Introduction
Gliomas are the most common type of intrinsic brain
tumor of the central nervous system in adults, and are highly
aggressive and insensitive to treatment. Current treatment
strategies have not significantly improved long-term survival of
patients (1); the median survival
is just 12–15 months for patients with glioblastoma (World Health
Organization (WHO) IV), 2–5 years for patients with anaplastic
glioma (WHO III), and 6–8 years for patients with low-grade glioma
(WHO II) (2). Surgery is the
preferred treatment option, which is followed by observation of
postoperative remission (3,4). However, even when surgery is combined
with chemo- or radiotherapy, relapse is inevitable (5–7).
Human gliomas contain a small population of cells
with stem cell-like features known as brain tumor stem cells
(BTSCs) (8). These cells are
hypothesized to initiate tumor growth and recurrence, since they
are resistant to standard anticancer therapies (9). BTSCs may be difficult to remove
surgically since they do not form an obvious tumor mass and may not
manifest tumor-specific histological features (9).
The stem cell surface antigen cluster of
differentiation (CD)133 (also known as AC133 or human prominin-1)
is expressed in a subset of neural stem/precursor cells in the
adult central nervous system and is a marker used to identify and
isolate BTSCs (10). CD133
expression increases with tumor grade and is correlated with glioma
patient survival (11); moreover,
glioma recurrence can be predicted by an increase in the fraction
of CD133-positive cells within the primary tumor (12). However, the regulation of
CD133 gene expression in gliomas is not well understood.
Promoter methylation is a mechanism underlying the
downregulation of CD133 in colon and ovarian cancers and
glioblastoma (13–15). DNA methylation can prevent binding
of transcription factors to their target binding sites, thereby
repressing transcription. CD133 is transcribed in a
tissue-specific manner from five alternative promoters (P1–P5)
(16): liver, kidney, pancreas,
placenta, lung, spleen, and colon express transcripts containing
exons 1A and 1B. On the other hand, exon 1B transcripts are
expressed in the brain and ovary, and exon 1A transcripts in the
prostate, fetal liver, and small intestine. All CD133 promoters are
TATA-less and P1, P2, and P3 are located in a CpG island (16). Recent evidence suggests that DNA
methylation of P1 and P2 plays a role in the regulation of
CD133 expression. P1 methylation is not tissue-specific, and
therefore is not likely to affect transcript levels; in contrast,
the tissue-specific methylation of P2 is inversely correlated with
CD133 expression (17).
In this study, we analyzed CD133 expression in pairs
of primary and recurrent human glioma specimens from 24 patients
and found that recurrent gliomas exhibited aberrantly upregulated
CD133 expression. To clarify the underlying mechanisms, we analyzed
P2 of the CD133 gene by bisulfite sequencing, and found that
CD133 P2 hypomethylation is associated with increased
CD133 gene expression and glioma recurrence.
Materials and methods
Glioma tissues
Surgical specimens of gliomas were obtained from 24
patients who had undergone tumor resection at Affiliated Hospital
of Nantong University from January 2004 to December 2014. The study
was approved by the local ethics committee, written informed
consent was obtained from all the patients. Recurrence of glioma
was defined as the presence of glioma at >3 months after surgery
for primary glioma. All patients received chemoradiotherapy after
the first surgical intervention. Pathological findings were
determined by more than 2 pathologists and classified according to
the WHO classification standard (Table
I). The paired primary and recurrent glioma specimens were from
the same patient. For immunohistochemistry, glioma tissue was fixed
with 4% formalin, dehydrated, and then embedded in paraffin.
Portions of the tumor tissues were rapidly frozen by liquid
nitrogen and stored at −80°C until RNA and DNA extraction for
real-time PCR, DNA methylation and western blot analysis.
 | Table IWHO grades of glioma patients. |
Table I
WHO grades of glioma patients.
| Patient | WHO grade
|
|---|
| Primary | Recurrent |
|---|
| 1 | II | II |
| 2 | IV | IV |
| 3 | II | IV |
| 4 | III | III |
| 5 | III | IV |
| 6 | II | IV |
| 7 | III | IV |
| 8 | II | II |
| 9 | III | IV |
| 10 | IV | IV |
| 11 | IV | IV |
| 12 | III | III |
| 13 | II | II |
| 14 | IV | IV |
| 15 | IV | IV |
| 16 | IV | IV |
| 17 | III | III |
| 18 | II | IV |
| 19 | II | III |
| 20 | II | IV |
| 21 | IV | IV |
| 22 | IV | IV |
| 23 | IV | IV |
| 24 | IV | IV |
Cell culture
The U251MG, U87MG, and A172MG glioblastoma cell
lines were purchased from the Shanghai Cell Institution of Chinese
Academic Sciences, which were cultured as described previously
(18). They were maintained in
Dulbecco's modified Eagle's medium: nutrient mixture F-12
(MDEM/F12) supplemented with 10% heat-inactivated fetal bovine
serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin
(Gibco). Cells were incubated at 37°C in a humidified 5%
CO2 air atmosphere. When cell culture reached 50%
confluence, U87MG, U251MG and A172MG cells were treated with
5-Aza-2′-deoxycytidine (5-Aza-dc, A3656; Sigma-Aldrich, St. Louis,
MO, USA) at the final concentration of 10 nM for 72 h,
respectively.
Western blot analysis
Cells or tissues were washed with cold PBS and lysed
in ice-cold RIPA buffer containing protein inhibitors. Cell lysates
were incubated on ice for 30 min and then centrifuged at 4°C for 10
min. Supernatants were collected, and protein concentrations were
measured. Equivalent amounts of protein in each sample were
separated on a 10% SDS-PAGE gel for separation and then
electrotransferred to PVDF membrane. Membranes were blocked and
then probed with primary CD133 antibodies (2 µg/ml, Abcam)
followed by the horseradish peroxidase (HRP)-conjugated goat
anti-mouse or rabbit IgG antibodies. Mouse monoclonal anti-β-actin
antibody (1:5,000, Sigma) was used as an internal control. The
membranes were developed using an ECL detection system (Pierce,
Rockford, IL, USA). The intensity of bands was determined using the
Image-Pro Plus 6.0 software. The western blot experiments were
repeated at least three times.
RNA extraction and real-time PCR
RNA expression levels of CD133 were determined using
quantitative real-time PCR with GAPDH as positive controls. Total
mRNA was isolated from glioma specimens and cell lines using mRNA
isolation kit (Roche, UK) following the manufacturer's
instructions. The concentration and purity of mRNA was determined
by ultraviolet spectrophotometry. Isolated mRNA (100 ng) from each
sample was transcribed to complementary DNA (cDNA) using a
First-Strand cDNA Synthesis kit Roche (Roche), which was then used
as a template for quantitative real-time PCR. Primers used for
CD133 were 5′-GCACTCTATACCAAAGCGTCAA-3′ (sense) and
5′-CTCCCATACTTCTTAGTTTCCTCA-3′ (antisense); and for GAPDH primers
were 5′-GGAAAGCTGTGGCGTGAT-3′ (sense) and
5′-AAGGTGGAAGAATGGGAGTT-3′ (antisense). The primers were designed
using Primer 5.0 software and manufactured by TIBmolbiol. After an
initial denaturation at 95°C for 5 min, the samples were subjected
to 40 cycles of RT-PCR (95°C for 10 sec, annealing temperature 59°C
for 15 sec, and 72°C for 20 sec). At the end of each cycle, the
fluorescence emitted was measured in a single step in channel F1
(gain 1). After the 40th cycle, the specimens were heated to 95°C
and rapidly cooled to 59°C for 15 sec. All heating and cooling
steps were performed with a slope of 20°C/sec. The temperature was
subsequently raised with a slope of 12°C/sec and fluorescence was
measured continuously (channel F1, gain 1) to obtain data for the
melting curve analysis. The PCR reaction was subjected to a melting
curve analysis to verify the presence of a single amplicon before
the PCR products were visualized on agarose gels using a gel
analyser (SynGene, UK). All PCR reactions were performed in
triplicate and a negative control was included that contained
primers without DNA.
Immunohistochemical analyses
Serial sections measuring 5 µm thick were cut
from paraffin blocks and mounted on glass slides coated with 10%
polylysine. Sections were dewaxed in xylene and rehydrated in
graded ethanols. Endogenous peroxidase activity was blocked by
immersion in 0.3% methanolic peroxide for 30 min. Immuno-reactivity
was enhanced by microwaving and incubating the tissue sections for
10 min in 0.1 mol/l citrate buffer. Tissues were then incubated
with an anti-CD133 antibody (1:200, Abcam) overnight at 4°C.
Immuno-staining was performed using the avidin-biotin peroxidase
complex method, and antigen-antibody reactions were visualized with
chromogen diaminobenzadine. Appropriate positive and negative
controls were used. Ten high-power fields were randomly chosen, and
≥300 tumor cells were counted per field. Percentage of cells
showing positive staining in cytoplasm/membrane was designated as
the CD133 labeling index, as a percentage (%). The staining
procedures were repeated at least three times.
DNA isolation and bisulfite
sequencing
With the proteinase K digestion and
phenole-chloroform method, genomic DNA was extracted from frozen
tissues (19). Sodium bisulfite
treatment of the extracted DNA was performed as previously
described (20). In brief, 10
µg DNA in 50 µl TE was incubated with 5.5 µl
of 0.3 M NaOH at 37°C for 15 min and 95°C for 2 min, and subjected
to sodium bisulfite chemical treatment (2.4 M sodium metabisulfite;
0.5 mM hydroquinone, pH 5.0, both from Sigma). Following incubation
at 55°C for 4 h, the treated DNA was purified using the SK1261 kit
(Shenggong, China), desulfo-nated in 0.3 M NaOH, neutralized to pH
7.0 using 3 M sodium acetate (pH 5.2). The neutralized DNA was
purified using SK1261 purification kit again, dissolved in TE
buffer (pH 8.0).
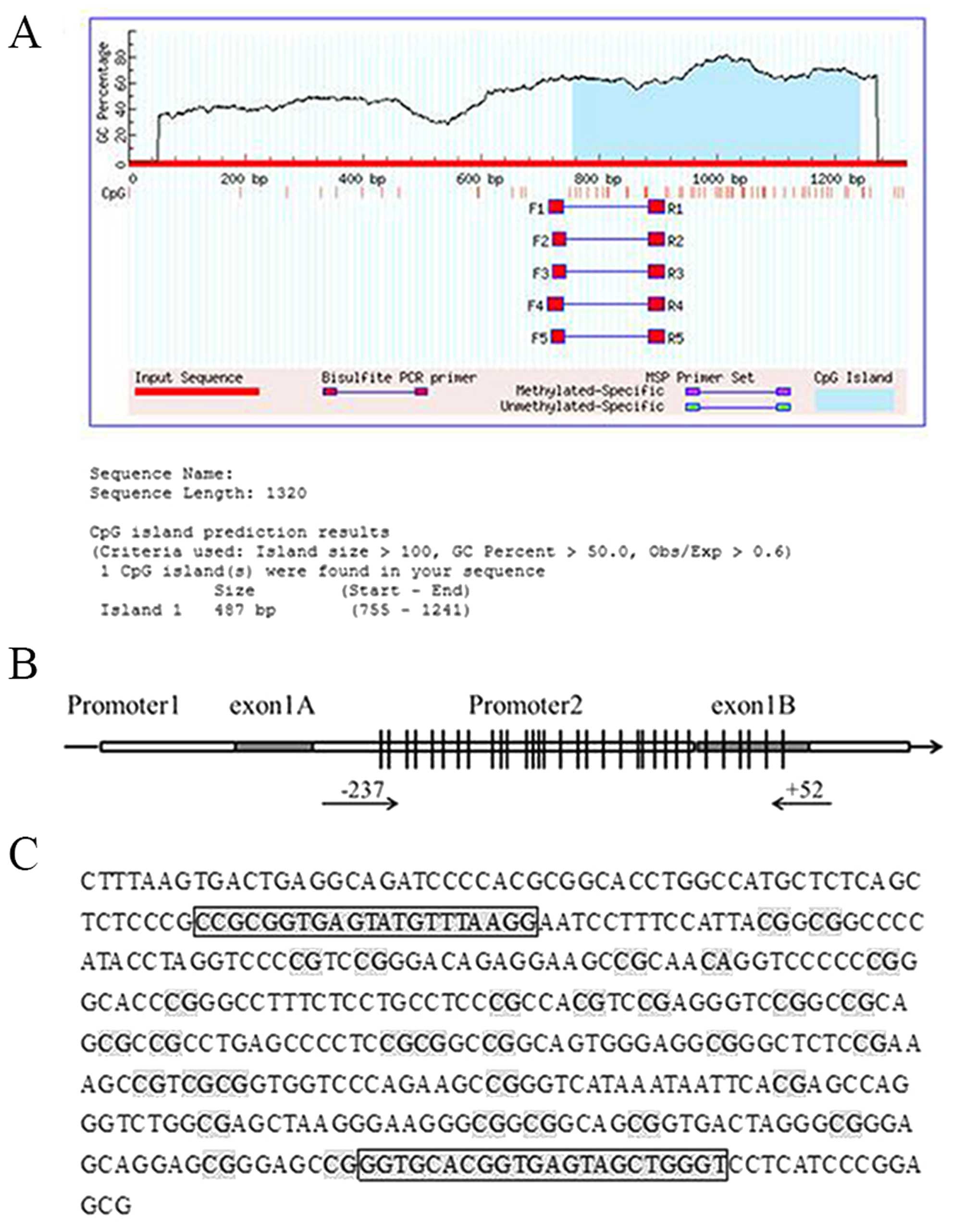
The primers (fwd: 5′-TYGYGGTGAGTATGTTTAAGG-3′, rev:
5′-ACCCAACTACTCACCRTACACC-3′) were designed to amplify the promoter
2 from −237 to +52 for bisulfite genomic sequencing. An initial
denaturation at 98°C for 4 min was followed by five PCR cycles of
94°C for 45 sec, 68°C for 45 sec and 72°C for 1 min. The PCR was
then completed with 35 cycles of 45 sec at 95°C, 45 sec at 58°C.
The amplified products were gel-purified using the SK1261 kit and
subjected to TA-cloning using pUC18-T vector (Shenggong,
Biotechnology Co.). Ten clones for each case were selected for
sequencing using BigDye version 3.1, and analyzed on automated DNA
sequence analyzer (ABI PRISM 3730; Applied Biosystems, Inc., Foster
City, CA, USA). The cytosine or thymine residues at the CpG sites
represented methylated or unmethylated status, respectively.
Statistical analyses
Statistical analysis was performed using SPSS 13.0
for Windows. Data are expressed as median and 25–75 percentiles
[median (25th percentile, 75th percentile)]. Mann-Whitney U test
was used to determine the differences of CD133 expression between
primary and recurrent tumor. Paired t-test was used to analyze the
differences of CD133 expression between the treated glioma cell
lines. The correlation between CD133 DNA methylation and mRNA
expression was analyzed by Pearson's correlation test. All
statistical tests were calculated two-sided and values of P<0.05
was considered to be statistically significant.
Results
CD133 expression is upregulated in
recurrent glioma
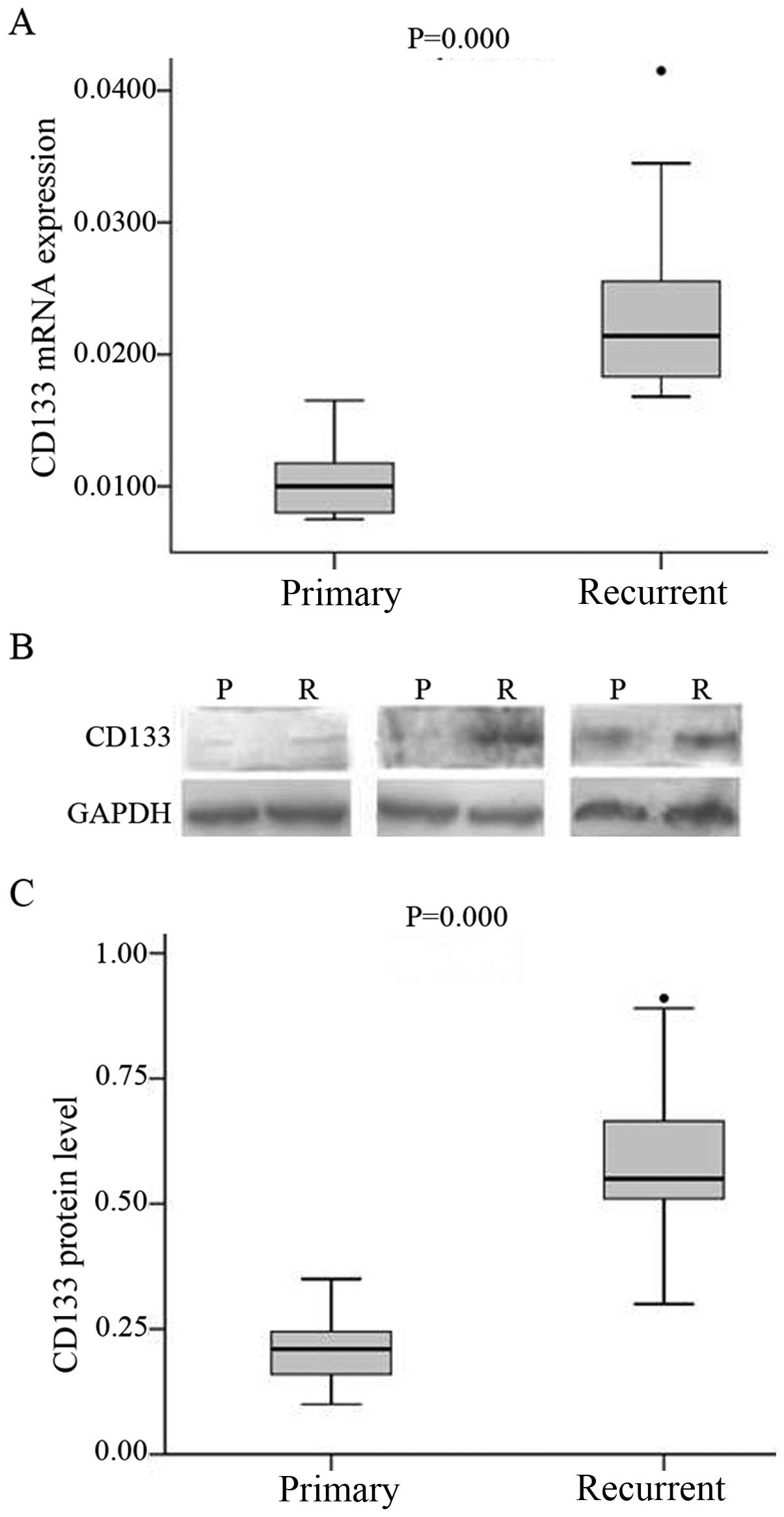
CD133 mRNA and protein expression in primary and
recurrent glioma specimens was assessed by real-time PCR and
western blotting, respectively. While CD133 transcript was
detected in all specimens, the levels were higher in recurrent
(median 0.0214; range, 0.0182–0.0249) than in primary glioma
(median 0.1,000; range, 0.0080–0.0119, P<0.001) (Fig. 1A). A similar trend was observed for
CD133 protein level (Fig. 1B and
C).
CD133-positive cell number is increased
in recurrent as compared to primary glioma
CD133 protein expression and localization in glioma
specimens were evaluated by immunohistochemistry. CD133-positive
cells were detected in all specimens, and expression was mainly
observed in the cytoplasm and plasma membrane (Fig. 2A). The percentage of CD133-positive
cells was significantly higher in recurrent (median 12.99%; range,
3.94–58.23%) as compared to primary glioma (median 4.35%; range,
2.43–14.35%, P<0.05) (Fig.
2B).
Hypomethylation of the CD133 promoter
increases CD133 expression in recurrent glioma
To clarify the mechanism underlying the upregulation
of CD133 level in recurrent glioma, we analyzed the methylation
status of CpG islands in the CD133 promoter. A sequence
analysis revealed that the CD133 P2 promoter contains a
487-bp CpG island, with a GC content >50% and a CpG ratio
(Obs/Exp) >0.6 (Fig. 3A). The P2
promoter region (−237 to +52) contained 32 CpG sites, as determined
by bisulfite genomic sequencing (Fig.
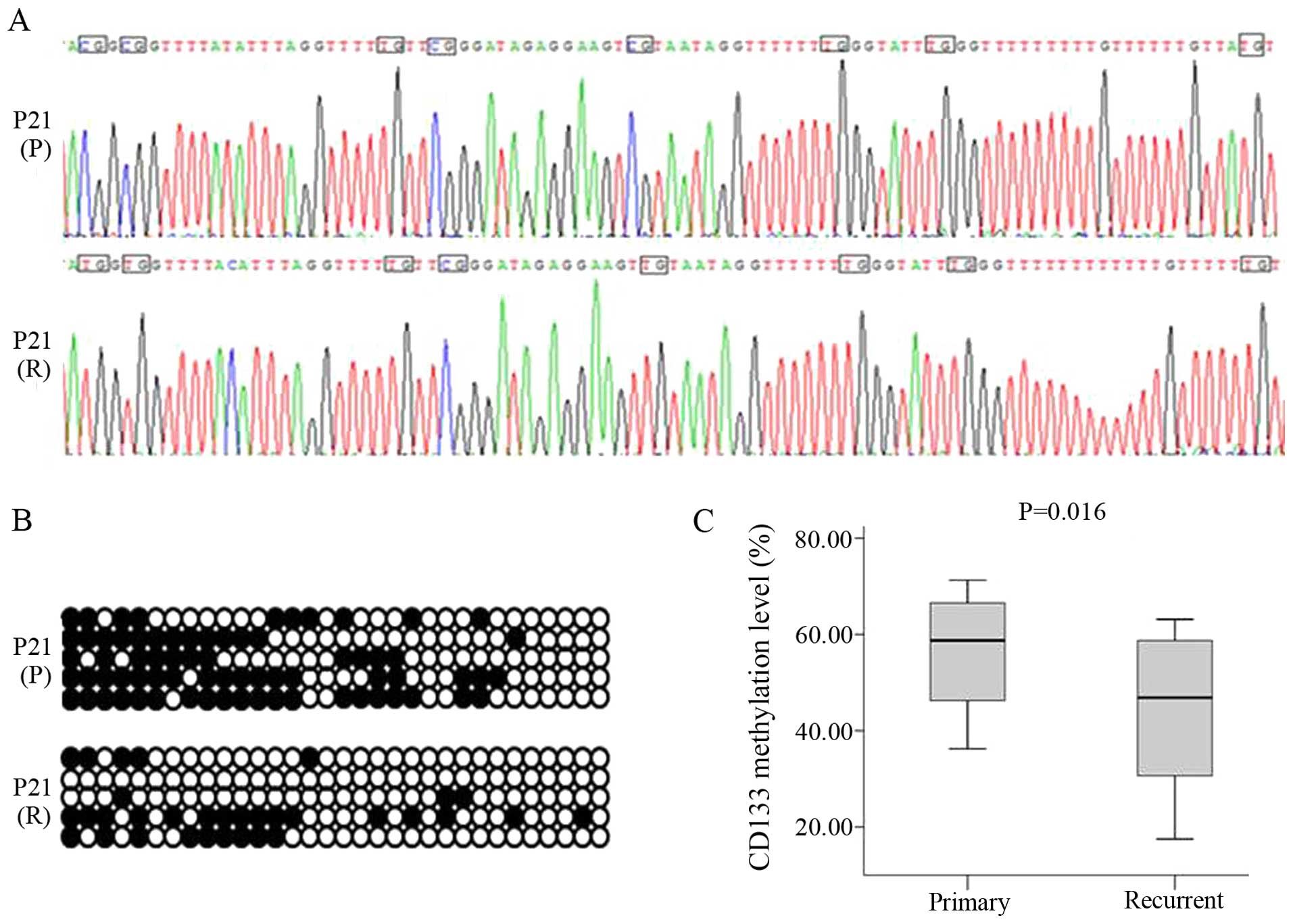
3B and C). The 332-bp CD133 target fragment was cloned and
sequenced, revealing unmethylated C residues that were completely
converted to U by bisulfite treatment; these were replaced by T
following PCR amplification. In contrast, methylated C residues
persisted after bisulfite treatment, indicating that CpG islands of
the CD133 promoter were methylated (Fig. 4A and B). However, DNA methylation
levels were lower in recurrent (median, 46.88%; range,
26.56–59.69%) as compared to primary glioma (median, 58.76%; range,
42.19–67.35%, P<0.05) (Fig. 4C).
Correlation analysis showed a negative relationship between DNA
methylation and CD133 expression levels (r = −0.715,
P<0.05).
Promoter demethylation leads to
upregulation of CD133 expression
To assess the effects of demethylation in
vitro, we treated U87, U251, and A172 glioma cells with the
demethylating agent 5-aza-2′-deoxycytidine (10 µM for 72 h).
CD133 mRNA expression was upregulated in these cells by
2.15, 2.56, and 3.03-fold, respectively (Fig. 5A). A similar trend was observed for
CD133 protein expression (Fig. 5B).
These results indicate that CD133 demethylation increases CD133
level in glioma.
Discussion
BTSCs are thought to be the main determinants of the
occurrence, development, metastasis, recurrence, and treatment
sensitivity of malignant glioma (21). BTSCs can form the same pathological
type of tumor as the parent tumor both in vitro and in
vivo (22,23). However, the direct study of BTSCs is
challenging due to the lack of specific markers. In theory,
expression levels of stem cell-related genes represent changes in
the BTSC population of gliomas. The most common stem cell-related
genes are nestin, CD133, ATP-binding cassette superfamily G
member 2, SRY box-containing gene 2, POU class 5 homeobox
1/OCT4, and musashi-1. In our previous study we reported
the expression and methylation status of OCT4 in glioma
(18). Based on these findings and
other previous reports, we hypothesized that BTSCs initiate gliomas
and are responsible for their recurrence.
CD133 is a cell membrane glycoprotein that has been
identified as a marker of a subset of BTSCs in the adult central
nervous system and in glioblastoma stem-like cells (10,11).
CD133-positive cells exhibit stem cell-like qualities in
vitro and are capable of tumor formation in vivo
(24). CD133-positive BTSCs in an
animal model showed an unlimited capacity for self-renewal and for
inducing tumor initiation and progression (25). CD133 has been isolated from
hematopoietic stem cells using an antibody that recognizes the
AC133 epitope, and is used as a marker for the isolation of brain
cancer stem cells (26).
In this study, we determined that CD133 expression
was upregulated in recurrent as compared to newly diagnosed glioma
specimens. This suggests that there were more residual BTSCs in
recurrent than in primary glioma. Surgery is an invasive procedure
that is accompanied by tissue injury, the production of
inflammatory cytokines, angiogenesis, and glioma cell
proliferation, which can lead to changes in the local
microenvironment and the recruitment of residual BTSCs to the
surgery site. The proliferation of BTSCs and their differentiation
into glioma cells (27,28) result in the upregulation of CD133
expression and glioma recurrence.
Changes in DNA methylation patterns are an important
hallmark of tumor development and progression (29). CD133 contains five alternative
promoters, three of which are partly regulated by methylation
(16). AC133 promoters are
TATA-less and three (P1-P3) are located within a CpG island. In
vitro methylation suppressed the activity of P1 and P2, and
recent studies have shown that DNA methylation is inversely
correlated with CD133 transcription (13,30,31).
Here, we found that CD133 transcription in glioblastoma is
dependent on DNA hypomethylation, which has been shown to promote
tumori-genesis by inducing the activation of oncogenes and/or
causing genomic instability (32).
Thus, CD133 promoter hypomethylation may be associated with
the maintenance of BTSCs in brain tumors. It has been proposed that
a reduction in overall genomic methylation, an important feature of
tumor cells, is responsible for decreased methylation of specific
genes (33).
Treatment of glioma cell lines with a demethylating
agent increased CD133 mRNA and protein expression, which was
consistent with changes reported for some oncogenes (34). DNA methylation status regulates the
transition from CD133 transcriptional activation to repression in
glioma. DNA methylation is reversible; as such, methylation status
and consequent dysregulation of target gene expression can be
altered by drug treatment (35). In
conclusion, the results of this study provide insight into the
mechanism of glioma recurrence and provide a basis for novel
therapies for glioma treatment.
Acknowledgments
This study was supported by the Youth Fund of the
National Natural Science Foundation of China (81201975, 81201349),
China Postdoctoral Science Foundation (2015M581845), the Six Major
Human Resources Project of Jiangsu Province (2013-WSN-077,
2014-WSW-031), the Health Department Project of Jiangsu Province
(H201422) and the Translational Medicine Foundation of Affiliated
Hospital of Nantong University (TDFzh2014015).
References
|
1
|
Braganza MZ, Kitahara CM, Berrington de
González A, Inskip PD, Johnson KJ and Rajaraman P: Ionizing
radiation and the risk of brain and central nervous system tumors:
A systematic review. Neuro-oncol. 14:1316–1324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gonzalez J and Gilbert MR: Treatment of
astrocytomas. Curr Opin Neurol. 18:632–638. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hentschel SJ and Lang FF: Current surgical
management of glioblastoma. Cancer J. 9:113–125. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wallner KE, Galicich JH, Krol G, Arbit E
and Malkin MG: Patterns of failure following treatment for
glioblastoma multiforme and anaplastic astrocytoma. Int J Radiat
Oncol Biol Phys. 16:1405–1409. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sneed PK, Gutin PH, Larson DA, Malec MK,
Phillips TL, Prados MD, Scharfen CO, Weaver KA and Wara WM:
Patterns of recurrence of glioblastoma multiforme after external
irradiation followed by implant boost. Int J Radiat Oncol Biol
Phys. 29:719–727. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aydin H, Sillenberg I and von Lieven H:
Patterns of failure following CT-based 3-D irradiation for
malignant glioma. Strahlenther Onkol. 177:424–431. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim SS, Pirollo KF and Chang EH: Isolation
and culturing of glioma cancer stem cells. Curr Protoc Cell Biol.
23.10.1–23.10.10. 2015. View Article : Google Scholar
|
|
9
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H and Li SY: Research progression of
CD133 as a marker of cancer stem cells. Clin J Cancer. 29:243–247.
2010.
|
|
11
|
Zeppernick F, Ahmadi R, Campos B, Dictus
C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B and
Herold-Mende CC: Stem cell marker CD133 affects clinical outcome in
glioma patients. Clin Cancer Res. 14:123–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Auffinger B, Tobias AL, Han Y, Lee G, Guo
D, Dey M, Lesniak MS and Ahmed AU: Conversion of differentiated
cancer cells into cancer stem-like cells in a glioblastoma model
after primary chemotherapy. Cell Death Differ. 21:1119–1131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeon YK, Kim SH, Choi SH, Kim KH, Yoo BC,
Ku JL and Park JG: Promoter hypermethylation and loss of CD133 gene
expression in colorectal cancers. World J Gastroenterol.
16:3153–3160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Min KJ, So KA, Ouh YT, Hong JH and Lee JK:
The effects of DNA methylation and epigenetic factors on the
expression of CD133 in ovarian cancers. J Ovarian Res. 5:282012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gopisetty G, Xu J, Sampath D, Colman H and
Puduvalli VK: Epigenetic regulation of CD133/PROM1 expression in
glioma stem cells by Sp1/myc and promoter methylation. Oncogene.
32:3119–3129. 2013. View Article : Google Scholar
|
|
16
|
Shmelkov SV, Jun L, St Clair R, McGarrigle
D, Derderian CA, Usenko JK, Costa C, Zhang F, Guo X and Rafii S:
Alternative promoters regulate transcription of the gene that
encodes stem cell surface protein AC133. Blood. 103:2055–2061.
2004. View Article : Google Scholar
|
|
17
|
Tabu K, Sasai K, Kimura T, Wang L,
Aoyanagi E, Kohsaka S, Tanino M, Nishihara H and Tanaka S: Promoter
hypomethylation regulates CD133 expression in human gliomas. Cell
Res. 18:1037–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi J, Shi W, Ni L, Xu X, Su X, Xia L, Xu
F, Chen J and Zhu J: OCT4 is epigenetically regulated by DNA
hypomethylation of promoter and exon in primary gliomas. Oncol Rep.
30:201–206. 2013.PubMed/NCBI
|
|
19
|
Bigner SH, Matthews MR, Rasheed BK,
Wiltshire RN, Friedman HS, Friedman AH, Stenzel TT, Dawes DM,
McLendon RE and Bigner DD: Molecular genetic aspects of
oligodendrogliomas including analysis by comparative genomic
hybridization. Am J Pathol. 155:375–386. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jansen GA, Mihalik SJ, Watkins PA, Moser
HW, Jakobs C, Denis S and Wanders RJ: Phytanoyl-CoA hydroxylase is
present in human liver, located in peroxisomes, and deficient in
Zellweger syndrome: Direct, unequivocal evidence for the new,
revised pathway of phytanic acid alpha-oxidation in humans. Biochem
Biophys Res Commun. 229:205–210. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Germano I, Swiss V and Casaccia P: Primary
brain tumors, neural stem cell, and brain tumor cancer cells: Where
is the link? Neuropharmacology. 58:903–910. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He H, Li MW and Niu CS: The pathological
characteristics of glioma stem cell niches. J Clin Neurosci.
19:121–127. 2012. View Article : Google Scholar
|
|
23
|
Achanta P, Sedora Roman NI and
Quiñones-Hinojosa A: Gliomagenesis and the use of neural stem cells
in brain tumor treatment. Anticancer Agents Med Chem. 10:121–130.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwab LP, Peacock DL, Majumdar D, Ingels
JF, Jensen LC, Smith KD, Cushing RC and Seagroves TN:
Hypoxia-inducible factor 1α promotes primary tumor growth and
tumor-initiating cell activity in breast cancer. Breast Cancer Res.
14:R62012. View
Article : Google Scholar
|
|
25
|
Chakraborty S, Kanakasabai S and Bright
JJ: Constitutive androstane receptor agonist CITCO inhibits growth
and expansion of brain tumour stem cells. Br J Cancer. 104:448–459.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kahlert UD, Maciaczyk D, Dai F, Claus R,
Firat E, Doostkam S, Bogiel T, Carro MS, Döbrössy M, Herold-Mende
C, et al: Resistance to hypoxia-induced, BNIP3-mediated cell death
contributes to an increase in a CD133-positive cell population in
human glioblastomas in vitro. J Neuropathol Exp Neurol.
71:1086–1099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem
cells in glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar
|
|
28
|
Liu JM, Mao BY, Hong S, Liu YH and Wang
XJ: The postoperative brain tumour stem cell (BTSC) niche and
cancer recurrence. Adv Ther. 25:389–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gómez-Díaz E, Jordà M, Peinado MA and
Rivero A: Epigenetics of host-pathogen interactions: The road ahead
and the road behind. PLoS Pathog. 8:e10030072012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baba T, Convery PA, Matsumura N, Whitaker
RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S, et al:
Epigenetic regulation of CD133 and tumorigenicity of
CD133+ ovarian cancer cells. Oncogene. 28:209–218. 2009.
View Article : Google Scholar
|
|
31
|
He J, Shan Z, Li L, Liu F, Liu Z, Song M
and Zhu H: Expression of glioma stem cell marker CD133 and
O6-methylguanine-DNA methyltransferase is associated with
resistance to radiotherapy in gliomas. Oncol Rep. 26:1305–1313.
2011.PubMed/NCBI
|
|
32
|
Mudbhary R, Hoshida Y, Chernyavskaya Y,
Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson
RT, et al: UHRF1 overexpression drives DNA hypomethylation and
hepatocellular carcinoma. Cancer Cell. 25:196–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hackett JA and Surani MA: DNA methylation
dynamics during the mammalian life cycle. Philos Trans R Soc Lond B
Biol Sci. 368:201103282013. View Article : Google Scholar :
|
|
34
|
Krutovskikh V and Partensky C: New
insights in oncology: Epigenetics and cancer stem cells. Cancer
Radiother. 15:716–722. 2011.In French. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Radpour R, Barekati Z, Kohler C,
Schumacher MM, Grussenmeyer T, Jenoe P, Hartmann N, Moes S, Letzkus
M, Bitzer J, et al: Integrated epigenetics of human breast cancer:
Synoptic investigation of targeted genes, microRNAs and proteins
upon demethylation treatment. PLoS One. 6:e273552011. View Article : Google Scholar : PubMed/NCBI
|