Digestive system cancers (DSCs) are common cancers
and the leading causes of cancer-related deaths worldwide (1). According to the classification by the
World Health Organization in 2010 (2), DSCs consist of alimentary tract
cancers of the esophagus, stomach, colorectum, and digestive gland
cancers of the liver, gallbladder, bile duct, and pancreas.
Although great advances in surgical techniques and chemotherapy
have been achieved, the prognosis of DSCs is still poor since they
are mostly diagnosed at advanced stage, which may be accompanied by
malignant proliferation, extensive invasion, and distant metastasis
(3,4). Genetic and epigenetic alterations have
been suggested to participate in the development and progression of
DSCs (5–7). The elucidation of the molecular
regulatory network in DSCs will provide novel biomarkers for early
diagnosis and more effective therapy.
Over the past decade, noncoding RNAs are emerging as
new players in various diseases. Long noncoding RNAs (lncRNAs) are
defined as transcripts of greater than 200 nucleotides that lack
protein-coding capability (8).
LncRNAs regulate gene expression at various levels, including
chromatin modification (9),
transcription (10), and
post-transcription (11,12). Increasing evidence suggests that
lncRNAs play important roles in cancer (13). LncRNAs are critically involved in
tumorigenesis, tumor growth, and tumor metastasis (14). In this review, we present an updated
view of the roles of lncRNAs in DSCs with an emphasis on the
underlying mechanisms. We also provide new insights into the
prospective of lncRNAs as potential diagnostic biomarkers and
therapeutic targets for DSCs.
To identify the lncRNAs most closely related to
DSCs, a systematic literature search was conducted to find all the
lncRNAs which have been reported in esophageal, stomach,
colorectal, liver, gallbladder, bile duct, and pancreatic cancers.
LncRNAs that are involved in at least three types of DSCs were
included for further review. A total of ten lncRNAs were included,
which are H19, Hox transcript antisense intergenic RNA (HOTAIR),
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),
highly upregulated in liver cancer (HULC), maternally expressed
gene 3 (MEG3), growth arrest-specific transcript 5 (GAS5),
antisense noncoding RNA in the INK4 locus (ANRIL), plasmacytoma
variant translocation 1 (PVT1), colon cancer associated transcript
1 (CCAT1), and lncRNA-ATB. The characteristics of these
DSCs-related lncRNAs are shown in Table
I. The functional roles and clinical correlations of these
lncRNAs in DSCs are presented in Table
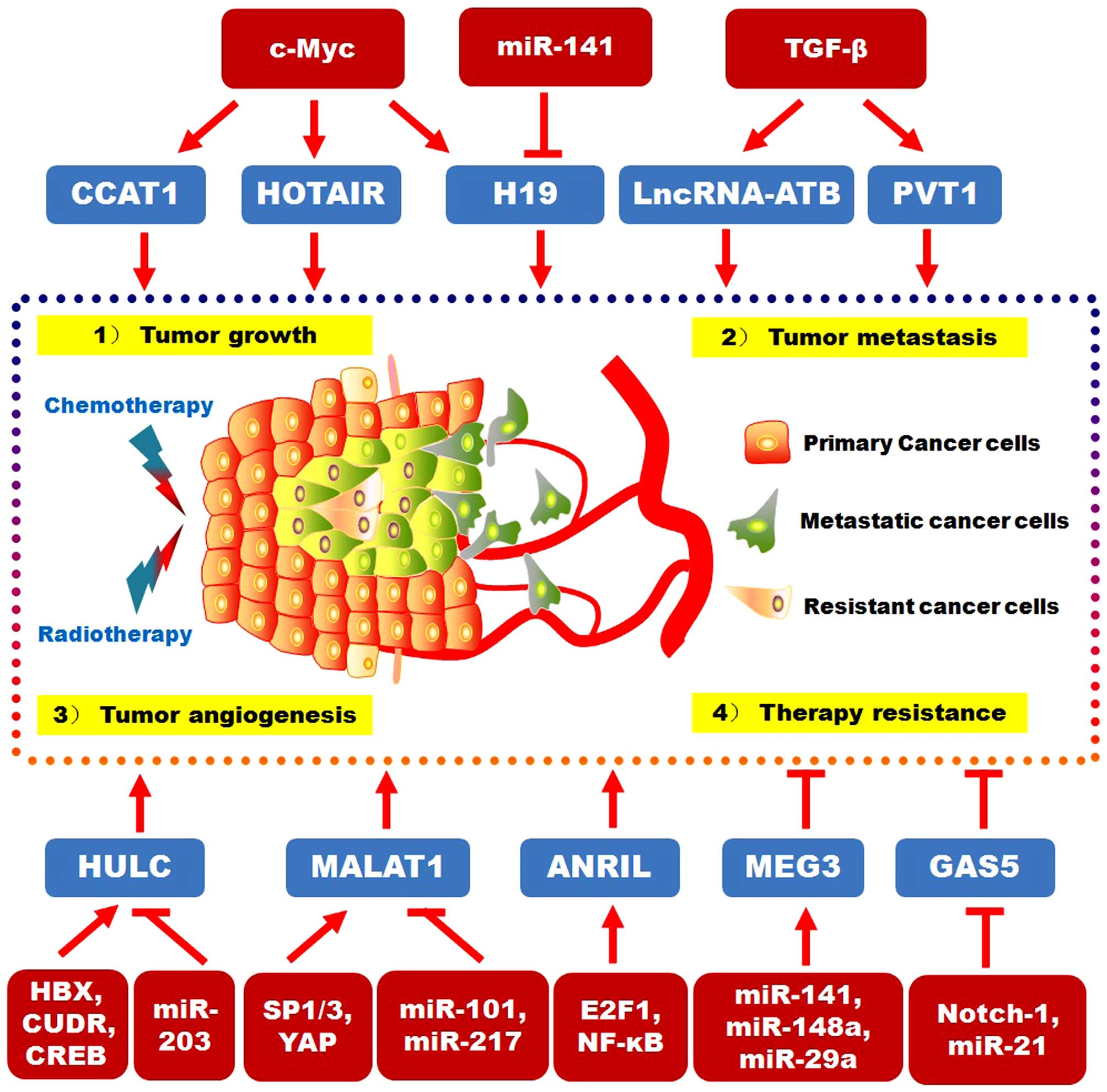
II. The upstream signaling pathways and downstream targets for
DSC-related lncRNAs are reviewed in Fig. 1.
The H19 gene, located at chromosome 11p15.5, belongs
to a highly conserved cluster of imprinted genes which involve the
insulin-like growth factor 2 gene (15). H19 is expressed exclusively from the
maternal allele and encodes a 2.3 kb lncRNA (16). During the past decade, extensive
research has been carried out on H19 in various cancers. Initially,
H19 was found to be tumor-suppressive because of its abilities to
repress tumorigenicity (17).
However, increasing evidence suggests that H19 is upregulated in
most cancers and has oncogenic properties (18–20).
In particular, the upregulated levels of H19 have been confirmed in
most DSCs, except the less studied gallbladder cancer (21–25).
The role of H19 in liver cancer is still controversial, with some
studies showing beneficial (26,27)
and the others showing detrimental (28,29)
effects to cancer progression.
The HOTAIR is expressed from the developmental HOXC
locus located on chromosome 12q13.13. It was first revealed in
breast cancer that lncRNA HOTAIR could promote cancer metastasis
through inducing polycomb repressive complex 2 (PRC2)-mediated
trimethylation of histone H3 lysine-27 (H3K27me3) (35). The enhanced expression of HOTAIR is
reported in the cell lines and tumor tissues of most DSCs (9,36–41).
In gastric cancer, the increased expression of HOTAIR is associated
with TNM stage, lymph node metastasis, venous invasion, and poor
survival (37,42). HOTAIR knockdown leads to a promotion
of radio-sensitivity in colorectal cancer cells (43). In esophageal squamous cell
carcinoma, HOTAIR promotes cell migration and invasion by directly
downregulating WIF-1 expression and activating the Wnt⁄β-catenin
pathway (44). Song et al
suggested that the enhanced expression of HOTAIR in gastric cancer
is associated with tumor escape by inhibiting miR-152 and
upregulating HLA-G (45). HOTAIR
could act as a ceRNA in gastric cancer cells by efficiently binding
to miR-331-3p and thereby reactivating HER2 to promote cancer cell
growth, migration, and invasion (46). HOTAIR induces the silence of miR-34a
via PRC2, promoting EMT in human gastric cancer cells (47). Kogo et al demonstrated that
HOTAIR is involved in a genome-wide reprogramming of PRC2 in
colorectal cancer. The upregulation of HOTAIR is critical to the
metastatic progression (9).
Recently, Zhang et al suggested that HOTAIR can promote the
proteasomal degradation of SUZ12 and ZNF198 during liver
carcinogenesis induced by hepatitis B virus (48). Ye et al demonstrated that
HOTAIR induces malignant transformation of liver normal stem cells
by downregulating E-cadherin and inducing EMT (49). In liver cancer stem cells, HOTAIR
has been shown to promote malignant growth through downregulating
SETD2 (50). HOTAIR negatively
regulates RNA binding motif protein 38 (RBM38) in hepatocellular
carcinoma cells to promote cell motility (38). Ma et al demonstrated that
c-Myc induces HOTAIR expression through direct interaction with the
promoter region of HOTAIR in gallbladder cancer. In addition, they
also suggested that the oncogenic role of HOTAIR functions in part
through negatively regulating miRNA-130a (39).
MALAT1, a metastasis-associated lncRNA, was
originally discovered to be a marker for metastasis development in
early stage lung adenocarcinoma (51). Recently, the upregulated expression
of MALAT1 was observed in both cell lines and clinical tissue
samples in all types of human DSCs (52–57).
Among them, colorectal cancer is most studied for MALAT1
dysregulation (54,58–62).
Overexpression of MALAT1 promotes colorectal cancer cell
proliferation, migration, and invasion in vitro, and
stimulates tumor growth and metastasis in vivo.
Reciprocally, the knockdown of MALAT1 inhibits tumor growth and
metastasis (60). The higher level
of MALAT1 significantly correlates with peritoneal metastasis in
gastric cancer patients (53). The
knockdown of MALAT1 significantly inhibits the proliferation and
metastasis of gallbladder carcinoma cell lines both in vitro
and in vivo (56). The
inhibition of MALAT-1 could suppress pancreatic cancer cell
proliferation, migration, and invasion in vitro (63). A recent study suggests that MALAT-1
overexpression could increase the proportion of cancer stem cells
in pancreatic cancer (57).
In esophageal cancer cells, miR-101 and miR-217
conduct an Ago2-dependent post-transcriptional regulation of
MALAT1, leading to reduced cell proliferation, migration, and
invasion. MALAT1 knockdown significantly represses the
proliferation of esophageal squamous cell carcinoma cells, which
may be associated with the upregulation of p21 and p27 expression
(52). It was suggested that MALAT1
may regulate esophageal squamous cell carcinoma cell proliferation
by inactivating the ATM-CHK2 pathway (64). In liver cancer, MALAT1 is
upregulated by the oncoprotein Yes-associated protein (YAP) through
the interaction with TCF/β-catenin on the MALAT1 promoter, which
can be inhibited by serine/arginine-rich splicing factor 1 (SRSF1)
(65). The knockdown of Sp1 and Sp3
in hepatocellular carcinoma jointly downregulates MALAT1 expression
(66). In gallbladder cancer cells,
MALAT1 knockdown significantly inhibits the proliferation and
metastasis of gallbladder cancer cells in vitro and in
vivo through the inactivation of ERK/MAPK signaling pathway
(56).
The lncRNA HULC is regarded as the first lncRNA with
highly specific upregulation in hepatocellular carcinoma (67). Compared with normal controls, HULC
expression is significantly higher in tumor cells, tumor tissues
and plasma of hepatocellular carcinoma patients (68–70).
The overexpression of HULC is also reported in gastric cancer and
pancreatic cancer and is correlated with advanced lymph node
metastasis (71,72). Hepatitis B virus X protein (HBx)
contributes to the development of hepatocellular carcinoma.
Intriguingly, HULC is involved in HBx-mediated
hepatocarcinogenesis. HBx could activate the HULC expression to
promote hepatocellular carcinoma cell proliferation through the
downregulation of p18 (70). HULC
is both a target and a regulator of CREB through its interaction
with microRNA-372 in liver cancer cells, providing a fine-tuned
auto-regulatory loop (73). HULC is
responsible for the perturbations in circadian rhythm through
upregulating circadian oscillator CLOCK in hepatoma cells,
resulting in the promotion of hepatocarcinogenesis (74). HULC contributes to abnormal lipid
metabolism to support the growth of hepatoma cells through the
regulation of miR-9-mediated RXRA signaling (10). Recently, Lu et al
demonstrated that HULC promotes angiogenesis in liver cancer by
upregulating sphingosine kinase 1 (SPHK1). HULC sequesters miR-107
from binding to E2F1 transcription factor and therefore activates
E2F1-dependent transcription of SPHK1 (75). Wan et al suggested that HULC
could be downregulated by miR-203, leading to the suppression of
hepatocellular carcinoma (76).
Moreover, HULC could be upregulated by another lncRNA, cancer
upregulated drug-resistant gene (CUDR), contributing to the
malignant differentiation of liver cancer stem cells (77).
MEG3 represents an imprinted gene belonging to the
imprinted DLK1-MEG3 locus located at human chromosome 14q32.2
(78). Accumulating evidence
suggests that the expression of MEG3 is decreased in DSCs (79–82).
MEG3 is decreased in gastric cancer cell lines and tissues, and its
expression is associated with the metastasis of gastric cancer. The
overexpression of MEG3 in gastric cancer cells inhibits cell
proliferation, migration, invasion, and promotes cell apoptosis
(79). MEG3 overexpression could
inhibit colorectal cancer cell proliferation both in vitro
and in vivo (80). The
enforced expression of MEG3 in hepatocellular carcinoma cells
significantly decreases both anchorage-dependent and -independent
cell growth, and induces apoptosis (81).
The mechanism responsible for the downregulation of
MEG3 in cancer remains unclear. The interactions between MEG3 and
other molecules in DSCs have been widely studied. Peng et al
suggested that lncRNA MEG3 competitively binds to the miR-181
family and functions as a ceRNA, upregulating downstream target
B-cell lymphoma-2 (Bcl-2), and then suppressing gastric cancer
progression (79). Zhou et
al recently demonstrated that there is a positive correlation
between MEG3 and miR-141 in gastric cancer tissues. They further
suggested that miR-141 activates MEG3 by targeting E2F3, which
inhibits the proliferation of gastric cancer cells (83). Yan et al demonstrated that
the ectopic expression of miR-148a activates MEG3 via modulation of
DNMT-1 (84). It has been shown
that methylation-dependent regulation of MEG3 by miR-29a is
involved in hepatocellular carcinoma (81,85,86).
Similarly, dendrosomal curcumin upregulates miR-29a and miR-185 and
induces DNA hypomethylation, thus promoting the expression of MEG3
in hepatocellular carcinoma (87).
The re-expression of MEG3 could lead to the accumulation of p53
protein and its target gene expression, inducing inhibition of
cancer cell growth (88). Sun et
al also suggested that MEG3 overexpression upregulates the
protein level of p53 in gastric cancer cells harboring wild-type
p53, indicating that MEG3 may function as a tumor suppressor
partially through the activation of p53 in gastric cancer (89). In colorectal and liver cancers, the
tumor suppressive role of MEG3 through the activation of p53 is
also revealed (80,90). In addition, Modali et al
identified MEG3 as a tumor suppressor lncRNA by targeting c-MET to
elicit Menin tumor-suppressor activity in pancreatic cancer
(82).
GAS5, encoded at chromosome 1q25, with about 630
nucleotides in length, was initially discovered during screening
for potential tumor suppressor genes (91). Previous studies have shown that GAS5
is downregulated in DSCs, including gastric, colorectal, liver, and
pancreatic cancers (92–96). The lower expression of GAS5 is
positively correlated with aggressive tumor behavior (92,93).
The underlying mechanism for the roles of GAS5 in DSCs is not yet
well characterized. In gastric cancer cells, GAS5 binds to the YBX1
protein and regulates its turnover. The downregulation of GAS5
decreases the YBX1 protein level, subsequently inhibiting
YBX1-transactivated p21 expression and G1 phase arrest (97). Sun et al also suggested that
GAS5 regulates the expression of p21, E2F1, and cyclin D1 in
gastric cancer cells through post-transcriptional mechanism
(93). In gastric cancer cells and
pancreatic cancer cells, GAS5 functions as a tumor suppressor by
suppressing the expression of cyclin-dependent kinase (CDK) 6
(96, 98). Chang et al revealed that GAS
inhibits hepatoma cell proliferation by the regulation of vimentin
(95). GAS5 could also act as ceRNA
by binding to miR-21, inhibiting the migration and invasion of
hepatocellular carcinoma cells (94).
ANRIL is transcribed as a 3.8 kb lncRNA from the
INK4b-ARF-INK4a gene cluster in the opposite direction (99,100).
ANRIL has been identified as a genetic susceptibility locus
associated with various diseases including cancer (101,102). Previous studies have shown that
ANRIL is upregulated in esophageal squamous cell carcinoma, gastric
cancer, colorectal cancer, and hepatocellular carcinoma (103–105). ANRIL is verified as a growth
stimulator in esophageal, gastric, and colorectal cancer cells
(103,104,106). The knockdown of ANRIL could not
only inhibit hepatocellular carcinoma cell proliferation, but also
induced cell apoptosis both in vitro and in vivo
(107). In esophageal squamous
cell carcinoma, the inhibition of ANRIL upregulates the expressions
of p15 and TGFβ1, suggesting a significant role of ANRIL in the
development of esophageal squamous cell carcinoma (103). In gastric cancer, ANRIL could
indirectly regulate mTOR and CDK6/E2F1 pathway through binding to
PRC2 and epigenetically silencing miR-99a/miR-449a, partially
accounting for ANRIL-mediated cell growth regulation. In addition,
the upregulated E2F1 promotes ANRIL expression and forms a positive
feedback loop to promote gastric cancer cell proliferation
(104). In hepatocellular
carcinoma, ANRIL could epigenetically silence the transcription of
Krüppel-like factor 2 (KLF2) by interacting with PRC2 and
recruiting it to the promoter region of KLF2, leading to increased
cell proliferation and decreased cell apoptosis (107).
CCAT1, a 2628 nucleotide-lncRNA located on
chromosome 8q24.21, was first discovered in colon cancer by Nissan
et al in 2012 (115). CCAT1
is strongly expressed in colon cancer tissues but not in normal
tissues (115). The upregulation
of CCAT1 is evident in pre-malignant conditions and through all
disease stages, including advanced metastatic colon cancer
(116). Emerging evidence suggests
that CCAT1 is also upregulated in other DSCs including gastric
cancer, hepatocellular cancer, and gallbladder cancer (12,117,118). The upregulation of CCAT1 is
correlated with the growth of primary tumor, lymph node metastasis,
and metastatic disease (117),
while patients with low expression of CCAT1 demonstrate better
overall and relapse-free survival (119,120). He and colleagues suggest that in
colon cancer cells, c-Myc could accelerate CCAT1 transcription by
direct binding to its promoter region, and the enhanced expression
of CCAT1 promotes cell proliferation and invasion (120). The regulation of CCAT1 by c-Myc is
also revealed in gastric cancer and hepatocellular carcinoma
(117,119). Moreover, CCAT1 competitively binds
to let-7 to antagonize its function and de-represses its targets
HMGA2 and c-Myc, thus promoting cell proliferation and migration in
hepatocellular carcinoma cells (121). Similarly, CCAT1 promotes the
proliferation and invasiveness of gallbladder cancer cells
partially through 'spongeing' miRNA-218-5p and inducing the
expression of oncogenic gene Bmi1 (12).
LncRNA-AL589182.3 (ENST00000493038) is identified in
hepatocellular carcinoma cells as lncRNA-activated by TGF-β
(lncRNA-ATB) (122). Yuan et
al suggested that lncRNA-ATB is a key regulator of TGF-β
signaling pathway. LncRNA-ATB can be activated by TGF-β, and
subsequently competitively binds to the miR-200 family, leading to
the upregulation of ZEB1 and ZEB2 and the induction of EMT. In
addition, lncRNA-ATB facilitates hepatocellular carcinoma cell
colonization by increasing autocrine induction of IL-11 and
activating STAT3 signaling (122).
Recent studies have demonstrated the pathological roles of
lncRNA-ATB in other DSCs including gastric cancer, colorectal
cancer, and pancreatic cancer. LncRNA-ATB is also upregulated in
gastric cancer and colorectal cancer and this overexpression is
correlated with clinical features and prognosis (123,124). Yue et al demonstrated that
lncRNA-ATB suppresses E-cadherin expression and promotes EMT
(125). However, lncRNA-ATB
expression is decreased in pancreatic cancer tissues and cell
lines. Downregulated expression level of lncRNA-ATB is
significantly correlated with lymph node metastases, neural
invasion, and clinical stage in pancreatic cancer (126). The roles of lncRNA-ATB in
interacting with other miRNAs and mRNA still need to be
investigated (127).
The early diagnosis is critical for the improvement
of patient survival. However, traditional tumor biomarkers, such as
carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9,
present low sensitivity and specificity. Recently, circulating RNA
have emerged as non-invasive diagnostic markers. Although the
earlier studies concentrated on microRNAs (128,129), the investigations on the
diagnostic role of lncRNAs are increasing. In DSCs, there are
growing numbers of lncRNAs which have the potential to serve as
diagnostic biomarkers. Zhou et al systematically
characterized the potential of circulating lncRNAs in plasma as
diagnostic markers for gastric cancer. They detected the expression
of eight lncRNAs in plasma from gastric cancer patients and healthy
subjects. Three lncRNAs (H19, HOTAIR, and MALAT1) were identified
with significantly elevated levels in plasma from gastric cancer
patients compared to normal controls. Moreover, H19 shows a good
stability in the blood and can distinguish early stage gastric
cancer, presenting higher sensitivity and specificity than
traditional biomarkers (130).
Circulating lncRNAs are thought to be unstable due to the high
RNase level in plasma, especially in cancer patients carrying more
RNase in plasma (131). However,
Zhou et al showed that circulating lncRNAs are very stable
even when digested directly with RNase A (130), in accordance with another study
(132). One possible explanation
is that lncRNAs are protected by packaging into microparticles such
as exosomes (133,134). Li et al suggested that
there is no significant difference in LINC00152 levels between
plasma and exosomes, indicating that LINC00152 may be released into
the blood by exosomes (135).
LINC00152 and HULC are screened out from eight lncRNAs for
evaluating the diagnostic value of plasma lncRNAs in hepatocellular
carcinoma. Circulating HULC and LINC00152 are significantly
upregulated in the plasma of hepatocellular carcinoma patients
(69). The combination of HULC,
LINC00152, and AFP shows a high prediction value of hepatocellular
carcinoma (69). Therefore, the
combined analysis of individual lncRNA with traditional biomarkers
may improve the sensitivity and specificity of diagnosis and
enhance the diagnostic value of lncRNAs.
The prognostic biomarkers can predict therapeutic
effects after certain treatments, thus providing valuable guidance
for therapy. There are a number of lncRNAs that have been well
characterized as prognostic biomarkers in DSCs (136). The performance of H19 as
prognostic marker is still elusive. It is evident that high H19
expression is an independent predictor of the poor overall survival
in gastric cancer patients (31).
However, in hepatocellular carcinoma patients, low ratio of H19
expression in intratumoral hepatocellular carcinoma tissues to that
in peritumoral tissues predicts poor prognosis (29). High ANRIL expression has been
suggested to serve as an independent predictor of poor prognosis in
both gastric cancer and hepatocellular carcinoma patients (104,105). HOTAIR may serve as an outstanding
biomarker for its prognostic value in DSCs including esophageal,
gastric, colorectal, liver, and pancreatic cancers (40,41,47,137,138). In particular, the upregulated
HOTAIR expression in blood of colorectal cancer patients is
associated with unfavorable prognosis, representing an alternative
prognostic marker for colorectal cancer (137). For esophageal squamous cell
carcinoma, HOTAIR is most commonly identified as negative prognosis
marker among these DSC-related lncRNAs (36,41,44,139).
Since identified in 2014 (121),
lncRNA-ATB has been well characterized for its prognostic value in
DSCs. The high level of lncRNA-ATB is correlated with poor
prognosis in liver cancer (121),
gastric cancer, and colorectal cancer (122,123). However, the patients with low
lncRNA-ATB expression exhibit markedly poor overall survival
(125), suggesting a tumor
type-dependent role of lncRNA-ATB. The decreased expression of
GAS5, a tumor-suppressive lncRNA, could independently predict poor
overall survival of gastric, colorectal, and liver cancer patients
(92,93,95,140).
Frequent recurrence is a major obstacle for long-term survival.
Several DSC-related lncRNAs could predict recurrence of
hepatocellular carcinoma patients, demonstrating their potential as
prognostic biomarkers for recurrence (55,86,111,119,122,138).
LncRNAs are considered as potential therapeutic
targets in cancer. The specific targeting of lncRNAs could greatly
affect the malignant phenotypes such as tumor growth and
metastasis. LncRNA-oriented therapies involving small interfering
RNAs (siRNAs) target specific oncogenic lncRNAs and show promising
anticancer effects. For example, Liu et al showed that
HOTAIR knockdown inhibits the metastasis of gastric cancer cells
both in vitro and in vivo. They also suggested that
therapies targeting HOTAIR lead to improvement for gastric cancer
treatment (47). The ectopic
overexpression of tumor-suppressive lncRNAs may also have
anticancer effects. For instance, GAS5 transfected hepatocellular
carcinoma cells exhibit decreased migration and invasion,
indicating the potential of GAS5 as a target for hepatocellular
carcinoma therapy (94). Another
strategy is to interfere with the functional molecules that
interact with DSC-related lncRNAs. PRKA A-kinase anchor protein 9
(AKAP-9) is a target of MALAT1 and highly expressed in human
primary colorectal cancer tissues with lymph node metastasis. The
knockdown of AKAP-9 inhibits MALAT1-mediated in vivo cell
proliferation, migration and invasion, which may become a potential
therapy for colorectal cancer (60). The development of multidrug
resistance (MDR) is a major reason for therapy failure in cancer,
leading to recurrence and metastasis. Recent studies revealed that
lncRNAs play an important role in MDR for DSCs. For example, the
cisplatin-resistant gastric cancer cells transfected with PVT-1
siRNA and treated with cisplatin exhibit significant higher
apoptosis and lower survival rate, suggesting that PVT1 may be an
effective target for reversing MDR in gastric cancer therapy
(141). Similarly, the lncRNAs
MRUL and HOTTIP also represent potential targets to reverse the MDR
phenotype in DSC (142,143). In addition, the intra-arterially
administered DTA-H19 plasmid (also known as BC-819), carrying the
diphtheria toxin A-chain gene under the regulation of the H19
promoter sequence, significantly repress the tumor growth of the
rat liver metastases from colon cancer (144). The combined treatment of BC-819
and gemcitabine show desirable antitumor activity in animal models
with pancreatic carcinoma (145),
suggesting a promising therapeutic value to be further evaluated in
clinical trial (146). Despite the
emerging studies, the research is still in its infancy for the
therapeutic role of lncRNAs. Further studies and large clinical
trials need to be conducted to investigate the clinical therapeutic
interventions of lncRNAs in DSCs.
Accumulating evidence suggests that lncRNAs
function as oncogenes or tumor suppressors in DSCs. A group of
lncRNAs including H19, HOTAIR, MALAT1, HULC, MEG3, GAS5, ANRIL,
PVT1, CCAT1, and lncRNA-ATB generally share consistent functions in
different types of DSCs. The dysregulated expression of these
lncRNAs affects tumor development, progression, and metastasis in
DSCs. Moreover, lncRNAs exhibit clinical significance in serving as
potential diagnostic and prognostic biomarkers and therapeutic
targets in DSCs.
The convenient detection of circulating lncRNAs in
blood and body fluids such as gastric juice can reflect the
malignancy of cancer, which is valuable in distinguishing DSC
patients from benign diseases and healthy individuals with the
advantages of non-invasiveness and cost-effectiveness. However, the
use of lncRNAs for diagnosis still has several issues of concern.
First, the concentration of measured circulating lncRNAs may not
represent the actual amount in the diagnosed patients. The
stability of circulating lncRNAs, which may result in unreliable
diagnostic accuracy, needs further validation. Second, as one
certain lncRNA can be involved in various types of DSCs, the
diagnostic specificity and sensitivity need to be explored and
improved. One feasible solution is combined diagnosis. The combined
diagnosis of different lncRNAs, or with other biomarkers such as
traditional biomarkers and circulating miRNAs, may increase the
diagnostic accuracy. Third, the cohort size presented in current
studies is generally too small for the validation steps. Therefore,
prospective studies with large cohort size should investigate the
practicability of using circulating lncRNAs, as well as combination
with miRNAs or other molecules, as diagnostic biomarkers for
DSCs.
The study of lncRNAs as therapeutic targets is
still in its early stage and has not reached the clinical practice
for DSCs. Several challenges are hindering the development of
lncRNA-oriented therapeutics. First, the large size of lncRNAs
frequently generates secondary structures, which makes it difficult
for the design of siRNAs. Besides, the obstacles for RNA therapies
such as side effects and lack of delivery methods and appropriate
vectors also impede the clinical use of lncRNAs (147). Moreover, most lncRNAs are not
conserved evolutionarily, which makes animal models unavailable
prior to clinical applications. Ultimately, the role of DSC-related
lncRNAs in interacting with other molecules, including DNA, RNA,
and proteins remains not completely understood. Identifying the
pivotal role of lncRNAs in DSCs and elucidating their mechanisms
will help to block malignant signaling and to specifically
eliminate DSC cells.
Therefore, great efforts should be continuously
made to further elucidate the lncRNA-based regulatory network in
DSCs. Large scaled clinical trials are required to select candidate
lncRNAs as novel biomarkers with sufficient sensitivity and
specificity, as well as to evaluate the effect of combined
diagnosis along with other biomarkers. Further studies focusing on
improving the therapeutic effect while minimizing the side effect
of lncRNA-based therapy should also be conducted. Thus, better
understanding of the functional roles of lncRNAs in DSCs would
provide new strategies for the early diagnosis and targeted therapy
of DSCs.
This study was funded by the National Natural
Science Foundation of China (81201660), the Natural Science
Foundation of Jiangsu Province (BK20141303), the Key Research and
Development Project of Zhenjiang (SH2015034), the Jiangsu Key
Laboratory of Medical Science and Laboratory Medicine Project
(JSKLM-2014-006), and the Key R&D Special Fund of Jiangsu
Province (BE2015666).
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO Classification of Tumours of the Digestive System.
4th edition. IARC Press; Geneva: 2010
|
|
3
|
Thiel A and Ristimäki A: Targeted therapy
in gastric cancer. APMIS. 123:365–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carr BI and Guerra V: HCC and its
microenvironment. Hepatogastroenterology. 60:1433–1437.
2013.PubMed/NCBI
|
|
5
|
Waraya M, Yamashita K, Ema A, Katada N,
Kikuchi S and Watanabe M: Exclusive association of p53 mutation
with super-high methylation of tumor suppressor genes in the p53
pathway in a unique gastric cancer phenotype. PLoS One.
10:e01399022015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakai E, Fukuyo M, Ohata K, Matsusaka K,
Doi N, Mano Y, Takane K, Abe H, Yagi K, Matsuhashi N, et al:
Genetic and epigenetic aberrations occurring in colorectal tumors
associated with serrated pathway. Int J Cancer. 138:1634–1644.
2016. View Article : Google Scholar :
|
|
7
|
Anwar SL, Krech T, Hasemeier B, Schipper
E, Schweitzer N, Vogel A, Kreipe H and Lehmann U: Loss of DNA
methylation at imprinted loci is a frequent event in hepatocellular
carcinoma and identifies patients with shortened survival. Clin
Epigenetics. 7:1102015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W,
Yan L, Wang K and Feng J: Downregulated long noncoding RNA BANCR
promotes the proliferation of colorectal cancer cells via
downregualtion of p21 expression. PLoS One. 10:e01226792015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meseure D, Drak Alsibai K, Nicolas A,
Bieche I and Morillon A: Long noncoding RNAs as new architects in
cancer epigenetics, prognostic biomarkers, and potential
therapeutic targets. Biomed Res Int. 2015:3202142015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin Z, Hu Y, Lai S, Xue M, Lin J, Qian Y,
Zhuo W, Chen S, Si J and Wang L: Long noncoding RNA: Its partners
and their roles in cancer. Neoplasma. 62:846–854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ayesh S, Matouk I, Schneider T, Ohana P,
Laster M, Al-Sharef W, De-Groot N and Hochberg A: Possible
physiological role of H19 RNA. Mol Carcinog. 35:63–74. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gabory A, Ripoche MA, Yoshimizu T and
Dandolo L: The H19 gene: Regulation and function of a non-coding
RNA. Cytogenet Genome Res. 113:188–193. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao Y, Crenshaw T, Moulton T, Newcomb E
and Tycko B: Tumour-suppressor activity of H19 RNA. Nature.
365:764–767. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Upregulated H19 contributes to bladder cancer cell proliferation
by regulating ID2 expression. FEBS J. 280:1709–1716. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vennin C, Spruyt N, Dahmani F, Julien S,
Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X and
Adriaenssens E: H19 non coding RNA-derived miR-675 enhances
tumorigenesis and metastasis of breast cancer cells by
downregulating c-Cbl and Cbl-b. Oncotarget. 6:29209–29223.
2015.PubMed/NCBI
|
|
21
|
Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou
Y, Li H, Gao M, Li W, Zhang Q, et al: Upregulation of H19 promotes
invasion and induces epithelial-to-mesenchymal transition in
esophageal cancer. Oncol Lett. 10:291–296. 2015.PubMed/NCBI
|
|
22
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The Interaction between miR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang Z, Lu Y, Xu Q, Tang B, Park CK and
Chen X: HULC and H19 played different roles in overall and
disease-free survival from hepatocellular carcinoma after curative
hepatectomy: A preliminary analysis from gene expression omnibus.
Dis Markers. 2015:1910292015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7's suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li H, Li J, Jia S, Wu M, An J, Zheng Q,
Zhang W and Lu D: miR675 upregulates long noncoding RNA H19 through
activating EGR1 in human liver cancer. Oncotarget. 6:31958–31984.
2015.PubMed/NCBI
|
|
27
|
Lv J, Yu YQ, Li SQ, Luo L and Wang Q:
Aflatoxin B1 promotes cell growth and invasion in hepatocellular
carcinoma HepG2 cells through H19 and E2F1. Asian Pac J Cancer
Prev. 15:2565–2570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv J, Ma L, Chen XL, Huang XH and Wang Q:
Downregulation of LncRNAH19 and MiR-675 promotes migration and
invasion of human hepatocellular carcinoma cells through
AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog
Med Sci. 34:363–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the miR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar
|
|
30
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang EB, Han L, Yin DD, Kong R, De W and
Chen J: c-Myc-induced, long, noncoding H19 affects cell
proliferation and predicts a poor prognosis in patients with
gastric cancer. Med Oncol. 31:9142014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhuang M, Gao W, Xu J, Wang P and Shu Y:
The long non-coding RNA H19-derived miR-675 modulates human gastric
cancer cell proliferation by targeting tumor suppressor RUNX1.
Biochem Biophys Res Commun. 448:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar
|
|
34
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hajjari M, Behmanesh M, Sadeghizadeh M and
Zeinoddini M: Up-regulation of HOTAIR long non-coding RNA in human
gastric adenocarcinoma tissues. Med Oncol. 30:6702013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du
C, Peng C, Xie H, Zhou L, Wu J, et al: Long non-coding RNA HOTAIR
promotes cell migration and invasion via down-regulation of RNA
binding motif protein 38 in hepatocellular carcinoma cells. Int J
Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
41
|
Lv XB, Lian GY, Wang HR, Song E, Yao H and
Wang MH: Long noncoding RNA HOTAIR is a prognostic marker for
esophageal squamous cell carcinoma progression and survival. PLoS
One. 8:e635162013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One. 8:e770702013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep.
35:479–487. 2016.
|
|
44
|
Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY,
Xue WQ, Chen YB, Zhang Y and Jia WH: HOTAIR, a prognostic factor in
esophageal squamous cell carcinoma, inhibits WIF-1 expression and
activates Wnt pathway. Cancer Sci. 104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song B, Guan Z, Liu F, Sun D, Wang K and
Qu H: Long non-coding RNA HOTAIR promotes HLA-G expression via
inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res
Commun. 464:807–813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu YW, Sun M, Xia R, Zhang EB, Liu XH,
Zhang ZH, Xu TP, De W, Liu BR and Wang ZX: LincHOTAIR
epigenetically silences miR34a by binding to PRC2 to promote the
epithelial-to-mesenchymal transition in human gastric cancer. Cell
Death Dis. 6:e18022015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang H, Diab A, Fan H, Mani SK, Hullinger
R, Merle P and Andrisani O: PLK1 and HOTAIR accelerate proteasomal
degradation of SUZ12 and ZNF198 during hepatitis B virus-induced
liver carcinogenesis. Cancer Res. 75:2363–2374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ye P, Wang T, Liu WH, Li XC, Tang LJ and
Tian FZ: Enhancing HOTAIR/miR-10b drives normal liver stem cells
toward a tendency to malignant transformation through inducing
epithelial-to-mesenchymal transition. Rejuvenation Res. 18:332–340.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu
H and Lu D: LncRNA HOTAIR promotes human liver cancer stem cell
malignant growth through downregulation of SETD2. Oncotarget.
6:27847–27864. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar :
|
|
53
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et
al: Metastasis-associated long non-coding RNA drives gastric cancer
development and promotes peritoneal metastasis. Carcinogenesis.
35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
55
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar
|
|
56
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT1 promotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jiao F, Hu H, Han T, Yuan C and Wang L,
Jin Z, Guo Z and Wang L: Long noncoding RNA MALAT-1 enhances stem
cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci.
16:6677–6693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu
YJ, Jian SF, Hung JY, Wang JY and Kuo PL: Chemokine (C-C Motif)
Ligand 5 is involved in tumor-associated dendritic cell-mediated
colon cancer progression through non-coding RNA MALAT-1. J Cell
Physiol. 230:1883–1894. 2015. View Article : Google Scholar
|
|
60
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar
|
|
61
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
62
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J, et al: Resveratrol inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/β-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar
|
|
63
|
Jiao F, Hu H, Yuan C and Wang L, Jiang W,
Jin Z, Guo Z and Wang L: Elevated expression level of long
noncoding RNA MALAT-1 facilitates cell growth, migration and
invasion in pancreatic cancer. Oncol Rep. 32:2485–2492.
2014.PubMed/NCBI
|
|
64
|
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y
and Yang K: Up-regulation of long noncoding RNA MALAT1 contributes
to proliferation and metastasis in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 34:72015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang J, Wang H, Zhang Y, Zhen N, Zhang L,
Qiao Y, Weng W, Liu X, Ma L, Xiao W, et al: Mutual inhibition
between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced
tumourigenesis in liver cancer. Cell Signal. 26:1048–1059. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Huang Z, Huang L, Shen S, Li J, Lu H, Mo
W, Dang Y, Luo D, Chen G and Feng Z: Sp1 cooperates with Sp3 to
upregulate MALAT1 expression in human hepatocellular carcinoma.
Oncol Rep. 34:2403–2412. 2015.PubMed/NCBI
|
|
67
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. BioMed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 Act as Novel Biomarkers in
Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhao Y, Guo Q, Chen J, Hu J, Wang S and
Sun Y: Role of long non-coding RNA HULC in cell proliferation,
apoptosis and tumor metastasis of gastric cancer: A clinical and in
vitro investigation. Oncol Rep. 31:358–364. 2014.
|
|
72
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cui M, Zheng M, Sun B, Wang Y, Ye L and
Zhang X: A long noncoding RNA perturbs the circadian rhythm of
hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia.
17:79–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu Z, Xiao Z1, Liu F, Cui M, Li W, Yang Z,
Li J, Ye L and Zhang X: Long non-coding RNA HULC promotes tumor
angiogenesis in liver cancer by up-regulating sphingosine kinase 1
(SPHK1). Oncotarget. 7:241–254. 2016.
|
|
76
|
Wan D, Shen S, Fu S, Preston B, Brandon C,
He S, Shen C, Wu J, Wang S, Xie W, et al: miR-203 suppresses the
proliferation and metastasis of hepatocellular carcinoma by
targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC.
Anticancer Agents Med Chem. 16:414–423. 2016. View Article : Google Scholar
|
|
77
|
Gui X, Li H, Li T, Pu H and Lu D: Long
Noncoding RNA CUDR regulates HULC and β-catenin to govern human
liver stem cell malignant differentiation. Mol Ther. 23:1843–1853.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wylie AA, Murphy SK, Orton TC and Jirtle
RL: Novel imprinted DLK1/GTL2 domain on human chromosome 14
contains motifs that mimic those implicated in IGF2/H19 regulation.
Genome Res. 10:1711–1718. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Peng W, Si S, Zhang Q, Li C, Zhao F, Wang
F, Yu J and Ma R: Long non-coding RNA MEG3 functions as a competing
endogenous RNA to regulate gastric cancer progression. J Exp Clin
Cancer Res. 34:792015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Modali SD, Parekh VI, Kebebew E and
Agarwal SK: Epigenetic regulation of the lncRNA MEG3 and its target
c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol.
29:224–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhou X, Ji G, Ke X, Gu H, Jin W and Zhang
G: MiR-141 inhibits gastric cancer proliferation by interacting
with long noncoding RNA MEG3 and down-regulating E2F3 expression.
Dig Dis Sci. 60:3271–3282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yan J, Guo X, Xia J, Shan T, Gu C, Liang
Z, Zhao W and Jin S: MiR-148a regulates MEG3 in gastric cancer by
targeting DNA methyltransferase 1. Med Oncol. 31:8792014.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu LX, Deng W, Zhou XT, Chen RP, Xiang
MQ, Guo YT, Pu ZJ, Li R, Wang GF and Wu LF: The mechanism of
adenosine-mediated activation of lncRNA MEG3 and its antitumor
effects in human hepatoma cells. Int J Oncol. 48:421–429. 2016.
|
|
86
|
Zhuo H, Tang J, Lin Z, Jiang R, Zhang X,
Ji J, Wang P and Sun B: The aberrant expression of MEG3 regulated
by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol
Carcinog. 55:209–219. 2016. View Article : Google Scholar
|
|
87
|
Zamani M, Sadeghizadeh M, Behmanesh M and
Najafi F: Dendrosomal curcumin increases expression of the long
non-coding RNA gene MEG3 via up-regulation of epi-miRs in
hepatocellular cancer. Phytomedicine. 22:961–967. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista
DL, Gejman R, Ansell PJ, Zhao J, Weng C and Klibanski A: Activation
of p53 by MEG3 non-coding RNA. J Biol Chem. 282:24731–24742. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
90
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y, et al: Long noncoding RNA MEG3 interacts
with p53 protein and regulates partial p53 target genes in hepatoma
cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yin D, He X, Zhang E, Kong R, De W and
Zhang Z: Long noncoding RNA GAS5 affects cell proliferation and
predicts a poor prognosis in patients with colorectal cancer. Med
Oncol. 31:2532014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y,
Yang X, Shen J, Liu Q and Zhang J: Long noncoding RNA GAS5
suppresses the migration and invasion of hepatocellular carcinoma
cells via miR-21. Tumour Biol. 37:2691–2702. 2016. View Article : Google Scholar
|
|
95
|
Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q
and Liu Z: Decreased expression of long non-coding RNA GAS5
indicates a poor prognosis and promotes cell proliferation and
invasion in hepatocellular carcinoma by regulating vimentin. Mol
Med Rep. 13:1541–15450. 2016.
|
|
96
|
Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng
X, Chen H, Jin J, Peng C, Li H, et al: Downregulation of gas5
increases pancreatic cancer cell proliferation by regulating CDK6.
Cell Tissue Res. 354:891–896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS5 enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Guo X, Deng K, Wang H, Xia J, Shan T,
Liang Z, Yao L and Jin S: GAS5 inhibits gastric cancer cell
proliferation partly by modulating CDK6. Oncol Res Treat.
38:362–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Pasmant E, Laurendeau I, Héron D, Vidaud
M, Vidaud D and Bièche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural system
tumor family: Identification of ANRIL, an antisense noncoding RNA
whose expression coclusters with ARF. Cancer Res. 67:3963–3969.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yap KL, Li S, Muñoz-Cabello AM, Raguz S,
Zeng L, Mujtaba S, Gil J, Walsh MJ and Zhou MM: Molecular interplay
of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by
polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell.
38:662–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Bochenek G, Häsler R, El Mokhtari NE,
König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S and Schaefer
AS: The large non-coding RNA ANRIL, which is associated with
atherosclerosis, periodontitis and several forms of cancer,
regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 22:4516–4527.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang W, Chen Y, Liu P, Chen J, Song L,
Tang Y, Wang Y, Liu J, Hu FB and Hui R: Variants on chromosome
9p21.3 correlated with ANRIL expression contribute to stroke risk
and recurrence in a large prospective stroke population. Stroke.
43:14–21. 2012. View Article : Google Scholar
|
|
103
|
Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin
Y, Wu S, Mou X and Zhu Y: ANRIL inhibits p15(INK4b) through the
TGFβ1 signaling pathway in human esophageal squamous cell
carcinoma. Cell Immunol. 289:91–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang EB, Kong R, Yin DD, You LH, Sun M,
Han L, Xu TP, Xia R, Yang JS, De W, et al: Long noncoding RNA ANRIL
indicates a poor prognosis of gastric cancer and promotes tumor
growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget.
5:2276–2292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ
and Ma WL: High expression of long non-coding RNA ANRIL is
associated with poor prognosis in hepatocellular carcinoma. Int J
Clin Exp Pathol. 8:3076–3082. 2015.PubMed/NCBI
|
|
106
|
Naemura M, Tsunoda T, Inoue Y, Okamoto H,
Shirasawa S and Kotake Y: ANRIL regulates the proliferation of
human colorectal cancer cells in both two- and three-dimensional
culture. Mol Cell Biochem. 412:141–146. 2016. View Article : Google Scholar
|
|
107
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol Oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: A rising star among oncogenic long noncoding RNAs. BioMed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Ding J, Li D, Gong M, Wang J, Huang X, Wu
T and Wang C: Expression and clinical significance of the long
non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther.
7:1625–1630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar :
|
|
111
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J, et al: Long non-coding RNA PVT1 is
associated with tumor progression and predicts recurrence in
hepatocellular carcinoma patients. Oncol Lett. 9:955–963.
2015.PubMed/NCBI
|
|
112
|
Huang C, Yu W, Wang Q, Cui H, Wang Y,
Zhang L, Han F and Huang T: Increased expression of the lncRNA PVT1
is associated with poor prognosis in pancreatic cancer patients.
Minerva Med. 106:143–149. 2015.PubMed/NCBI
|
|
113
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W, et al: Oncofetal long noncoding
RNA PVT1 promotes proliferation and stem cell-like property of
hepatocellular carcinoma cells by stabilizing NOP2. Hepatology.
60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A,
Dayanc BE, Ritter G, Gomceli I, et al: Colon cancer associated
transcript-1: A novel RNA expressed in malignant and pre-malignant
human tissues. Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar
|
|
116
|
Alaiyan B, Ilyayev N, Stojadinovic A,
Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink
B, et al: Differential expression of colon cancer associated
transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence.
BMC Cancer. 13:1962013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar
|
|
118
|
Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C
and Lu J: CCAT1 promotes hepatocellular carcinoma cell
proliferation and invasion. Int J Clin Exp Pathol. 8:5427–5434.
2015.PubMed/NCBI
|
|
119
|
Zhu HQ, Zhou X, Chang H, Li HG, Liu FF, Ma
CQ and Lu J: Aberrant expression of CCAT1 regulated by c-Myc
predicts the prognosis of hepatocellular carcinoma. Asian Pac J
Cancer Prev. 16:5181–5185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
He X, Tan X, Wang X, Jin H, Liu L, Ma L,
Yu H and Fan Z: C-Myc-activated long noncoding RNA CCAT1 promotes
colon cancer cell proliferation and invasion. Tumour Biol.
35:12181–12188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Saito T, Kurashige J, Nambara S, Komatsu
H, Hirata H, Ueda M, Sakimura S, Uchi R, Takano Y, Shinden Y, et
al: A long non-coding RNA activated by transforming growth factor-β
is an independent prognostic marker of gastric cancer. Ann Surg
Oncol. 22(Suppl 3): S915–S922. 2015. View Article : Google Scholar
|
|
124
|
Iguchi T, Uchi R, Nambara S, Saito T,
Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et
al: A long noncoding RNA, lncRNA-ATB, is involved in the
progression and prognosis of colorectal cancer. Anticancer Res.
35:1385–1388. 2015.PubMed/NCBI
|
|
125
|
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu
F, Peng Z and Yan D: LncRNA-ATB mediated E-cadherin repression
promotes the progression of colon cancer and predicts poor
prognosis. J Gastroenterol Hepatol. 31:595–603. 2016. View Article : Google Scholar
|
|
126
|
Qu S, Yang X, Song W, Sun W, Li X, Wang J,
Zhong Y, Shang R, Ruan B, Zhang Z, et al: Downregulation of
lncRNA-ATB correlates with clinical progression and unfavorable
prognosis in pancreatic cancer. Tumour Biol. 37:3933–3938. 2016.
View Article : Google Scholar
|
|
127
|
Li W and Kang Y: A new Lnc in metastasis:
Long noncoding RNA mediates the prometastatic functions of TGF-β.
Cancer Cell. 25:557–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X,
Yu L, Wang L, Wang J, Wu Y, et al: A plasma microRNA panel for
early detection of colorectal cancer. Int J Cancer. 136:152–161.
2015. View Article : Google Scholar
|
|
129
|
Paik WH, Song BJ, Kim HW, Kim HR and Hwang
JH: MicroRNA-200c as a prognostic biomarker for pancreatic cancer.
Korean J Gastroenterol. 66:215–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Quackenbush JF, Cassidy PB, Pfeffer LM,
Boucher KM, Hawkes JE, Pfeffer SR, Kopelovich L and Leachman SA:
Isolation of circulating microRNAs from microvesicles found in
human plasma. Methods Mol Biol. 1102:641–653. 2014. View Article : Google Scholar
|
|
132
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
133
|
Orozco AF and Lewis DE: Flow cytometric
analysis of circulating microparticles in plasma. Cytometry A.
77:502–514. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Record M, Subra C, Silvente-Poirot S and
Poirot M: Exosomes as intercellular signalosomes and
pharmacological effectors. Biochem Pharmacol. 81:1171–1182. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
136
|
Paoletti X, Oba K, Burzykowski T, Michiels
S, Ohashi Y, Pignon JP, Rougier P, Sakamoto J, Sargent D, Sasako M,
et al GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research
International Collaboration) Group: Benefit of adjuvant
chemotherapy for resectable gastric cancer: A meta-analysis. JAMA.
303:1729–1737. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Li X, Wu Z, Mei Q, Li X, Guo M, Fu X and
Han W: Long non-coding RNA HOTAIR, a driver of malignancy, predicts
negative prognosis and exhibits oncogenic activity in oesophageal
squamous cell carcinoma. Br J Cancer. 109:2266–2278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
141
|
Zhang XW, Bu P, Liu L, Zhang XZ and Li J:
Overexpression of long non-coding RNA PVT1 in gastric cancer cells
promotes the development of multidrug resistance. Biochem Biophys
Res Commun. 462:227–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang Y, Zhang D, Wu K, Zhao Q, Nie Y and
Fan D: Long noncoding RNA MRUL promotes ABCB1 expression in
multidrug-resistant gastric cancer cell sublines. Mol Cell Biol.
34:3182–3193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Sorin V, Ohana P, Mizrahi A, Matouk I,
Birman T, Hochberg A and Czerniak A: Regional therapy with DTA-H19
vector suppresses growth of colon adenocarcinoma metastases in the
rat liver. Int J Oncol. 39:1407–1412. 2011.PubMed/NCBI
|
|
145
|
Sorin V, Ohana P, Gallula J, Birman T,
Matouk I, Hubert A, Gilon M, Hochberg A and Czerniak A:
H19-promoter-targeted therapy combined with gemcitabine in the
treatment of pancreatic cancer. ISRN Oncol.
2012:3517502012.PubMed/NCBI
|
|
146
|
Hanna N, Ohana P, Konikoff FM, Leichtmann
G, Hubert A, Appelbaum L, Kopelman Y, Czerniak A and Hochberg A:
Phase 1/2a, dose-escalation, safety, pharmacokinetic and
preliminary efficacy study of intratumoral administration of BC-819
in patients with unresectable pancreatic cancer. Cancer Gene Ther.
19:374–381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Burnett JC and Rossi JJ: RNA-based
therapeutics: Current progress and future prospects. Chem Biol.
19:60–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang
M, Wang CJ, Cao H and Xu J: HOTAIR long noncoding RNA promotes
gastric cancer metastasis through suppression of poly r(C)-binding
brotein (PCBP) 1. Mol Cancer Ther. 14:1162–1170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Konishi H, Ichikawa D, Yamamoto Y, Arita
T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et
al: Plasma level of metastasis-associated lung adenocarcinoma
transcript 1 is associated with liver damage and predicts
development of hepatocellular carcinoma. Cancer Sci. 107:149–154.
2016. View Article : Google Scholar :
|
|
150
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar
|
|
151
|
Mizrahi I, Mazeh H, Grinbaum R, Beglaibter
N, Wilschanski M, Pavlov V, Adileh M, Stojadinovic A, Avital I,
Gure AO, et al: Colon cancer sssociated transcript-1 (CCAT1)
expression in adenocarcinoma of the stomach. J Cancer. 6:105–110.
2015. View Article : Google Scholar :
|
|
152
|
Cao X, Zhao R, Chen Q, Zhao Y, Zhang B,
Zhang Y, Yu J, Han G, Cao W, Li J, et al: MALAT1 might be a
predictive marker of poor prognosis in patients who underwent
radical resection of middle thoracic esophageal squamous cell
carcinoma. Cancer Biomark. 15:717–723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Wang J, Su L, Chen X, Li P, Cai Q, Yu B,
Liu B, Wu W and Zhu Z: MALAT1 promotes cell proliferation in
gastric cancer by recruiting SF2/ASF. Biomed Pharmacother.
68:557–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
You L, Chang D, Du HZ and Zhao YP:
Genome-wide screen identifies PVT1 as a regulator of gemcitabine
sensitivity in human pancreatic cancer cells. Biochem Biophys Res
Commun. 407:1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|