 cells transfected with lentiviruses with miR-375
mimics; miR-206 group that contained Hep-2 cells transfected with
lentiviruses with miR-206 mimics; and miR-375 + miR-206 group that
contained Hep-2 cells transfected with lentiviruses with miR-375
and miR-206 mimics.
As shown in Fig. 2
after transfection with the miRNAs, the proliferation rates of the
Hep-2 cells in the GFP control group did not show any obvious
alteration during the time course. However, the viability of the
miR-375 group Hep-2 cells was evidently decreased at each time
point (12, 24, 48 and 72 h). The viability of the miR-206 group
Hep-2 cells was also decreased compared with the blank control
group, but the proliferation rate curve had a smaller decrease
after 24 h, and its downward trend was less pronounced than that of
miR-375. Additionally, an initial evident decrease in the survival
rate curve of the miR-375 + miR-206 group was noted, which was
similar to the miR-375 group. This was followed by a relative
stable state after 24 h, and its downward trend was always between
the levels of the miR-375 and miR-206 groups. These findings
indicated that reconstitution of miR-375, miR-206 or both miRNAs
could inhibit the viability of Hep-2 cells in vitro.
Furthermore, transfection with miR-375 alone was found to be the
most effective.
Overexpression of miR-375 and miR-206
promotes early apoptosis in Hep-2 cells
Flow cytometric analysis revealed that transfection
of Hep-2 cells with miRNAs for 72 h significantly induced higher
levels of apoptosis, compared with cells of the GFP control or the
uninfected cells. As shown in Fig.
3 the apoptosis rate of the GFP control group (2.8%) and blank
control group cells (2.6%) were nearly the same. However, the
apoptosis rate of miR-375 group cells (67.2%) was much higher than
the blank control group cells. Compared with the blank group, the
apoptosis rate of miR-206 group cells (55.7%) was also apparently
higher than that noted in the untreated cells, but lower than that
noted in the miR-375 group. The apoptosis rate of co-transfection
group cells (62.5%) was between the rated observed in the miR-375
and miR-206 group. These results confirmed the strong pro-apoptotic
effect of miR-375 and miR-206, however, the effect of miR-375 on
the promotion of apoptosis was stronger than the effect by miR-206,
and this effect was even stronger than miR-375 combined with
miR-206.
miR-375 and miR-206 overexpression
affects cell cycle progression in the Hep-2 cells
miR-375-transfected Hep-2 cells (72.1%) showed the
highest percentage of cells in the G1 phase compared to the GFP
control cells (60.1%) and blank control cells (57.5%) at 72 h
post-transfection (P<0.05). This percentage in the
miR-206-transfected cells (67.8%) was higher than that noted in the
GFP control cells or untreated group cells, but lower than that in
the miR-375 group cells. The percentage of cells arrested in the G1
phase in the miR-375 + miR-206-transfected cells (71.9%) was
between the percentage of cells in the miR-375 and miR-206 groups
(Fig. 4). This demonstrates that
miR-375 and miR-206 upregulation can both produce G1 phase arrest
since the number of cells in the G1 phase increased by >10% when
compared with the control groups. In addition, an increase in
miR-375 alone had the strongest effect on the G1 phase arrest.
miR-375 and miR-206 overexpression
suppresses Hep-2 cell invasion in vitro
To investigate whether miR-375 and miR-206
overexpression is beneficial to the Hep-2 cell invasive phenotype,
invasion assays were performed using 24-well Boyden chambers coated
with Matrigel. The number of Hep-2 cells that passed through the
filter after transfection with miRNAs for 72 h was much lower than
that observed for the GFP-control group cells (84±1.15) and blank
control cells (87±2.03). Among the three miRNA-transfected groups,
cells that were transfected with miR-375 (32.3±2.33) showed less
robust invasion than the group transfected with miR-206 (53±3.51)
and miR-375 + miR-206 (42±0.33). The number of invasive cells of
the miR-206 group was higher than that noted in the miR-375 group
as well as the co-transfection group. In addition, the number of
invasive cells in the co-transfection group was between this number
noted in the miR-375 and miR-206 groups (Fig. 5). However, the inhibitory effect on
invasion by miR-375 was stronger than that of miR-206 and miR-375 +
miR-206, and the inhibitory effect of miR-206 was the weakest.
Overexpression of miR-375 and miR-206
suppresses the expression of KLF4
KLF4 is an important gene with effective
transcriptional regulation on various downstream genes involved in
cell cycle, apoptosis, proliferation and invasion. To compare the
expression of KLF4 in LSCC and adjacent normal tissues, KLF4 mRNA
in 60 patients with LSCC samples and the corresponding adjacent
non-neoplastic tissues were performed using qRT-PCR. As shown in
Fig. 6A, the expression level of
KLF4 mRNA in LSCC was ~4-fold higher than that of the corresponding
matched samples.
To detect the expression of KLF4 following
regulation of miR-375 and miR-206 in the Hep-2 cells after
transfection, KLF4 mRNA and protein expression was examined using
qRT-PCR and western blotting, respectively.
As shown in Fig. 6B,
after transfection, the level of KLF4 mRNA in the GFP control group
did not show an obvious change as compared with the blank control
group. However, the KLF4 mRNA level in the miR-375 group showed a
much lower level than that in the blank control group. Expression
of KLF4 mRNA in the miR-206 group was also lower than that in the
blank control group, but higher than that in the miR-375 group. In
addition, the KLF4 mRNA level in the miR-375 + miR-206 group was
lower than that in the miR-206 group but higher than that in the
miR-375 group (P<0.05).
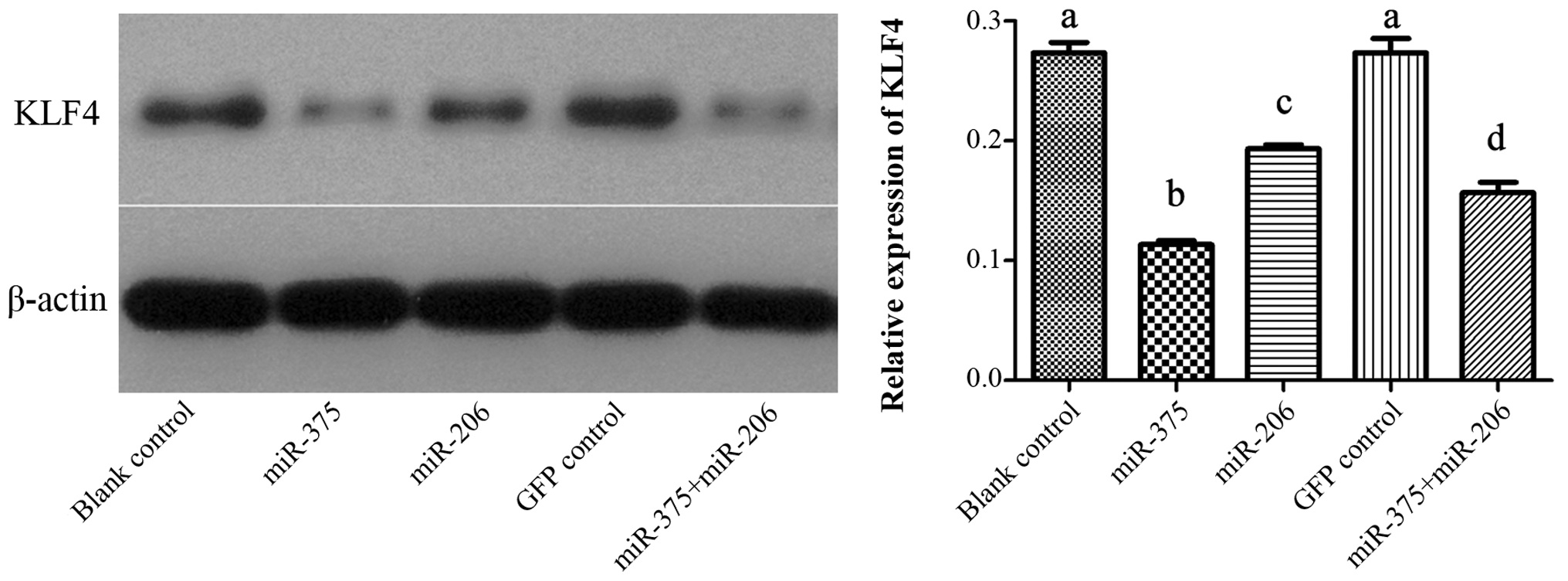
Results of western blotting showed that the
expression of KLF4 protein was significantly downregulated after
forced expression of miR-375. The level of KLF4 in the miR-206
group was also decreased compared with the control groups, but not
as much as miR-375. In addition, the expression of KLF4 in the
co-transfection group was lower than that in the miR-206 group but
higher than that in the miR-375 group (P<0.05). However, cells
in the GFP control group did not show any significant changes in
the expression of KLF4 protein as compared to the untreated Hep-2
cells (P>0.05; Fig. 7).
Discussion
Increasing evidence suggests that miRNAs play key
roles in diverse biological processes, including development, cell
proliferation, differentiation and apoptosis (31,32).
Furthermore, miRNAs have also been reported to play essential roles
in carcinogenesis and tumor progression (33,34).
Numerous published studies have reported that miR-375 is
downregulated in several types of cancer (35,36).
In the present study, it was confirmed that miR-375 was
downregulated in LSCC, which is consistent with the results of Luo
et al (37). It was found
that lower expression of miR-375 was closely correlated with lymph
node metastasis, advanced clinical stage and high T classification.
Considering the association of these clinicopathological parameters
with the poor prognosis of patients with LSCC, these results imply
that miR-375 may play a role in the progression and influence the
prognosis in LSCC. To explore the effect of the overexpression of
miR-375 on the metastasis and progression of LSCC, we examined the
alterations in cell growth and behavior following miR-375
overexpression in the Hep-2 cells. Elevated expression of miR-375
suppressed proliferation, induced cell cycle arrest in the G1
phase, and suppressed cell migration and invasion in the Hep-2
cells. Taken together, these results suggest that miR-375 is a
tumor-suppressor in the growth and progression of LSCC. Various
previous studies have proposed that the co-regulation of one target
gene or even an entire biological module by combinations of miRNAs
may lead to potent synergistic effects (38,39).
In the present study, both miR-375 and miR-206 were overexpressed
to test the effects on tumor cell function. However, the results
indicated that the inhibitory effect in the co-infected LSCC cells
was not stronger than that noted in the LSCC cells transfected with
miR-375 alone, although it was better than that in the Hep-2 cells
transfected with miR-206 alone.
KLF4 was of particular interest to us due to its
role in proliferation during tumorigenesis and its dual functions
in the context of the tumor microenvironment. For instance, KLF4
appears to suppress the formation of tumors in tissues such as the
gut (26), while it can promote
malignant properties in other tissues, such as the breast and skin
(27–29). KLF4 plays an important role in
regulating multiple biological functions, including proliferation,
survival, epithelial-mesenchymal transition, migration, invasion
and capillary tube formation (40,41).
Consistent with Suer et al (42), KLF4 was found to be overexpressed in
LSCC tumor tissues compared with its level in adjacent normal
tissues. In the present study, an inverse correlation was found
between the expression of miR-375 and KLF4 in the LSCC tissues.
Furthermore, expression of KLF4 at both the mRNA and protein levels
was significantly downregulated by overexpression of miR-375 using
miR-375 mimic transfection. KLF4 was also inhibited following
over-expression of miR-206, but the inhibitory effect of miR-375 on
KLF4 was more obvious than that of miR-375 + miR-206, and the
inhibitory effect of miR-206 was the weakest. Taken together, these
results suggest that decreased KLF4, induced by restoring miR-375,
participates in the progression of LSCC.
Previous studies have indicated that KLF4 can
upregulate the expression of miR-206, and the latter can also
promote the expression of KLF4 by an autoregulatory feedback loop
formed by KLF4 and miR-206 (25),
the regulatory mechanism related to tumor initiation (43). Thus, while both miR-375 and miR-206
can act on KLF4 (7,30), the regulatory pathways are
different. Furthermore, the two miRNAs may depend on other crucial
target genes which participate in the mechanisms of blockade of
proliferation of LSCC cells or increase in apoptosis. Hence, no
synergistic effects were noted in the LSCC cells by simple
co-transfection of miR-375 and miR-206.
Another possible reason concerning the results of
the present study is as follows. KLF4 can bind to β-catenin and
decrease mRNA levels, and can also inhibit β-catenin function and
signaling. Furthermore, KLF4 shows inhibitory activity in
β-catenin-mutant cells (44).
β-catenin is overexpressed in LSCC (45). Downregulation of miR-375 is reported
to be associated with β-catenin mutants, suggesting that β-catenin
signaling can repress miR-375 (46,47).
It was demonstrated that miR-375 suppresses the expression of KLF4.
Thus, KLF4, β-catenin and miR-375 may mutually restrict only to
form a circle. When both miR-375 and miR-206 are overexpressed,
exogenous miR-206 may decrease KLF4 mRNA levels, thereby disrupting
the mutual restriction relationship among miR-375, KLF4 and
β-catenin, ultimately impairing the inhibitory function of miR-375
toward KLF4, such that the combination of miR-375 and miR-206 did
not strengthen the inhibitory effect of miR-375. Such interaction
could partially explain the finding that the co-transfection did
not have the strongest effect in LSCC cells.
In conclusion, the expression of miR-375 was
downregulated in LSCC tissues and was correlated with neck lymph
node metastasis, clinical stage and other poor prognostic
clinicopathological parameters. The increased expression of miR-375
suppressed cell proliferation, invasion and promoted cell apoptosis
of LSCC cells through KLF4 in vitro. Taken together, these
results suggest that the overexpression of miR-375 induced the
decrease in the expression of target gene KLF4, and consequently
inhibited cell malignant behaviors of LSCC. These results also
suggest that the miRNA-regulatory network is further complicated by
the fact that any single miRNA can regulate hundreds of targets,
while multiple miRNAs may converge to control the same process
(48).
Unfortunately, the complete mechanisms by which
miRNAs regulate the function of KLF4 in LSCC and the doses of
miRNAs to KLF4 are not known yet, but the present findings clearly
indicate that simply combining several miRNAs with a common target
gene does not always yield additive effects. miR-375 presents an
exciting new opportunity for the treatment of laryngeal cancer in
the future. In addition, assessing the effects of combinatorial
miRNA treatment on different tumor types remains necessary, as this
information could be useful for developing therapeutic agents
targeting LSCC.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of China (nos. 81402234, 81572647 and
81372902), the National Science Foundation of China, and the
National Science Foundation of Heilongjiang Province (QC2013C117,
ZD201215/H1302).
References
|
1
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parasramka MA, Ho E, Williams DE and
Dashwood RH: MicroRNAs, diet, and cancer: New mechanistic insights
on the epigenetic actions of phytochemicals. Mol Carcinog.
51:213–230. 2012. View
Article : Google Scholar
|
|
4
|
Guo X, Chen Y, Xu Z, Xu Z, Qian Y and Yu
X: Prognostic significance of VEGF-C expression in correlation with
COX-2, lymphatic microvessel density, and clinicopathologic
characteristics in human non-small cell lung cancer. Acta Biochim
Biophys Sin. 41:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Genden EM, Ferlito A, Silver CE, Jacobson
AS, Werner JA, Suárez C, Leemans CR, Bradley PJ and Rinaldo A:
Evolution of the management of laryngeal cancer. Oral Oncol.
43:431–439. 2007. View Article : Google Scholar
|
|
6
|
Ma J, Liu Y, Huang XL, Zhang ZY, Myers JN,
Neskey DM and Zhong LP: Induction chemotherapy decreases the rate
of distant metastasis in patients with head and neck squamous cell
carcinoma but does not improve survival or locoregional control: A
meta-analysis. Oral Oncol. 48:1076–1084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mao Q, Quan T, Luo B, Guo X, Liu L and
Zheng Q: miR-375 targets KLF4 and impacts the proliferation of
colorectal carcinoma. Tumour Biol. 37:463–471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, et
al: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar
|
|
11
|
Li M, Tian L, Wang L, Yao H, Zhang J, Lu
J, Sun Y, Gao X, Xiao H and Liu M: Down-regulation of miR-129-5p
inhibits growth and induces apoptosis in laryngeal squamous cell
carcinoma by targeting APC. PLoS One. 8:e778292013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren J, Zhu D, Liu M, Sun Y and Tian L:
Downregulation of miR-21 modulates Ras expression to promote
apoptosis and suppress invasion of laryngeal squamous cell
carcinoma. Eur J Cancer. 46:3409–3416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang T, Liu M, Wang C, Lin C, Sun Y and
Jin D: Down-regulation of miR-206 promotes proliferation and
invasion of laryngeal cancer by regulating VEGF expression.
Anticancer Res. 31:3859–3863. 2011.PubMed/NCBI
|
|
14
|
Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M
and Xiao H: miR-203 is downregulated in laryngeal squamous cell
carcinoma and can suppress proliferation and induce apoptosis of
tumours. Tumour Biol. 35:5953–5963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian L, Zhang J, Ge J, Xiao H, Lu J, Fu S,
Liu M and Sun Y: MicroRNA-205 suppresses proliferation and promotes
apoptosis in laryngeal squamous cell carcinoma. Med Oncol.
31:7852014. View Article : Google Scholar
|
|
16
|
Wu TY, Zhang TH, Qu LM, Feng JP, Tian LL,
Zhang BH, Li DD, Sun YN and Liu M: MiR-19a is correlated with
prognosis and apoptosis of laryngeal squamous cell carcinoma by
regulating TIMP-2 expression. Int J Clin Exp Pathol. 7:56–63.
2013.
|
|
17
|
Ren J, Sun Y, Zhao X, Wang X, Feng J, Liu
M and Zhu D: Downregulation of miR-21 regulates MMP-2 expression
and suppress migration of Laryngeal squamous cell carcinoma. Head
Neck Oncol. 4:65–69. 2012.
|
|
18
|
Boll K, Reiche K, Kasack K, Mörbt N,
Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn
F, et al: MiR-130a, miR-203 and miR-205 jointly repress key
oncogenic pathways and are downregulated in prostate carcinoma.
Oncogene. 32:277–285. 2013. View Article : Google Scholar
|
|
19
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting α subunit of protein kinase CKII in human
colorectal cancer cells. Biochem Biophys Res Commun. 429:173–179.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baroukh NN and Van Obberghen E: Function
of microRNA-375 and microRNA-124a in pancreas and brain. FEBS J.
276:6509–6521. 2009. View Article : Google Scholar
|
|
21
|
Kachakova D, Mitkova A, Popov E, Popov I,
Vlahova A, Dikov T, Christova S, Mitev V, Slavov C and Kaneva R:
Combinations of serum prostate-specific antigen and plasma
expression levels of let-7c, miR-30c, miR-141, and miR-375 as
potential better diagnostic biomarkers for prostate cancer. DNA
Cell Biol. 34:189–200. 2015. View Article : Google Scholar :
|
|
22
|
Fu C, Dong W, Wang Z, Li H, Qin Q and Li
B: The expression of miR-21 and miR-375 predict prognosis of
esophageal cancer. Biochem Biophys Res Commun. 446:1197–1203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim HK, Lee YS, Sivaprasad U, Malhotra A
and Dutta A: Muscle-specific microRNA miR-206 promotes muscle
differentiation. J Cell Biol. 174:677–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geiman DE, Ton-That H, Johnson JM and Yang
VW: Transactivation and growth suppression by the gut-enriched
Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic
amino acid residues and protein-protein interaction. Nucleic Acids
Res. 28:1106–1113. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin CC, Liu LZ, Addison JB, Wonderlin WF,
Ivanov AV and Ruppert JM: A KLF4-miRNA-206 autoregulatory feedback
loop can promote or inhibit protein translation depending upon cell
context. Mol Cell Biol. 31:2513–2527. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghaleb AM, McConnell BB, Nandan MO, Katz
JP, Kaestner KH and Yang VW: Haploinsufficiency of Krüppel-like
factor 4 promotes adenomatous polyposis coli dependent intestinal
tumorigenesis. Cancer Res. 67:7147–7154. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Foster KW, Ren S, Louro ID, Lobo-Ruppert
SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT and
Ruppert JM: Oncogene expression cloning by retroviral transduction
of adenovirus E1A-immortalized rat kidney RK3E cells:
Transformation of a host with epithelial features by c-MYC and the
zinc finger protein GKLF. Cell Growth Differ. 10:423–434.
1999.PubMed/NCBI
|
|
28
|
Liu Z, Teng L, Bailey SK, Frost AR, Bland
KI, LoBuglio AF, Ruppert JM and Lobo-Ruppert SM: Epithelial
transformation by KLF4 requires Notch1 but not canonical Notch1
signaling. Cancer Biol Ther. 8:1840–1851. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pandya AY, Talley LI, Frost AR, Fitzgerald
TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA,
Krontiras H, et al: Nuclear localization of KLF4 is associated with
an aggressive phenotype in early-stage breast cancer. Clin Cancer
Res. 10:2709–2719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Parasramka MA, Dashwood WM, Wang R, Saeed
HH, Williams DE, Ho E and Dashwood RH: A role for low-abundance
miRNAs in colon cancer: The miR-206/Krüppel-like factor 4 (KLF4)
axis. Clin Epigenetics. 4:162012. View Article : Google Scholar
|
|
31
|
Sassen S, Miska EA and Caldas C: MicroRNA:
Implications for cancer. Virchows Arch. 452:1–10. 2008. View Article : Google Scholar
|
|
32
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
96(Suppl): R40–R44. 2007.PubMed/NCBI
|
|
33
|
Shen Y, Tang D, Yao R, Wang M, Wang Y, Yao
Y, Li X and Zhang H: microRNA expression profiles associated with
survival, disease progression, and response to gefitinib in
completely resected non-small-cell lung cancer with EGFR mutation.
Med Oncol. 30:750–757. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi B, Piazza GA, Su X and Xi Y: MicroRNA
and cancer chemo-prevention. Cancer Prev Res. 6:401–409. 2013.
View Article : Google Scholar
|
|
35
|
Shi ZC, Chu XR, Wu YG, Wu JH, Lu CW, Lü
RX, Ding MC and Mao NF: MicroRNA-375 functions as a tumor
suppressor in osteosarcoma by targeting PIK3CA. Tumour Biol.
36:8579–8584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Xing R, Zhang X, Dong W, Zhang J,
Yan Z, Li W, Cui J and Lu Y: miR-375 targets the p53 gene to
regulate cellular response to ionizing radiation and etoposide in
gastric cancer cells. DNA Repair. 12:741–750. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS,
Yang W, Fu QL and Lei W: miR-375 suppresses IGF1R expression and
contributes to inhibition of cell progression in laryngeal squamous
cell carcinoma. Biomed Res Int. 2014:3745982014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dong CG, Wu WK, Feng SY, Wang XJ, Shao JF
and Qiao J: Co-inhibition of microRNA-10b and microRNA-21 exerts
synergistic inhibition on the proliferation and invasion of human
glioma cells. Int J Oncol. 41:1005–1012. 2012.PubMed/NCBI
|
|
39
|
Noguchi S, Yasui Y, Iwasaki J, Kumazaki M,
Yamada N, Naito S and Akao Y: Replacement treatment with
microRNA-143 and -145 induces synergistic inhibition of the growth
of human bladder cancer cells by regulating PI3K/Akt and MAPK
signaling pathways. Cancer Lett. 328:353–361. 2013. View Article : Google Scholar
|
|
40
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng X, Li A, Zhao L, Zhou T, Shen Q, Cui
Q and Qin X: Key role of microRNA-15a in the KLF4 suppressions of
proliferation and angiogenesis in endothelial and vascular smooth
muscle cells. Biochem Biophys Res Commun. 437:625–631. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Suer I, Karatas OF, Yuceturk B, Yilmaz M,
Guven G, Buge O, Cansiz H and Ozen M: Characterization of stem-like
cells directly isolated from freshly resected laryngeal squamous
cell carcinoma specimens. Curr Stem Cell Res Ther. 9:347–353. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin CC, Sharma SB, Farrugia MK, McLaughlin
SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D and
Ruppert JM: Kruppel-like factor 4 signals through microRNA-206 to
promote tumor initiation and cell survival. Oncogenesis.
4:e1552015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang W, Chen X, Kato Y, Evans PM, Yuan S,
Yang J, Rychahou PG, Yang VW, He X, Evers BM, et al: Novel cross
talk of Kruppel-like factor 4 and beta-catenin regulates normal
intestinal homeostasis and tumor repression. Mol Cell Biol.
26:2055–2064. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Greco A, De Virgilio A, Rizzo MI, Pandolfi
F, Rosati D and de Vincentiis M: The prognostic role of E-cadherin
and β-catenin overexpression in laryngeal squamous cell carcinoma.
Laryngoscope. 126:E148–E155. 2016. View Article : Google Scholar
|
|
46
|
Wang Y, Huang C, Reddy Chintagari N,
Bhaskaran M, Weng T, Guo Y, Xiao X and Liu L: miR-375 regulates rat
alveolar epithelial cell trans-differentiation by inhibiting
Wnt/β-catenin pathway. Nucleic Acids Res. 41:3833–3844. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bracken CP, Khew-Goodall Y and Goodall GJ:
Network-based approaches to understand the roles of miR-200 and
other microRNAs in cancer. Cancer Res. 75:2594–2599. 2015.
View Article : Google Scholar : PubMed/NCBI
|