Introduction
Lung cancer is the most common malignancy worldwide
(1), and approximately 1.82 million
new cases and 1.56 million deaths were reported in 2012 (2). As the most common sub-type of
non-small cell lung cancer (NSCLC), lung adenocarcinoma accounts
for nearly 40% of cases (3).
Although many advances in clinical management have been achieved
recently, e.g. surgery, radiotherapy, chemotherapy, and targeted
therapy, the 5-year survival rate of lung adenocarcinoma is still
less than 15%, in part because a majority of cases are diagnosed at
a more malignant stage (4). Hence,
screening individuals with high risk of developing lung
adenocarcinoma at an early stage has the potential to improve
clinical outcome.
MicroRNAs (miRNAs) belong to a new class of
endogenous small non-coding RNAs, and they generally play key roles
in regulating the translation and degradation of their target mRNAs
and thus participate in many biological and pathological processes
(5,6). The differential expression and
potential diagnostic values of miRNAs have been widely investigated
in human lung adenocarcinoma (7,8). For
instance, Patnaik et al reported miRNAs that were
differentially expressed between whole blood samples from lung
adenocarcinoma patients and healthy individuals, e.g., let-7e,
miR-22, miR-30a-5p, miR-185, miR-210 and miR-423-5p, using miRNA
expression profiles (9). A study in
a Chinese population showed that high level of miR-155 in serum
specimens had a high sensitivity for diagnosing lung adenocarcinoma
(10). Additionally, combinations
of 4 miRNAs (miR-21, miR-486, miR-375 and miR-200b) and 7 miRNAs
(miR-486, miR-126, miR-145, miR-21, miR-182, miR-200b and miR-375)
in sputum were screened as marker panels to distinguish patients
with stage I lung adenocarcinoma from healthy individuals (11). In addition, an increased level of
miR-21 expression in surgical resected lung adenocarcinoma tissues
was found to be related to tumor node metastasis (TNM) stage II-IV
compared to stage I (12). In spite
of these efforts, using aberrant miRNA expression levels to
diagnose lung adenocarcinoma or predict the clinicopathological
features of lung adenocarcinoma has not been fully investigated,
especially in Chinese cases.
Hence, in the present study, we compared miRNA
expression profiles between surgical resected lung adenocarcinoma
tissues and the corresponding paired non-cancerous tissues, as well
as identified the potential miRNAs with which to predict the
clinicopathological features of lung adenocarcinoma. Furthermore,
bioinformatics approaches such as prediction of target genes and
enrichment analysis were performed to explore the underlying
molecular mechanisms.
Materials and methods
Statement of ethics
All samples used in this study were obtained from
the tissue bank of the 117th Hospital of the People's Liberation
Army (PLA), and written informed consent was obtained from all
subjects. This study was approved by the Ethics Committee of the
117th Hospital of PLA.
Patients and study design
miRNA microarray data were obtained using the tumor
tissues of 10 patients (gender, 4 males and 6 females; age,
50.0±10.8 years) with lung adenocarcinoma, as well as the paired
control samples from their adjacent normal tissues. Thereafter,
differentially expressed miRNAs (DEMs) were identified, and were
verified using real-time quantitative reverse
transcription-polymerase chain reaction (qRT-PCR) from the 10
sample pairs. Furthermore, the clinical information of the 10
patients and an additional 39 patients were collected to analyze
the correlations between miRNA expression and clinicopathological
features, including gender, age, pathological stage, histological
grade of differentiation, tumor diameter and lymph node
metastasis.
All tumor specimens used in this study were
histologically classified as lung adenocarcinoma based on the World
Health Organization (WHO) classification of lung tumors (13). The pathological tumor stages were
determined based on the TNM classification criteria established by
the International Union Against Cancer (IUAC) (14). The histological grades were
classified as well/moderately/poorly differentiated (15).
RNA extraction
Total RNA was isolated using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA). RNA quality was determined using a
Bioanalyzer (UV spectrophotometer Q3000; Quawell, San Jose, CA,
USA) and RNA concentration was assessed using NanoDrop™ 2000
(Thermo Scientific, Waltham, MA, USA). RNA samples were stored at
−80°C until used for miRNA microarray or qRT-PCR.
miRNA expression microarray
For each sample, 4–8 μg of RNA was used to
perform miRNA microarray assay. Briefly, the RNA sample was size
fractionated (<200 nucleotides) and 3′-polyadenylated by using
poly(A) polymerase. Then, an oligonucleotide tag conjugating Cy3
dye was ligated to the poly(A) tail for later fluorescent dye
staining. Hybridization was performed overnight at 34°C on a
μParaflo™ microfluidic chip (LC Sciences, Houston, TX, USA)
using 6X SSPE buffer (0.90 M NaCl, 60 mM
Na2HPO4, 6 mM EDTA, pH 6.8) plus 25%
formamide (16). Each of the probes
on the microfluidic chip contained a chemically modified nucleotide
segment complementary to a certain miRNA, as well as a long
non-nucleotide molecule spacer. After washing, the chip was scanned
using a laser scanner (GenePix 4000B; Molecular Devices, Sunnyvale,
CA, USA) and the fluorescent images were digitized using Array-Pro
image analysis software (Media Cybernetics, Carlsbad, CA, USA).
Idetification of DEMs based on the miRNA
microarray
The digitized data were analyzed by first
subtracting the background and then normalizing the signals using
locally-weighted regression (LOWESS) filter (17). miRNAs with microarray signal <500
were excluded, and two-tailed paired-sample t-test (18) was used to identify the DEMs between
lung adenocarcinoma and normal tissues. To obtain more potential
miRNA signatures associated with lung adenocarcinoma, miRNAs with
P<0.1 and fold change (FC) ≠1 were considered to be DEMs in the
microarray analysis.
Verification of DEMs based on
qRT-PCR
Expression levels of DEMs were detected using
qRT-PCR experiment. Briefly, PrimeScript® RT reagent kit
(DRR037A; Takara, Tokyo, Japan) was used to reverse transcribe 40
ng of RNA using miRNA-specific oligonucleotides. Subsequently, PCR
amplification was performed using Platinum SYBR Green qPCR
SuperMix-UDG kit (11733-038; Invitrogen) and the ABI StepOne Plus
Real-Time PCR system (ABI PRISM® 7900HT; Applied
Biosystems, Foster City, CA, USA). PCR protocol was: 50°C for 2
min, 95°C for 2 min, 39 cycles of 95°C for 15 sec and 60°C for 30
sec. Expression levels of miRNAs in each sample were normalized to
the internal control (RNU6B), and relative expression values were
calculated based on the 2−ΔΔCt method. When comparing
miRNA expression levels between cancer and normal groups, P<0.05
was considered as a criterion of significance.
miRNA target prediction, enrichment
analysis and miRNA-TF-target network construction
Target genes of the DEMs were predicted using
TargetScan (version 6.2, http://www.targetscan.org) (19), a target-prediction program. Based on
the Kyoto Encylopedia of Genes and Genomes (KEGG) database
(20), pathway enrichment analysis
of the predicted target genes was carried out using Database for
Annotation, Visualization and Integrated Discovery (DAVID, version
6.7, https://david.ncifcrf.gov/) (21). P-value <0.05 and gene count ≥2
were used as the thresholds for pathway enrichment analysis. Then,
transcription factors (TFs) were extracted from the target genes
based on the TRANScription FACtor (TRANSFAC, version 7.0,
http://www.gene-regulation.com/pub/databases.html)
(22) and ENCyclopedia Of DNA
Elements (ENCODE, http://encodeproject.org) (23) databases. Interactions between the
protein products of the target genes were extracted from Search
Tool for the Retrieval of Interacting Genes (STRING) database
(24), using combination score
>0.7 as the criterion. Finally, the miRNA-TF-target network was
constructed and visualized using Cytoscape software (version 3.2.1,
http://cytoscape.org) (25) to show the interactions among miRNAs,
TFs and target genes.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 19.0; SPSS Inc., Chicago, IL, USA). For
non-normally distributed variables, data are presented using median
and interquartile range (IQR), and Mann-Whitney U test was used to
analyze the correlations between miRNA expression and
clinicopathological features of patients with lung adenocarcinoma
(n=49). To further determine whether miRNA signatures have the
capability of predicting clinicopathological features, receiver
operating characteristics (ROC) were generated, and area under the
curve and feasible threshold values were calculated. P<0.05 was
set as the cut-off criterion for statistical significance.
Results
DEMs in lung adenocarcinoma
Ten samples of lung adenocarcinoma and their paired
adjacent normal tissues were analyzed via miRNA expression
microarray. Overall, 119 miRNAs were differentially expressed
between two groups, among them, 99 miRNAs were excluded due to
their weak microarray signal (signal <500). As a result, 26
miRNAs were identified as DEMs between lung adenocarcinoma and
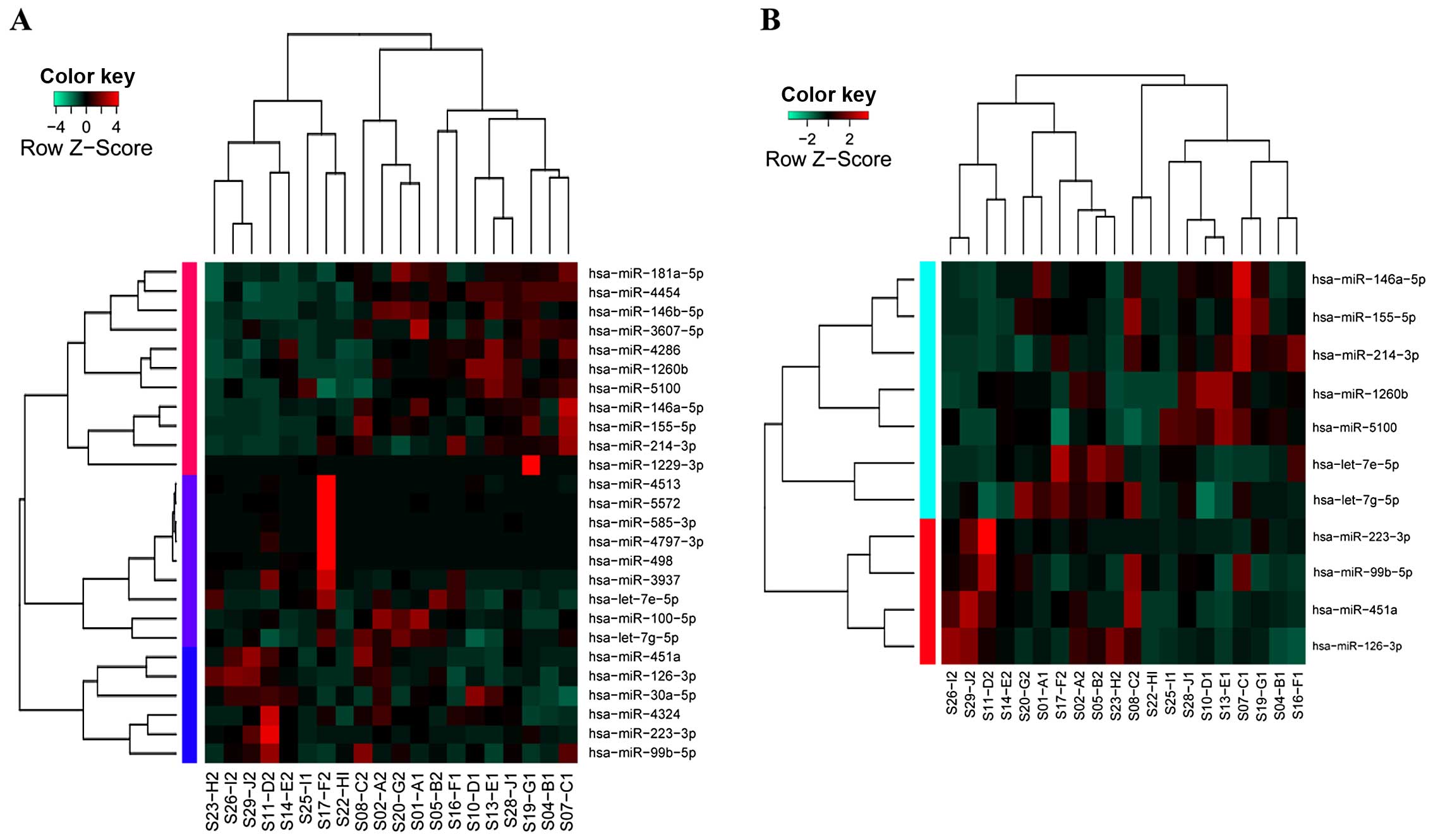
normal tissues. Hierarchical clustering analysis showed that 26
DEMs were unable to clearly distinguish these two types of samples
(sensitivity, 80%; specificity, 60%; Fig. 1A), while the top 11 DEMs displayed
satisfactory sensitivity (90%) and specificity (90%) (Fig. 1B). Therefore, qRT-PCR was further
performed for the 11 DEMs.
Validation of DEMs based on qRT-PCR
Expression levels of the 11 miRNAs were detected
using qRT-PCR. Results indicated that miR-126-3p (P=1.95E-05) and
miR-451a (P=1.56E-04) were significantly downregulated (Table I) in lung adenocarcinoma, and this
was consistent with the results of the microarray analysis. In
addition, changes in expression of 6 miRNAs including miR-214-3p,
miR-223-3p, miR-155-5p, miR-1260b, miR-5100 and miR-146a-5p showed
the same tendency, although the differences were not statistically
significant (Table I; P=0.0587,
0.0767, 0.0856, 0.218, 0.247 and 0.257, respectively). Hence,
miR-126-3p and miR-451a were defined as candidate miRNA signatures
of lung adenocarcinoma, and were thus further analyzed.
 | Table IValidation of miRNA microarray
results using qRT-PCR. |
Table I
Validation of miRNA microarray
results using qRT-PCR.
| miRNA microarray
| qRT-PCR
|
|---|
| FC |
log2FC | P-value | FC |
log2FC | P-value |
|---|
|
hsa-miR-126-3pa,b | 0.46 | −1.12 | 4.68E-04 | 0.29 | −0.56 | 1.95E-05 |
|
hsa-miR-451aa,b | 0.23 | −2.11 | 1.12E-03 | 0.26 | −0.51 | 1.56E-04 |
| hsa-miR-99b-5p | 0.73 | −0.45 | 3.39E-02 | 1.51 | 1.68 | 2.32E-02 |
|
hsa-miR-214-3pa | 1.73 | 0.79 | 2.59E-03 | 2.53 | 0.75 | 5.87E-02 |
|
hsa-miR-223-3pa | 0.48 | −1.06 | 6.46E-02 | 0.76 | −2.53 | 7.67E-02 |
|
hsa-miR-155-5pa | 1.53 | 0.61 | 4.45E-02 | 2.26 | 0.85 | 8.56E-02 |
| hsa-let-7g-5p | 0.62 | −0.69 | 1.27E-02 | 1.19 | 3.98 | 1.56E-01 |
| hsa-let-7e-5p | 0.55 | −0.87 | 9.92E-02 | 1.31 | 2.57 | 1.76E-01 |
|
hsa-miR-1260ba | 2.17 | 1.12 | 3.12E-02 | 1.42 | 1.98 | 2.18E-01 |
|
hsa-miR-5100a | 1.77 | 0.82 | 4.97E-03 | 1.50 | 1.71 | 2.47E-01 |
|
hsa-miR-146a-5pa | 1.97 | 0.98 | 6.16E-02 | 2.34 | 0.82 | 2.57E-01 |
Correlations between candidate miRNA
signatures and clinicopathological features of lung
adenocarcinoma
We next assessed the correlation of miR-126-3p and
miR-451a expression (qRT-PCR) with the clinicopathological features
of 49 patients with lung adenocarcinoma (gender, 24 males and 25
females; age, 59.76±12.35 years). A total of 25 cases in
pathological stage I and 24 cases in pathological stage II-IV were
included, and the tumor diameter ranged from 0.8 to 12 cm.
Additionally, 32 cases were well or moderately differentiated,
while 17 cases were poorly differentiated. Lymph node metastasis
occurred in 22 patients. In order to show the specificity and
significance of miR-126-3p and miR-451a, we utilized a negative
control, miR-214-3p, which had a tendency to be significant in the
qRT-PCR validation.
As a consequence, no significant correlation was
observed between the expression levels of miR-126-3p (or miR-451a)
and 3 clinicopathological features including gender, age and
histological grade of differentiation (P>0.05; Table II). However, lower expression
levels of miR-126-3p and miR-451a were detected in pathological
stage II-IV cases when compared with stage I cases (P=0.010 and
0.004, respectively), in cases with tumor diameter >3 cm when
compared with those with tumor diameter ≤3 cm (P=0.038 and 0.030,
respectively), as well as in cases with lymph node metastasis when
compared with those without lymph node metastasis (P=0.023 and
0.003, respectively) (Table II).
Additionally, no correlation was observed between miR-214-3p
expression level and any tested clinicopathological feature
(P>0.05).
 | Table IICorrelation between
clinicopathological features and expression levels of miR-126-3p,
miR-451a and miR-214-3p in 49 patients with lung
adenocarcinoma. |
Table II
Correlation between
clinicopathological features and expression levels of miR-126-3p,
miR-451a and miR-214-3p in 49 patients with lung
adenocarcinoma.
| Parameter | N (%) | miR-126-3p
| miR-451a
| miR-214-3p
|
|---|
| Median (IQR) | P-value | Median (IQR) | P-value | Median (IQR) | P-value |
|---|
| Gender |
| Male | 24 (49) | 11.96 (6.77,
39.59) | 0.603 | 3.45 (1.18,
10.16) | 0.689 | 1.00 (0.646,
1.88) | 0.203 |
| Female | 25 (51) | 22.46 (7.07,
33.67) | | 3.45 (1.25,
9.38) | | 1.71 (0.77,
2.23) | |
| Age (years) |
| ≤60 | 25 (51) | 17.99 (6.39,
34.95) | 0.795 | 3.45 (1.21,
9.28) | 0.920 | 1.61, (0.67,
2.18) | 0.484 |
| >60 | 24 (49) | 16.76 (7.62,
35.25) | | 3.40 (1.23,
11.10) | | 1.13, (0.65,
1.92) | |
| Pathological
stage |
| I | 25 (51) | 24.45 (9.26,
57.34) | 0.010a | 4.71 (3.29,
13.31) | 0.004b | 1.79 (0.65,
2.53) | 0.379 |
| II-IV | 24 (49) | 8.81 (5.50,
22.14) | | 1.67 (0.90,
7.15) | | 1.23 (0.67,
1.81) | |
| Histologic grade of
differentiation |
|
Well/moderately | 32 (65) | 16.81 (6.98,
29.06) | 0.900 | 3.52 (1.60,
9.43) | 0.721 | 1.53 (0.69,
2.10) | 0.366 |
| Poorly | 17 (37) | 17.87 (6.23,
46.71) | | 2.47 (1.04,
10.69) | | 1.17 (0.59,
1.92) | |
| Tumor diameter
(cm) |
| ≤3 | 29 (59) | 23.54 (8.53,
56.01) | 0.038a | 4.09 (2.10,
13.52) | 0.030a | 1.53 (0.69,
2.08) | 0.502 |
| >3 | 20 (41) | 10.60 (4.91,
23.03) | | 2.40 (0.87,
7.46) | | 1.32 (0.56,
1.92) | |
| Lymph node
metastasis |
| Negative | 27 (55) | 23.54 (8.86,
53.03) | 0.023a | 4.28 (3.26,
11.16) | 0.003b | 1.79 (0.61,
2.25) | 0.688 |
| Positive | 22 (45) | 8.82 (5.87,
21.61) | | 1.58 (0.83,
5.63) | | 1.24 (0.68,
1.80) | |
The ROC results suggested that both miR-126-3p and
miR-451a had the capability to predict pathological stage, tumor
diameter and occurrence of lymph node metastasis (Fig. 2). The optimal cut-off values of
miR-126-3p were 20.29, 17.93 and 18.44 for predicting pathological
stage (AUC=0.715, P=0.010), tumor diameter (AUC=0.676, P=0.038) and
lymph node metastasis (AUC=0.690, P=0.023). The corresponding
sensitivity and specificity were 64 and 75%, 62 and 70%, and 67 and
77%, respectively. At a cut-off value of 3.02, 3.02 and 2.40,
miR-451a showed the best potential to predict pathological stage
(AUC=0.742, P=0.004), tumor diameter (AUC=0.684, P=0.030) and lymph
node metastasis (AUC=0.749, P=0.003). The corresponding sensitivity
and specificity were 84 and 67%, 72 and 60%, and 89 and 64%,
respectively. In addition, there was no difference between
miR-126-3p and miR-451a in the capability of predicting pathologic
stage (P=0.317), tumor diameter (P=0.090) or lymph node metastasis
(P=0.438).
miRNA-TF-target network associated with
lung adenocarcinoma and pathway enrichment analysis of target
genes
Totally, 154 and 397 target genes of miR-126-3p and
miR-451a were predicted, respectively, and 10 genes were
co-regulated by miR-126-3p and miR-451a, including AMMECR1L,
FBXO33, GATAD2B, HIP1, KCMF1, KIAA1456, PCDH7, SAMD12, TSC1 and
ZADH2. Target genes of miR-126-3p were significantly enriched in 29
KEGG pathways such as 'apoptosis', 'prostate cancer', 'focal
adhesion', 'mTOR signaling pathway', 'endometrial cancer' and
'non-small cell lung cancer' (the top 15 pathways are shown in
Table III), while target genes of
miR-451a were markedly enriched in 5 KEGG pathways such as 'PPAR
signaling pathway' (e.g., PPARA) (Table III).
 | Table IIIPathway enrichment analysis of the
target genes of miR-126-3p and miR-451a. |
Table III
Pathway enrichment analysis of the
target genes of miR-126-3p and miR-451a.
| miRNAs | ID | Terms | Count | P-value | Gene symbols |
|---|
| miR-126-3p | 4722 | Neurotrophin
signaling pathway | 8 | 2.00E-05 | BCL2, CRK, FOXO3,
FRS2, IRS1, IRS2, PIK3CD, PIK3R2 |
| 4960 |
Aldosterone-regulated sodium
reabsorption | 5 | 3.92E-05 | IRS1, IRS2, KCNJ1,
PIK3CD, PIK3R2 |
| 4210 | Apoptosis | 6 | 1.44E-04 | BCL2, DFFB, PIK3CD,
PIK3R2, PPP3CB, TNFRSF10B |
| 4930 | Type II diabetes
mellitus | 4 | 9.89E-04 | IRS1, IRS2, PIK3CD,
PIK3R2 |
| 4910 | Insulin signaling
pathway | 6 | 1.70E-03 | CRK, IRS1, IRS2,
PIK3CD, PIK3R2, TSC1 |
| 4730 | Long-term
depression | 4 | 4.00E-03 | GNA13, IGF1R, NOS1,
PLA2G12A |
| 4370 | VEGF signaling
pathway | 4 | 5.37E-03 | PIK3CD, PIK3R2,
PLA2G12A, PPP3CB |
| 533 | Glycosaminoglycan
biosynthesis-keratan sulfate | 2 | 8.37E-03 | B4GALT4, CHST6 |
| 5215 | Prostate
cancer | 4 | 9.35E-03 | BCL2, IGF1R,
PIK3CD, PIK3R2 |
| 4510 | Focal adhesion | 6 | 1.05E-02 | BCL2, CRK, IGF1R,
PARVA, PIK3CD, PIK3R2 |
| 4150 | mTOR signaling
pathway | 3 | 1.24E-02 | PIK3CD, PIK3R2,
TSC1 |
| 5213 | Endometrial
cancer | 3 | 1.24E-02 | FOXO3, PIK3CD,
PIK3R2 |
| 5014 | Amyotrophic lateral
sclerosis (ALS) | 3 | 1.31E-02 | BCL2, NOS1,
PPP3CB |
| 5223 | Non-small cell lung
cancer | 3 | 1.38E-02 | FOXO3, PIK3CD,
PIK3R2 |
| 5210 | Colorectal
cancer | 3 | 1.99E-02 | BCL2, PIK3CD,
PIK3R2 |
| miR-451a | 3320 | PPAR signaling
pathway | 5 | 1.21E-02 | ACADL, ADIPOQ,
CYP8B1, GK, PPARA |
| 561 | Glycerolipid
metabolism | 4 | 1.66E-02 | AKR1B1, DAK, GK,
LIPG |
| 51 | Fructose and
mannose metabolism | 3 | 3.35E-02 | AKR1B1, C12orf5,
PMM2 |
| 360 | Phenylalanine
metabolism | 2 | 4.34E-02 | ALDH3B2, MIF |
| 620 | Pyruvate
metabolism | 3 | 4.38E-02 | ACACA, ACYP2,
AKR1B1 |
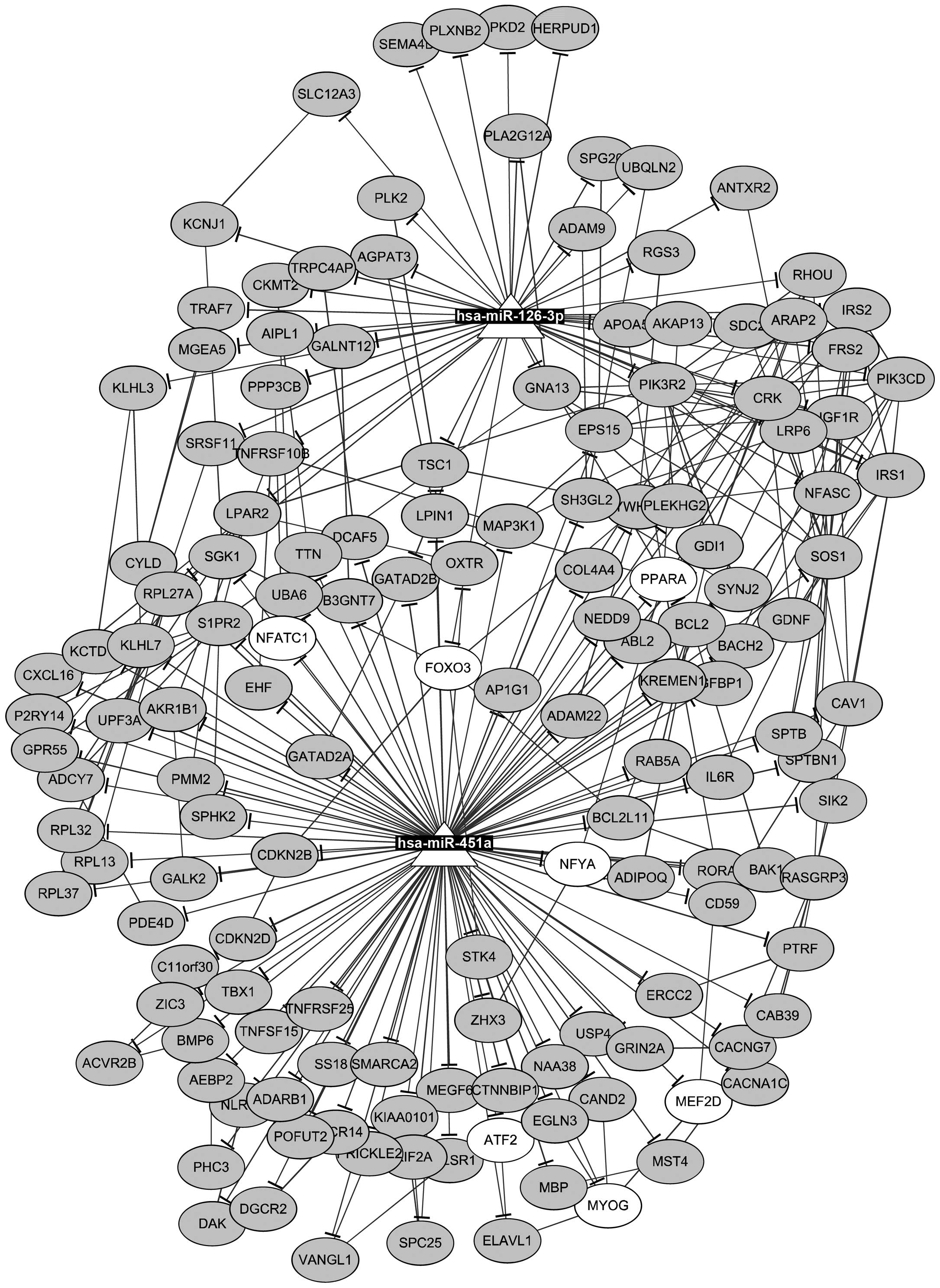
Furthermore, 14 TFs and 181 protein-protein
interactions were found among the target genes of miR-126-3p and
miR-451a. Then, an miRNA-TF-target network was constructed
(Fig. 3), containing 2 miRNAs, 7
TFs, 148 targets, as well as 150 miRNA-target pairs, 181
protein-protein pairs, and 21 TF-target pairs. For instance, PLXNB2
was targeted by miR-126-3p, PPARA was targeted by miR-451a, and
TSC1 was targeted by both miR-126-3p and miR-451a. In addition, a
sub-network was identified in the miRNA-TF-target network,
including 2 miRNAs, 15 targets, and 7 TFs (Fig. 4). Only target genes which were
targeted by both miRNAs and TFs were included in this sub-network.
The regulations between miRNAs and target genes (n=22) as well as
regulations between TFs and target genes (n=19) were clearly
revealed in the sub-network.
Discussion
A number of studies have directly profiled miRNA
expression in lung cancers, and some miRNAs have been identified
for cancer diagnosis and prognosis. However, limited studies have
focused on predicting clinicopathological features of lung cancers
(26–28), especially lung adenocarcinoma. In
this study, we screened the miRNA signatures using miRNA expression
profiling and qRT-PCR and identified two miRNAs (miR-126-3p and
miR-451a), which were significantly correlated with pathological
stage, tumor diameter and lymph node metastasis. In addition, ROC
analysis showed that both miR-126-3p and miR-451a were
significantly correlated with pathological stage, tumor diameter
and occurrence of lymph node metastasis.
Jusufovic et al reported that the miR-126
level was lower in NSCLC (including squamous and adenocarcinoma)
tissues in comparison with normal controls (29). However, it is unknown whether
miR-126 is a special signature for the severity of lung
adenocarcinoma. We also found that miR-126-3p was downregulated in
lung adenocarcinoma tissues, and low expression of miR-126-3p was
associated with large tumor size, as well as high pathological
stage. Liu et al showed that exogenous expression of miR-126
could inhibit the proliferation of human lung adenocarcinoma
epithelial cells and reduce the tumor weight in mice (30). Therefore, we proposed that the
downregulation of miR-126-3p in lung adenocarcinoma could promote
tumor cell proliferation and consequently increase tumor
severity.
Bioinformatics analysis was conducted to
preliminarily explore the possible molecular mechanism underlying
the effects of miR-126-3p on lung adenocarcinoma. A total of 154
genes were predicted to be regulated by miR-126-3p, and these
target genes were mainly enriched in cancer-related pathways, e.g.,
'apoptosis', 'prostate cancer', 'focal adhesion', 'endometrial
cancer' and 'non-small cell lung cancer'. Especially, miR-126-3p
was predicted to target plexin B2 (PLXNB2), which plays a role in
RhoA activation (31). Reportedly,
RhoA was found to promote cell proliferation in breast cancer and
esophageal squamous cell carcinoma cell lines (32,33).
Additionally, Pillé et al found that miR-126 could suppress
cell proliferation via inhibiting the RhoA/ROCK signaling pathway
in colon cancer cell lines (34).
Hence, we supposed that a similar mechanism may exist in lung
adenocarcinoma, i.e., miR-126-3p could restrain cell proliferation
via inhibiting PLXNB2 expression and the subsequent RhoA/ROCK
signaling pathway. In lung adenocarcinoma, miR-126-3p was
significantly downregulated, and thus its target PLXNB2 might be
upregulated, resulting in increased tumor cell proliferation and
tumor severity.
Moreover, we found that decreased expression of
miR-126-3p was related to the occurrence of lymph node metastasis.
Reportedly, RhoA is a key regulator of tumor cell motility
(35), and its activation is
associated with PLXNB2, a target of miR-126-3p. In addition,
over-activation of RhoA promotes non-physiological breakage of
cell-cell and cell-substrate contacts partly by regulating
cytoskeletal activity, including actin assembly, microtubule
dynamics and myosin II-dependent contractility of the actin-rich
cortex (36). Hence,
PLXNB2-activated RhoA signaling might be involved in the role of
miR-126-3p in lymph node metastasis of lung adenocarcinoma.
miR-451a has been confirmed as a downregulated miRNA
in lung cancer by using meta-analyses (37,38).
Wang et al reported that low expression of miR-451 in NSCLC
patients was associated with poor tumor differentiation, high
pathological stage and occurrence of lymph node metastasis, as well
as reduced overall survival of NSCLC patients (39). In the present study, it was found
that low expression of miR-451a was significantly associated with
high pathological stage II-IV and lymph node metastasis of lung
adenocarcinoma, and this was consistent with a previous study
(39). Furthermore, the ROC results
suggested that miR-451a could sensitively and specifically predict
pathological stage, tumor diameter and occurrence of lymph node
metastasis of lung adenocarcinoma, indicating that miR-451a is a
potential signature and biomarker of lung adenocarcinoma.
Reportedly, miR-451 suppressed the in vitro proliferation of
NSCLC cells and NSCLC development via downregulating ras-related
protein 14 (RAB14). In this study, RAB14 was also predicted as a
target of miR-451, showing the accuracy of the TargetScan tool.
Moreover, the target genes of miR-451a were most significantly
enriched in the peroxisome proliferator-activated receptor (PPAR)
signaling pathway, e.g., PPARA (PPARα). Although there is no direct
evidence for the involvement of PPARα in metastasis, downregulation
of PPARγ contributes to gastric carcinoma and lymph node metastasis
(40). Reportedly, PPARγ is
strongly expressed in lung cancer tissues (41), and its agonists inhibit human lung
cancer cell growth via inducting apoptosis (42). Hence, miR-451a might regulate the
progression and lymph node metastasis of lung adenocarcinoma via
modulating the PPAR signaling pathway.
In the present study, tuberous sclerosis 1 (TSC1)
was co-regulated by miR-126-3p and miR-451a. Reportedly, loss of
heterozygosity in TSC1-gene-region on chromosome 9q34 is frequently
observed in lung adenocarcinoma (43), and Tsc1 loss can synergize with Kras
mutation to enhance lung tumorigenesis in mouse via activating
mammalian target of rapamycin (mTOR) (44). In this study, miR-126-3p and
miR-451a were both downregulated in lung adenocarcinoma, reducing
the suppression on TSLC1 expression. This result was inconsistent
with previous studies (43,44), indicating that TSC1 expression might
also be regulated by other miRNAs or TFs.
miR-214 is also a cancer-related miRNA but functions
as an oncogene or a tumor suppressor in different types of cancers.
miR-214 is downregulated in cervical cancer and negatively
regulates HeLa cell proliferation (45), while upregulated miR-214 has been
reported in pancreatic cancer (46)
and gastric cancer (47) and is
associated with poor overall survival of patients. Higher
expression level of miR-214 was also found in lung cancer tissues
vs. non-cancerous lung tissues according to microarray data, which,
however, was not validated by qRT-PCR (48). Ishimura et al reported that
miR-214 is overexpressed in cancerous lung tissues from primary
NSCLC (49). We firstly reported
that although the expression level of miR-214 was slightly higher
in lung adenocarcinoma when compared with that in the paired
noncancerous tissues, the difference was not significant. These
findings further indicate the different functions of miR-214 in
different types of lung carcinoma.
In conclusion, in this study, miR-126-3p and
miR-451a were found to be significantly downregulated in human lung
adenocarcinoma. The low levels of miR-126-3p and miR-451a in lung
adenocarcinoma were significantly correlated with pathological
stage, tumor diameter and occurrence of lymph node metastasis. In
addition, miR-126-3p and miR-451a might play their roles in lung
adenocarcinoma via regulating the expression of target genes, e.g.,
PLXNB2 and PPARA. In our future study, the target relationships
between miR-126-3p and PLXNB2, as well as miR-451a and PPARA will
be validated using luciferase assay, and their roles will be
further investigated using shRNA silencing and vector
overexpressing technologies.
Acknowledgments
This study was supported by Science and Technology
Plan Project of Hangzhou City (20130633829, 20140633840), Public
Welfare Project of Zhejiang Science and Technology Department
(2013C33209, 2014C33277) and Natural Science Foundation of Zhejiang
Province (LY12H16001).
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
DEMs
|
differentially expressed microRNAs
|
|
TFs
|
transcription factors
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reaction
|
|
ROC
|
receiver operating characteristics
|
References
|
1
|
Teh E and Belcher E: Lung cancer:
Diagnosis, staging and treatment. Surgery. 32:242–248. 2014.
|
|
2
|
Stewart BW and Wild CP: World Cancer
Report 2014. World Health Organization; Lyon: 2014
|
|
3
|
Lu C, Onn A and Vaporciyan A: 78: Cancer
of the Lung. Holland-Frei Cancer Medicine. 8th edition. People's
Medical Publishing House; 2010
|
|
4
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frazier TP and Zhang B: Identification of
plant microRNAs using expressed sequence tag analysis. Plant
Reverse Genetics. Springer; pp. 13–25. 2011, View Article : Google Scholar
|
|
6
|
Schneider MR: MicroRNAs as novel players
in skin development, homeostasis and disease. Br J Dermatol.
166:22–28. 2012. View Article : Google Scholar
|
|
7
|
Fu SW, Chen L and Man YG: miRNA biomarkers
in breast cancer detection and management. J Cancer. 2:116–122.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nicolas FE, Lopez-Gomollon S,
Lopez-Martinez AF and Dalmay T: Silencing human cancer:
Identification and uses of microRNAs. Recent Pat Anticancer Drug
Discov. 6:94–105. 2011. View Article : Google Scholar
|
|
9
|
Patnaik SK, Yendamuri S, Kannisto E,
Kucharczuk JC, Singhal S and Vachani A: MicroRNA expression
profiles of whole blood in lung adenocarcinoma. PLoS One.
7:e460452012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao F, Chang J, Wang H and Zhang G:
Potential diagnostic value of miR-155 in serum from lung
adenocarcinoma patients. Oncol Rep. 31:351–357. 2014.
|
|
11
|
Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu
Z, Fang H, Zhang J, Katz RL and Jiang F: Early detection of lung
adenocarcinoma in sputum by a panel of microRNA markers. Int J
Cancer. 127:2870–2878. 2010. View Article : Google Scholar
|
|
12
|
Saito M, Schetter AJ, Mollerup S, Kohno T,
Skaug V, Bowman ED, Mathé EA, Takenoshita S, Yokota J, Haugen A, et
al: The association of microRNA expression with prognosis and
progression in early-stage, non-small cell lung adenocarcinoma: A
retrospective analysis of three cohorts. Clin Cancer Res.
17:1875–1882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Travis WD, Brambilla W, Mueller-Hermelink
HK and Harris CC: World Health Organization classification of
tumors: Pathology and Genetics of Tumors of the Lung, Pleura,
Thymus and Heart. IARC Press; Lyon: 2004
|
|
14
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours. 7th edition.
Wiley-Blackwell; 2009
|
|
15
|
Ohtsuka T, Nomori H, Watanabe K, Kaji M,
Naruke T, Suemasu K and Uno K: Prognostic significance of
[(18)F]fluorodeoxyglucose uptake on positron emission tomography in
patients with pathologic stage I lung adenocarcinoma. Cancer.
107:2468–2473. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Gulari E and Zhou X: In situ
synthesis of oligonucleotide microarrays. Biopolymers. 73:579–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Streichert T, Otto B and Lehmann U:
MicroRNA profiling using fluorescence-labeled beads: data
acquisition and processing. MicroRNA and Cancer. Springer; pp.
253–268. 2011, View Article : Google Scholar
|
|
19
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
21
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matys V, Kel-Margoulis OV, Fricke E,
Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M,
Hornischer K, et al: TRANSFAC and its module TRANSCompel:
Transcriptional gene regulation in eukaryotes. Nucleic Acids Res.
34:D108–D110. 2006. View Article : Google Scholar
|
|
23
|
Rosenbloom KR, Sloan CA, Malladi VS,
Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R,
Heitner SG, et al: ENCODE data in the UCSC Genome Browser: Year 5
update. Nucleic Acids Res. 41:D56–D63. 2013. View Article : Google Scholar :
|
|
24
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
25
|
Smoot ME, Ono K, Ruscheinski J, Wang P-L
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
26
|
Barshack I, Lithwick-Yanai G, Afek A,
Rosenblatt K, Tabibian-Keissar H, Zepeniuk M, Cohen L, Dan H, Zion
O, Strenov Y, et al: MicroRNA expression differentiates between
primary lung tumors and metastases to the lung. Pathol Res Pract.
206:578–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Raponi M, Zhang Y, Yu J, Chen G, Lee G,
Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, et al: Gene
expression signatures for predicting prognosis of squamous cell and
adenocarcinomas of the lung. Cancer Res. 66:7466–7472. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim MK, Jung SB, Kim JS, Roh MS, Lee JH,
Lee EH and Lee HW: Expression of microRNA miR-126 and miR-200c is
associated with prognosis in patients with non-small cell lung
cancer. Virchows Arch. 465:463–471. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jusufovic E, Keser D, Zukic E, Sejdinovic
R and Mrsic D: Downregulated anti-angiogenic miR-19a, miR-126 and
let-7b in non-small lung cancer have poor but different prognostic
values in squamous and adenocarcinoma subtypes. Eur Respir J.
42:46422013.
|
|
30
|
Liu B, Peng XC, Zheng X-L, Wang J and Qin
YW: miR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perrot V, Vázquez-Prado J and Gutkind JS:
Plexin B regulates Rho through the guanine nucleotide exchange
factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol
Chem. 277:43115–43120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li N, Tang A, Huang S, Li Z, Li X, Shen S,
Ma J and Wang X: miR-126 suppresses colon cancer cell proliferation
and invasion via inhibiting RhoA/ROCK signaling pathway. Mol Cell
Biochem. 380:107–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Faried A, Faried LS, Kimura H, Nakajima M,
Sohda M, Miyazaki T, Kato H, Usman N and Kuwano H: RhoA and RhoC
proteins promote both cell proliferation and cell invasion of human
oesophageal squamous cell carcinoma cell lines in vitro and in
vivo. Eur J Cancer. 42:1455–1465. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pillé JY, Denoyelle C, Varet J, Bertrand
JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP and Malvy C:
Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and
invasiveness of MDA-MB-231 breast cancer cells in vitro and in
vivo. Mol Ther. 11:267–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vial E, Sahai E and Marshall CJ: ERK-MAPK
signaling coordinately regulates activity of Rac1 and RhoA for
tumor cell motility. Cancer Cell. 4:67–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vasiliev JM, Omelchenko T, Gelfand IM,
Feder HH and Bonder EM: Rho overexpression leads to
mitosis-associated detachment of cells from epithelial sheets: A
link to the mechanism of cancer dissemination. Proc Natl Acad Sci
USA. 101:12526–12530. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Võsa U, Vooder T, Kolde R, Vilo J,
Metspalu A and Annilo T: Meta-analysis of microRNA expression in
lung cancer. Int J Cancer. 132:2884–2893. 2013. View Article : Google Scholar
|
|
38
|
Guan P, Yin Z, Li X, Wu W and Zhou B:
Meta-analysis of human lung cancer microRNA expression profiling
studies comparing cancer tissues with normal tissues. J Exp Clin
Cancer Res. 31:542012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He Q, Chen J, Lin HL, Hu PJ and Chen MH:
Expression of peroxisome proliferator-activated receptor gamma,
E-cadherin and matrix metalloproteinases-2 in gastric carcinoma and
lymph node metastases. Chin Med J (Engl). 120:1498–1504. 2007.
|
|
41
|
Inoue K, Kawahito Y, Tsubouchi Y, Yamada
R, Kohno M, Hosokawa Y, Katoh D, Bishop-Bailey D, Hla T and Sano H:
Expression of peroxisome proliferator-activated receptor
(PPAR)-gamma in human lung cancer. Anticancer Res. 21:2471–2476.
2001.PubMed/NCBI
|
|
42
|
Tsubouchi Y, Sano H, Kawahito Y, Mukai S,
Yamada R, Kohno M, Inoue K, Hla T and Kondo M: Inhibition of human
lung cancer cell growth by the peroxisome proliferator-activated
receptor-gamma agonists through induction of apoptosis. Biochem
Biophys Res Commun. 270:400–405. 2000. View Article : Google Scholar
|
|
43
|
Takamochi K, Ogura T, Yokose T, Ochiai A,
Nagai K, Nishiwaki Y, Suzuki K and Esumi H: Molecular analysis of
the TSC1 gene in adenocarcinoma of the lung. Lung Cancer.
46:271–281. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liang MC, Ma J, Chen L, Kozlowski P, Qin
W, Li D, Goto J, Shimamura T, Hayes DN, Meyerson M, et al: TSC1
loss synergizes with KRAS activation in lung cancer development in
the mouse and confers rapamycin sensitivity. Oncogene.
29:1588–1597. 2010. View Article : Google Scholar :
|
|
45
|
Yang Z, Chen S, Luan X, Li Y, Liu M, Li X,
Liu T and Tang H: MicroRNA-214 is aberrantly expressed in cervical
cancers and inhibits the growth of HeLa cells. IUBMB Life.
61:1075–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang XJ, Ye H, Zeng CW, He B, Zhang H and
Chen YQ: Dysregulation of miR-15a and miR-214 in human pancreatic
cancer. J Hematol Oncol. 3:462010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar
|
|
48
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ishimura M, Sakurai-Yageta M, Maruyama T,
Ando T, Fukayama M, Goto A and Murakami Y: Involvement of miR-214
and miR-375 in malignant features of non-small-cell lung cancer by
down-regulating CADM1. J Cancer Ther. 3:379–387. 2012. View Article : Google Scholar
|