Introduction
Lung cancer is the leading cause of cancer deaths
worldwide, and its incidence continues to increase (1). Non-small cell lung cancer (NSCLC)
accounts for ~85% of all lung cancers. Histologically, NSCLC is
divided into lung adenocarcinoma (LAD), squamous cell carcinoma
(SCC), and large cell carcinoma. Although there has been some
progress in chemotherapy, radiation and surgery, lung cancer
remains very aggressive and usually rapidly fatal (2). The average 5-year survival of lung
cancer is <15% (3–6). In recent years, a growing proportion
of LAD is due to socioeconomic development and environmental
problems. However, the mechanisms of LAD have not been
elucidated.
Studies have shown that lncRNAs are abnormally
expressed in tumor cells or tissues and regulate coding gene
expression. The altered expression of lncRNAs results in the
development, invasion, and metastasis of many cancers with a series
of mechanisms (7,8). The regulation of gene expression by
lncRNAs at the epigenetic level, transcriptional and
post-transcriptional level have been reported (9–11).
LncRNAs have been shown to be involved in the development and
progression of lung cancer. However, lung cancer-associated lncRNAs
are few including HOTAIR, H19, ANRIL, MALAT1 (12,13),
SCAL1 (14), AK126698 (15), and GAS6-AS1 (16), so it is very important to identify
additional lung cancer-associated lncRNAs and unveil their
mechanism of action.
We found that lncRNA RPLP0P2 was downregulated in
LAD by high-throughput microarray and real-time quantitative
reverse transcription-polymerase chain reaction (qPCR) method in
our previous study. Bioinformation analysis showed that LRRC10B
might be a target gene regulated by RPLP0P2. However, the clinical
roles and biological function of RPLP0P2 are not well understood in
LAD. In this study, the expression level of RPLP0P2 was estimated
by quantitative PCR in 57 pairs of LAD and NT samples and the
relation of RPLP0P2 to clinical data of LAD patients was analyzed.
We overexpressed RPLP0P2 based on the human LAD A549 cell line by
lentivirus-mediated technology and observed oncological behavior
change of A549 cells.
Materials and methods
Patient samples
The 57 LAD samples and corresponding NT samples were
prospectively collected from patients of the First Affiliated
Hospital of Wenzhou Medical University, China, from August 2013 to
August 2014. The clinical data of these cases are shown in Table I. The diagnosis of adenocarcinoma
was confirmed by histopathology. TNM clinical stage based on the
American Joint Committee on Cancer (AJCC) and the Union for
International Cancer Control (UICC) in 2002. The LAD and matched NT
samples were snap-frozen in liquid nitrogen immediately after
resection. We have followed prognosis of 35 LAD patients by
telephone or other means, the longest follow-up time was 28 months.
According to the expression level of RPLP0P2, the survival data are
divided into the high and low expression group. This study was
approved by the Institutional Ethics Review Board of the First
Affiliated Hospital of Wenzhou Medical University, and all patients
provided written informed consent for this study.
 | Table IThe clinical features of 57 LAD
patients and the relative expression levels of RPLP0P2. |
Table I
The clinical features of 57 LAD
patients and the relative expression levels of RPLP0P2.
| Term | Case (n) | RPLP0P2 relative
expression level | Kruskal-Wallis or
Mann-Whitney U test | P-value |
|---|
| Gender | | | 287.00 | 0.423 |
| Male | 28 | 0.792
(0.160–1.130) | | |
| Female | 29 | 1.064
(0.028–1.675) | | |
| TNM stage | | | 7.124 | 0.154 |
| Ia | 12 | 0.757
(0.098–1.293) | | |
| Ib | 28 | 0.604
(0.041–1.487) | | |
| IIa | 7 | 0.458
(0.097-1.359) | | |
| IIb | 2 | 0.893
(0.908–1.193) | | |
| IIIa | 8 | 1.200
(0.016-1.772) | | |
| Histological
degree | | | 3.235 | 0.676 |
| Poor | 11 | 1.000
(0.160–1.412) | | |
| Poor-moderate | 7 | 1.014
(0.097–1.134) | | |
| Moderate | 17 | 0.463
(0.064–1.773) | | |
| Moderate-high | 9 | 0.732
(0.318–1.004) | | |
| High | 13 | 1.117
(0.092–1.273) | | |
| Lymph node
metastasis | | | 9.102 | 0.011 |
| Yes | 14 | 0.130
(0.081–0.387) | | |
| No | 43 | 1.659
(0.362–2.96) | | |
| Smoking | | | 321.00 | 0.165 |
| Yes | 20 | 0.732
(0.154–1.266) | | |
| No | 37 | 0.917
(0.104–1.004) | | |
Quantitative PCR
Total RNA was extracted from frozen LAD tissues by
using TRIzol reagent (Invitrogen). According to the manufacturer's
instructions, total RNA was reverse-transcribed into cDNA using an
RT Reagent kit (Takara, Shanghai, China). RPLP0P2 and GAPDH mRNA
expression in LAD tissues were measured by quantitative PCR by
using SYBR Premix Ex Taq in ABI 7000 instrument. RPLP0P2 sense,
5′-AAAAACGATCAACGAACCTT-3′ and antisense,
5′-AATCGTCTCTGCTTTTCTTG-3′; GAPDH sense,
5′-TGACTTCAACAGCGACACCCA-3′ and antisense,
5′-CACCCTGTTGCTGTAGCCAAA-3′; LRRC10B sense, 5′-AAGCCACCGTGCCTCCA-3′
and antisense, 5′-TCCCTCGTCCCGTTATTGC-3′. Total RNA (2 mg) was
transcribed to cDNA. PCR was performed in a total reaction volume
of 20 µl, including 10 µl of SYBR Premix Ex Taq (2X),
2 µl of cDNA template, 1 µl of PCR forward primer (10
mM), 1 µl of PCR reverse primer (10 mM), and 6 µl of
double-distilled water. The quantitative real-time PCR reaction
included an initial denaturation step of 10 min at 95°C; 40 cycles
of 5 sec at 95°C, 30 sec at 60°C; and a final extension step of 5
min at 72°C. All experiments were performed in triplicate, and all
samples were normalized to GAPDH. The median in each triplicate was
used to calculate relative lncRNA concentrations (ΔCt = Ct median
lncRNA - Ct median GAPDH), and 2−ΔΔCt in expression was
calculated (17).
Cell culture
Five human LAD cell lines (SPCA-1, NCI-H1299, A549,
NCI-H441, LTEP-a2) were all purchased from the Cell Bank of the
Chinese Academy of Sciences and were cultured with complete medium
(containing 10% fetal serum and 90% RPMI-1640) set at 37°C, 5%
CO2 and complete medium was changed at least once every
two days.
Lentivirus-mediated overexpression vector
transfection
A549 cells were transfected overexpression vector
targeting RPLP0P2 as well as a negative control (GeneChem,
Shanghai, China). Transfection was accomplished by seeding
2x105 cells into a 6-well plate, and after 24 h, the
medium was aspirated and incubated with transfection complex
according to the manufacturer's instructions and MOI values
(MOI=20). The A549 cells were infected by lentivirus for 72 h and
the overexpression efficiency was detected by qPCR.
Cell migration and invasion assays
Migration and invasion assay was performed with
8.0-µm pore inserts (Millipore, USA) in a 24-well plate. For
migration assay, 2x104 cells were seeded into the upper
compartment of the Transwell inserts. The invasion assay was
performed with Matrigel-coated filters (Sigma Corp., USA). Cells
were allowed to incubate for 24 and 48 h, respectively. Migrated
and invaded were fixed by methanol and stained by 0.1 % (w/v)
crystal violet, then bleached with 33% acetic acid and absorbance
value measured at 570 nm on a microplate reader. Each experiment
was performed in triplicate.
Cell viability assay
Cell viability was evaluated by Cell Counting kit-8
(CCK-8; Corning, Inc., USA) abiding by the manufacturer's
instructions. Briefly, 3,000 cells were resuspended and seeded into
a 96-well plate supplemented in the presence of 10% FBS and
cultured for a week. The next day, the RPLP0P2 overexpression cells
were incubated with CCK-8 for 1 h and the absorbance was measured
at 450 nm using a multifunctional microplate reader (Tecan) at day
1, 3, 5 and 7. This experiment was done in quadruplicate cells.
Cell cycle assay
The cells were harvested by centrifugation and fixed
by 70% ethanol at 4°C overnight. The cells were resuspended with
400 µl PBS (containing 2 mg/ml RNA enzymes) and incubated at
37°C for 30 min, then added 400 µl propidium iodide (0.1
mg/ml) for 10 min and detected DNA content by a flow cytometry
analyzer (Cytomics FC 500; Beckman Coulter). The results were
analyzed using MultiCycle software.
Adhesion assay
The 96-well plates were processed with 50 µl
FN (50 µg/ml), and no processed wells were the CON group.
These wells were added into 2x104 cells/well and stained
by 0.1% (w/v) crystal violet, then dissolved with 2% SDS and
detected at OD550 nm. This experiment was done in quadruplicate
cells.
Statistical methods
Differences in variables among groups were tested
using the one-way ANOVA for the normal distribution or
Kruskal-Wallis test for the non-normal distribution. A comparison
between the two groups was performed by least significant
difference (LSD) test or Student's t-test or Mann-Whitney U test.
Survival analysis was performed using Chi-square test. P<0.05
was considered to be statistically significant.
Results
The expression level of RPLP0P2 in lung
cancer and adjacent tissues and analysis of its relationship with
clinical data
According to Table
I, RPLP0P2 expression level of LAD is 0.287 (0.131–2.96) and
significantly lower than its adjacent cancer tissues (Mann-Whitney
U =2.120, P=0.0029). We showed that the RPLP0P2 level of LAD with
lymph node metastasis was significantly lower than that of LAD
without lymph node metastasis group (Mann-Whitney U=9.102,
P=0.011). RPLP0P2 expression levels among different clinical stages
were not different (Kruskal-Wallis test =7.124, P=0.154). The
expression of RPLP0P2 was not relative to the histology
differentiation (Kruskal-Wallis test=3.235, P=0.676), smoking
(Mann-Whitney U=321.00, P=0.165), or gender (Mann-Whitney U=287.00,
P=0.423). LRRC10B mRNA expression level of LAD was significantly
higher than its adjacent cancer tissues (Mann-Whitney U=1.530,
P=0.000). Pearson correlation analysis showed that RPLP0P2
expression levels were negatively correlated to LRRC10B mRNA levels
(Pearson correlation =−0.754, P=0.0021) (Fig. 1).
RPLP0P2 expression in lung cancer
prognosis
The overall survival time of LAD low expression
RPLP0P2 group (median 10 months) was significantly lower than that
of the high expression (median 26 months) (χ2=18.81,
P<0.0001) (Fig. 2).
The expression level of RPLP0P2 fom five
LAD cells
Compared to normal human bronchial epithelial
BEAS-2B cell line, we detected the expression levels of RPLP0P2
from five LAD cell lines (including A549, NCI-H441, NCI-H1299,
SPCA-1, LETP-a2) by qPCR. It was shown that the expression levels
of RPLP0P2 from LETP-a2, SPCA-1, NCI-H441 cells were highly
expressed wherein LETP-a2 was the highest and that of SPCA-1,
NCI-H441 were moderately expressed, while that of A549 and
NCI-H1299 cells the lowest (Fig.
3). Therefore, A549 cells were lentivirus-mediated transfection
RPLP0P2 overexpression cells.
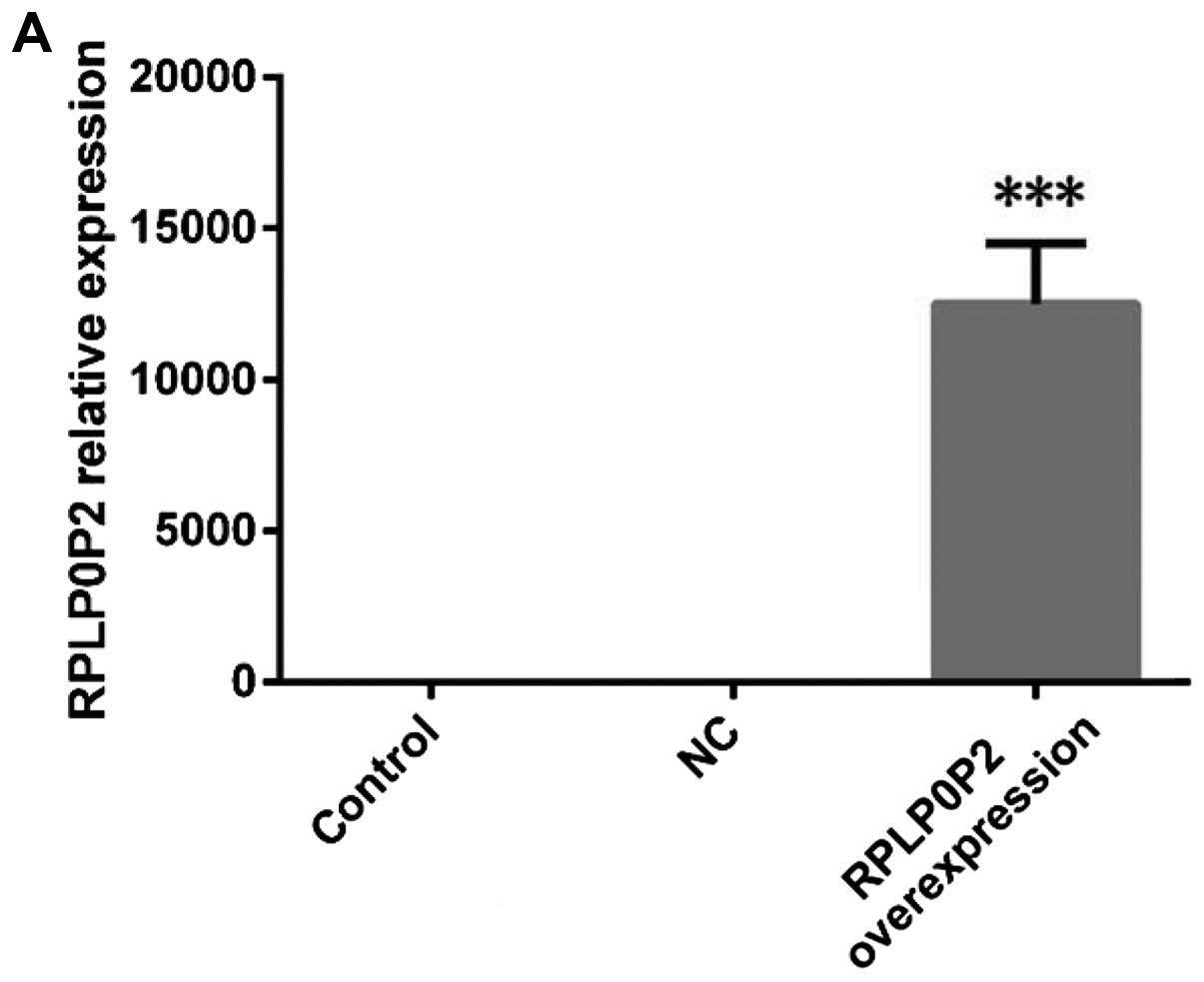
The expression levels of RPLP0P2 and
LRRC10B in three A549 cell groups
The expression levels of RPLP0P2 in three A549 cell
groups were different (F=117.00, P<0.0001) and that of
overexpression A549 cells was higher than that of the NC group
(t=10.34, P=0.0005), control group (t=10.81, P=0.0004) (Fig. 4). The LRRC10B mRNA expression levels
of three A549 cell groups were different (F=29.11, P=0.0008) and
that of RPLP0P2 overexpression A549 cells was lower than that of NC
group (t=5.909, P=0.0042), or control group (t=6.056, P=0.0037).
While RPLP0P2 expression levels were not changed significantly
after LRRC10B siRNA (F=0.4489, P=0.6582). These experiments hinted
that LRRC10B may be a downstream gene regulated by RPLP0P2.
RPLP0P2 is not related to cell migration
and invasion
The OD570 value of three A549 cell groups was not
different (F=1.262, P=0.3488), the OD570 value of RPLP0P2
overexpression A549 cells was similar to that of NC group (t=1.715,
P=0.153), and control group (t=0.7292, P=0.506) (Fig. 5). Thus, the cell migration ability
of A549 cells did not change after RPLP0P2 was overexpressed.
According to Fig. 6, the OD570
value of the three A549 cell groups was not different (F=0.9129,
P=0.4507) the OD570 value of RPLP0P2 overexpression A549 cells was
similar to that of NC group (t=0.9879, P=0.3791), and control group
(t=1.196, P=0.2979).
RPLP0P2 expression level is associated
with cell proliferation and adhesion
Fig. 7 shows the
OD450 nm of different A549 groups gradually increased with the
change of time. Compared with day 1, the OD450 nm of day 3
(P<0.05, P<0.05, P<0.05), day 5 (P<0.001, P<0.001,
P<0.01), day 7 (P<0.001, P<0.001, P<0.001) were
significantly increased. Compared with appropriate days of control
and NC group, the OD450 nm of 1 and 3 days in RPLP0P2
overexpression group had no statistically significant difference
(P>0.05), while that of the 5 days (P<0.05) and the 7 days
(P<0.01) significantly reduced, it indicates that cell
proliferation ability of A549 was significantly reduced after
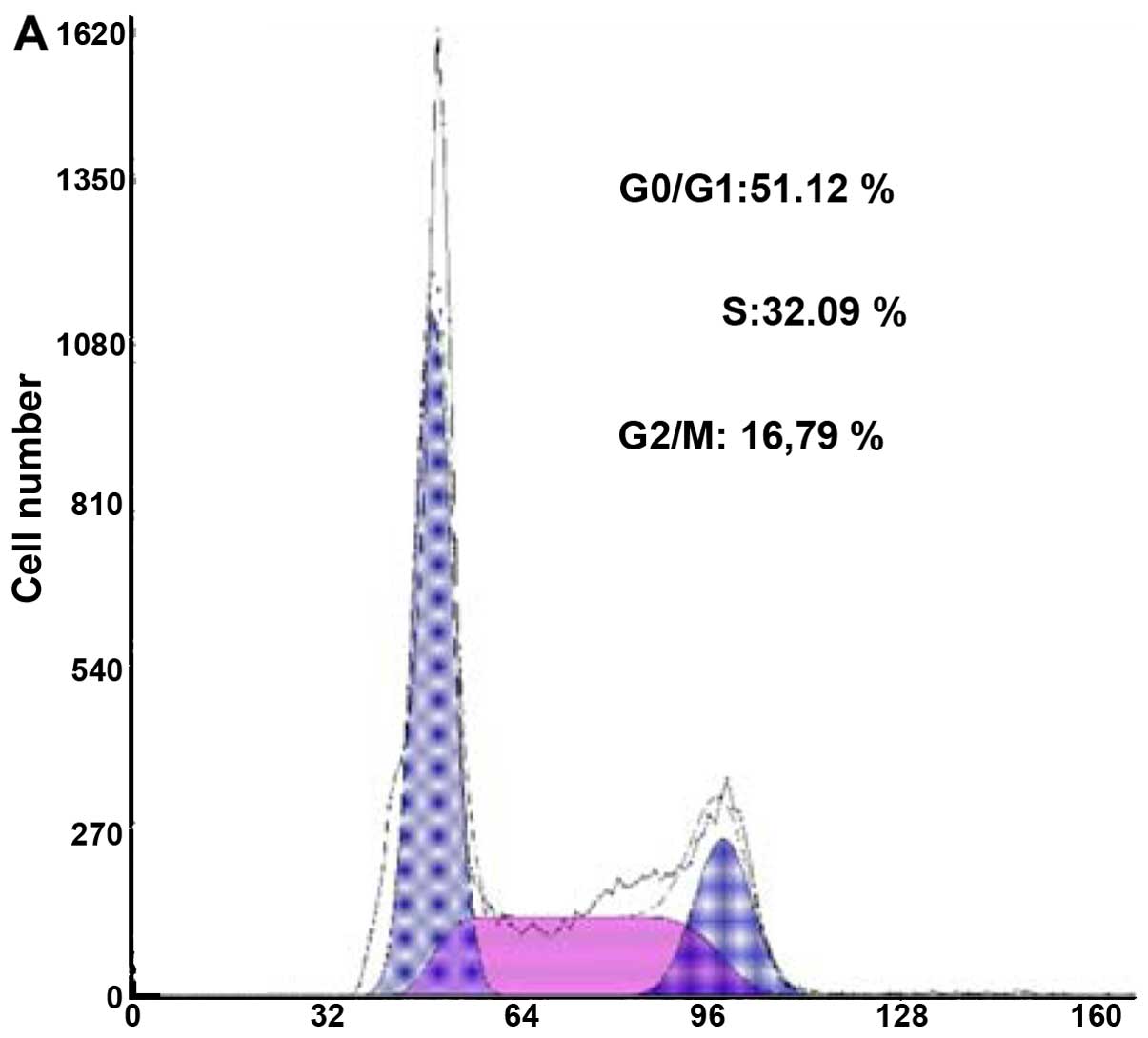
RPLP0P2 overexpression. Fig. 8
shows that after RPLP0P2 was overexpressed, S phase (P=0.0002 and
P=0.0001) and G2/M phase cells (P=0.0004 and P=0.0006) of A549
cells significantly reduced, while apoptosis and G0/G1 phase cells
(P=0.0003 and P=0.0007) obviously increased compared to control and
NC groups. The cell cycle results further confirmed that the
RPLP0P2 expression level was associated with the ability of cell
proliferation. The OD550 of control group (P<0.05), NC group
(P<0.05) were higher than RPLP0P2 overexpression group among CON
group in Fig. 9. Compared to CON
group, the OD550 of control group, NC group from FN processing
significantly increased while that of RPLP0P2 overexpression group
was not different. Therefore, it was shown that RPLP0P2 lowered the
adhesion capacity of A549 cells after overexpression.
 | Figure 7The cell proliferation results of
different A549 groups. (A) The different OD values on days 1, 3, 5
and 7. (B) The OD450 nm line boxplots of different A549 groups at
four time points. It shows that the OD450 nm of different A549
groups gradually increased over time. Compared with day 1, the
OD450 nm of days 3, 5 and 7 were significantly increased. Compared
with appropriate days of control and NC group, the OD450 nm of days
1 and 3 in the RPLP0P2 overexpression group had no statistically
significant difference, while that of days 5 and 7 was
significantly reduced. *P<0.05,
**P<0.01, ***P<0.001,
#P<0.05, ##P<0.01. |
Discussion
LncRNAs play an important role in many biological
processes, including X chromosome inactivation, gene imprinting
(18,19) and also control gene expression and
accelerate the development and progression in cancers (8,20).
Promoters bind to many transcription factors with mechanisms such
as chromosomal rearrangements and transfer elements (21). An important function of lncRNAs can
change the expression of nearby encoding genes by affecting the
process of transcription (22) or
directly playing an enhancer-like role (23,24).
Our bioinformation analysis showed that LRRC10B might be a target
gene regulated by RPLP0P2.
In this study, we uncovered the potential role of
RPLP0P2 in the pathogenesis of LAD. We found that RPLP0P2 was lower
expressed in LAD by qPCR. The expression of RPLP0P2 in lymph node
metastasis of LAD group was significantly lower than LAD without
lymph node metastasis group, while it was no relative to TNM stage,
degree of tissue differentiation, gender, age, or smoking. Survival
time of high expression RPLP0P2 was significantly longer than low
RPLP0P2 level in LAD patients, while LRRC10B mRNA level was higher
in LAD than NT by qPC. RPLP0P2 expression level negatively
correlated to LRRC10B mRNA level. These results hinted that RPLP0P2
is a tumor suppressor and abnormally expressed in LAD.
Compared to normal human bronchial epithelial
BEAS-2B cell line, we detected the expression levels of RPLP0P2
from five LAD cell lines. It was shown that the expression levels
of RPLP0P2 were highly expressed in LETP-a2, SPCA-1 and NCI-H441,
while in A549 and NCI-H1299 cells lowly expressed. In order to
further study the mechanism of RPLP0P2 we established RPLP0P2
overexpression of A549 cell line by lentivirus-mediated technology.
After RPLP0P2 was overexpressed, the proliferation rate, adhesion
ability, S and G2/M phase cells and LRRC10B mRNA significantly
reduced, while apoptosis and G0/G1 phase cells obviously increased,
but migration ability and invasion did not significantly
change.
To summarize, our study ascertained that the
expression of RPLP0P2 is downregulated in LAD and is associated
with poor prognosis and decreased proliferation and adhesion
ability of tumor cells. LRRC10B may be a downstream gene regulated
by RPLP0P2.
Acknowledgments
This study was financially supported by the National
Natural Science Foundation of China (8140736), the Zhejiang
Provincial Health Department (2014KYA133), and the Zhejiang
Provincial Natural Science Foundation (LQ16H160020).
References
|
1
|
Jemal A, Murray T, Ward E, Samuels A,
Tiwari RC, Ghafoor A, Feuer EJ and Thun MJ: Cancer statistics,
2005. CA Cancer J Clin. 55:10–30. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gridelli C, Rossi A and Maione P:
Treatment of non-small-cell lung cancer: State of the art and
development of new biologic agents. Oncogene. 22:6629–6638. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stewart DJ: Tumor and host factors that
may limit efficacy of chemotherapy in non-small cell and small cell
lung cancer. Crit Rev Oncol Hematol. 75:173–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen CH, Lai JM, Chou TY, Chen CY, Su LJ,
Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al: VEGFA
upregulates FLJ10540 and modulates migration and invasion of lung
cancer via PI3K/AKT pathway. PLoS One. 4:e50522009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ogawa E, Takenaka K, Katakura H, Adachi M,
Otake Y, Toda Y, Kotani H, Manabe T, Wada H and Tanaka F:
Perimembrane Aurora-A expression is a significant prognostic factor
in correlation with proliferative activity in non-small-cell lung
cancer (NSCLC). Ann Surg Oncol. 15:547–554. 2008. View Article : Google Scholar :
|
|
6
|
Rachet B, Woods LM, Mitry E, Riga M,
Cooper N, Quinn MJ, Steward J, Brenner H, Estève J, Sullivan R, et
al: Cancer survival in England and Wales at the end of the 20th
century. Br J Cancer. 99(Suppl 1): S2–S10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for long
non-coding RNA-associated diseases. Nucleic Acids Res. 41(D1):
D983–D986. 2013. View Article : Google Scholar
|
|
12
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thai P, Statt S, Chen CH, Liang E,
Campbell C and Wu R: Characterization of a novel long noncoding
RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer
cell lines. Am J Respir Cell Mol Biol. 49:204–211. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X,
Cui Z, Zhang J, Yi K, Xu W, et al: RNA-seq analysis of prostate
cancer in the Chinese population identifies recurrent gene fusions,
cancer-associated long noncoding RNAs and aberrant alternative
splicings. Cell Res. 22:806–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Drobnik W, Liebisch G, Biederer C, Tr
mbach B, Rogler G, Müller P and Schmitz G: Growth and cell cycle
abnormalities of fibroblasts from Tangier disease patients.
Arterioscler Thromb Vasc Biol. 19:28–38. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khachane AN and Harrison PM: Mining
mammalian transcript data for functional long non-coding RNAs. PLoS
One. 5:e103162010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W,
Chen X, George J, Leong B, Liu J, et al: The Oct4 and Nanog
transcription network regulates pluripotency in mouse embryonic
stem cells. Nat Genet. 38:431–440. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mattick JS and Gagen MJ: The evolution of
controlled multi-tasked gene networks: The role of introns and
other noncoding RNAs in the development of complex organisms. Mol
Biol Evol. 18:1611–1630. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mattick JS: Lincing Long noncoding RNAs
and enhancer function. Dev Cell. 19:485–486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|