Introduction
Hepatocellular carcinoma (HCC) is one of the fatal
cancers threatening people worldwide, with over half of them being
Chinese. Late diagnosis of HCC leads to poor prognosis by
restricting the selection of treatment protocols. Therefore,
finding tumor markers for early diagnosis is crucial (1,2).
Although AFP is an important marker of detecting and monitoring
HCC, its individual detection in small HCC patients has a 40%
chance of indicating false negative. Thus, it is imperative to find
other markers that are more sensitive and earlier detectable to
clarify the development mechanism of HCC and to improve therapeutic
effects and prognosis (3).
MicroRNA (miRNA), as a non-protein-coding RNA family
that can negatively regulate gene expression, can repress protein
translation or induce RNA degradation by interacting with mRNA.
miRNAs also function as effective small-molecule drugs. Mature
miRNA, which is a single-strand RNA molecule length of about 19025
nt, can inhibit the translation of target gene and participate in
cell differentiation, proliferation and apoptosis by binding 3′-UTR
of mRNA through complete or incomplete base pairing (4). The miRNA-101 expression profiles of
tumor cells and paracancerous tissues differ significantly, thus
being able to distinguish HCC (5),
breast carcinoma (6,7), lung carcinoma (8), pancreatic carcinoma (9) and leukemia (10) from adjoining normal tissues.
We have previously found that miR-101 was evidently
highly expressed in HCC patients, which is associated with the
unique regulatory effects of miRNA on phosphatase and tensin
homolog deleted on chromosome 10 (PTEN) reported by Karreth et
al (11,12). Although anti-miR-101 and PTEN have
previously been correlated, in vitro carcinogenesis
experiment has seldom been combined with clinical study results.
Thereby motivated, we explored the influence of miR-101 on the
onset of HCC as an oncogene and its direct effect on PTEN, aiming
to verify the results of Karreth et al, miRNA and gene
expression profiles in tumor and paracancerous tissues were
detected. The correlation between differentiated anti-miR-101
expressions and malignant behavior of HCC, prognosis, as well as
total and recurrence-free survival rates were summarized. The
target gene corresponding to miR-101 was determined to
preliminarily postulate the role of miR-101 in the onset and
development of HCC. The results indicate anti-miR-101 is a feasible
marker for the diagnosis and prognosis evaluation of HCC.
Materials and methods
Compliance with ethical requirements
Written consent was received from all the patients.
All information and sample collection as well as experimental
protocols accorded with the 'Ethics Committee Standard Operating
Procedures (SOP)', and all the experiments have been approved by
the Ethics Committee of our hospital.
General information and tissue
samples
Seventy-eight HCC patients who received consecutive
hepatectomies in our hospital from January 2009 to June 2015 were
selected. Inclusion criteria: i) Isolated or multiple tumors (no
more than 3); ii) without portal vein, hepatic vein or postcava
tumor thrombi; iii) without extrahepatic metastasis; iv) Child-Pugh
scores ranging between 5 and 7; and v) first visit patients who had
not received any transcatheter arterial chemoembolization, local
treatment or radiotherapy (13,14).
Tumor and the corresponding paracancerous tissues were subjected to
RNA later treatment after resection, archived and stored in tissue
bank, and stored at −80°C. No tumor tissues succumbed to necrosis,
and normal hepatic tissues located 3 cm from the edge of tumor
tissues were used as paracancerous tissues. Tissues of each case
were sampled in triplicate to prevent potential contamination.
Clinical data were collected from the medical records, and no
patients had received chemotherapy prior to the surgeries. Written
consent was received from all the patients. All of them were
diagnosed as HCC, as confirmed by H&E staining.
The patients aged 48–82 years (median, 53 years)
included 63 males (80.8%) and 15 females (19.2%). There were 61
serum HBsAg-positive cases (78.2%), and serum AFP levels were
0.78-115600 µg/l (median, 230 pg/l). Fifty-eight cases were
complicated with cirrhosis (74.4%), and the others were not
(25.6%). Maximum tumors were sized 1.4–15.4 cm (median, 7.3 cm).
Tumor differentiation degrees were determined according to
Edmondson-Steiner grading standard (40 cases of grade I and II, 38
cases of grade III and IV) (15).
The patients were classified into 37 cases of stage I, 24 cases of
stage II and 17 cases of stage III according to the TNM staging
standard stipulated by UICC and AJCC (16). They were last followed-up on 30th
June, 2015.
Cell lines
Human hepatocarcinoma cell lines HEPG2, HEP3B,
Sk-Hepl, Huh7, SMMC-772l, PLC, MHCC-97H and MHCC-97L were purchased
from Zhongyuan Union Stem Cell Bioengineering Co. (Zhejiang,
China). Immortalized hepatocytes L-02 and human kidney epithelial
cells 293T were provided by the Molecular Biotechnology Center of
our hospital.
Main reagents and solutions
miRNA expression vector anti-miR-101 (inhibitor),
pre-miR-101 (mimics) and control miR-NC were purchased from Ambion
(Carlsbad, CA, USA). Wild-type pcDNA 3.1-PTEN-WT and mutant pcDNA
3.1-PTEN-MU plasmids were purchased from GeneCopoeia, Inc.
(Rockville, MD, USA). Luciferase reporter assay vector pGL3M,
pcDNA3.1 plasmid, EcoRI and XbaI were obtained from
Promega Corp. (Madison, WI, USA). Lipofectamine™ 2000 liposome,
E. coli strain DH5α and quantitative PCR kit
Platinum® SYBR® Green qPCR SuperMix-UDG
reagent were purchased from Invitrogen (Carlsbad, CA, USA).
Dual-luciferase reporter assay kit Dual-Light® system
(P/N T1003) was obtained from Applied Biosystems Life Technologies
(Foster City, CA, USA). B-type ultrapure plasmid extraction kit was
bought from BioDev-Tech Co. (Beijing, China). Restriction
endonuclease HindIII and SpeI, as well as DNA T4
ligase were purchased from New England BioLabs, Inc. (Ipswich, MA,
USA). The First Strand cDNA Synthesis kit was purchased from
Fermentas (Waltham, MA, USA). Western blotting-related polyclonal
antibodies were obtained from Cell Signaling Technology, Inc.
(Danvers, MA, USA). Other reagents were purchased from
Sigma-Aldrich (St. Louis, MO, USA) or Sinopharm Chemical Reagent
Co., Ltd. (Shanghai, China) (analytically pure). Primers were
designed and synthesized by Shanghai Sangon Biological Engineering
Technology and Services Co., Ltd. (Shanghai, China).
Cell culture
All cell lines were cultured in high-glucose
Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal
bovine serum, and were incubated in a 5% CO2 incubator
at 37°C with saturated humidity. After being digested with 0.25%
trypsin, the cells growing to 80–90% confluence were then passaged.
The cells growing logarithmically were finally used (17).
Total RNA extraction
Total RNA was extracted by TRIzol reagent according
to the manufacturer's instructions. The tumor and paracancerous
tissues of 9 cases were compared by digital miRNA expression
profiling. They were then classified into stage I (4 cases), stage
II (3 cases) and stage III (2 cases) (16), including 7 males and 2 females.
Culture medium was discarded after cells fully adhered to the
bottom, to which 1 ml of TRIzol reagent was added per
0.5−1×107 cells. The following procedure was the same as
that of tissue RNA extraction (18).
Gene and digital miRNA expression
profiling
Illumina HiSeq™ 2000 high-throughput small-RNA
sequencing and synthesis were performed simultaneously, aiming to
minimize the secondary structure-induced local missing. The
screened high-quality sequences were classified and annotated to
obtain information on components and expression levels. All miRNA
segments were annotated, and target gene prediction was conducted
for new miRNA (19,20).
Detection of hsa-miR-101 expression
levels before and after transfecting miRNA precursor by
qRT-PCR
miRNA precursor was transfected by the RNA transient
transfection method according to the instruction of Lipofectamine™
2000 (Invitrogen). Diluted anti-miR-101, pre-miR-101 and miR-NC
were mixed with liposomes in culture medium. Total RNA was
extracted by mirVana™ miRNA isolation kit 24 and 48 h later to
measure miR-101 expression levels. After RNA purification, cDNA was
subjected to reverse transcription with the reaction systems and
primer sequences reported previously (21–25).
Primer sequences: miR-101 upstream, 5′-TCACACTATATCACATTGCCAGG-3′
and downstream, 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′; CXCL12 upstream,
5′-CAGTCAACCTGGGCAAAGCC-3′ and downstream,
5′-CCTGAGAGTCCTTTTGCGGG-3′; IL6R upstream,
5′-GGCAACCGAGCAAGACTCTC-3′ and downstream,
5′-GCGAGGACAGAAGATTTG-3′; FOXO3 upstream, 5′-CTTAAGGATAAGGGCGACA-3′
and downstream, 5′-CGACTATGCAGTGACAGGTTG-3′; PTEN upstream, 5′ -
AGACAGATCTGTGGGGTGCGGGGTAGGAGT-3′ and downstream,
5′-AGACAAGCTTGACGAAGAGGAGGCGAGA-3′; internal reference U6 upstream,
5′-ATTGGAACGATACAGAGAAGATT-3′ and downstream,
5′-GGAACGCTTCACGAATTTG-3′; internal reference GAPDH upstream,
5′-GTCAGTGGTGGACCTGACCT-3′ and downstream,
5′-TGAGGAGGGGAGATFCAGTG-3′.
Western blotting
Cells collected at different time intervals were
lysed, from which cytoplasm and nucleus protein supernatants were
extracted for SDS-PAGE at 120 V for 2 h. Then the products were
electrotransferred onto PVDF membrane and put in TBST blocking
buffer containing 5% defatted milk at room temperature for 1 h.
After TBST washing, primary antibody was added, and then the
membrane was shaken at 4°C overnight. After several times of TBST
washing, the membrane was shaken at room temperature for 1 h after
adding rabbit anti-human polyclonal antibody, washed three times
and colored by ECL (26).
Plasmid transfection and luciferase
reporter assay
Empty vector pcDNA3.1 and luciferase reporter vector
pGL3M, as well as recombinant reporter vector pcDNA 3.1-PTEN-WT (or
MUT/NC) were transfected with competent host bacteria DH5a.
Plasmids were extracted according to the manufacturer's
instructions, identified by XbaI and EcoRI double
digestion, and sequenced by Hohhot Mole Chemical Reagent Co., Ltd.
(Hohhot, China) (27).
With the transfection method mediated by the
Lipofectamine 2000 liposome, 293T cells were transfected with
pGL3M-PTEN-3′UTR (50 ng) and control plasmid pcDNA3.1 (10 ng), and
referred to as experimental group pGL3M-WT-PTEN-3′UTR, positive
control group pGL3M-MUT-PTEN-3′UTR and negative control group
pGL3M. Dual-luciferase reporter assay results were expressed as the
ratio of firefly luciferase activity to Renilla luciferase
activity.
Statistical analysis
Digital gene and miRNA expression profiles were
analyzed by non-monitoring clustering with SAM and TIGR Multiple
Array Viewer software package (TMeV version 4.0). Real-time
fluorescent quantitative PCR was performed by sequence detection
system (SDS) 2.3, and miRNA expression levels were expressed as ΔCt
values (Ct-miRNA-Ct U6). Biological data were expressed as mean ±
SD, and the differences between two groups were compared by
Student's t-test. Numeration data were compared by Chi-square test
or Fisher's exact test. Survival time was calculated by month, and
overall survival time was calculated by that from surgery to death
or last follow-up. Survival rate was calculated by Kaplan-Meier
method. Single variables were subjected to log-rank test, and
significant variables were analyzed by introducing the Cox's model.
P<0.05 was considered statistically significant. Data were
analyzed by SPSS 15.0.
Results
Correlation between miR-101 expression in
HCC tissues, clinical pathological factors and prognosis
The miRNAs with significantly changed expressions
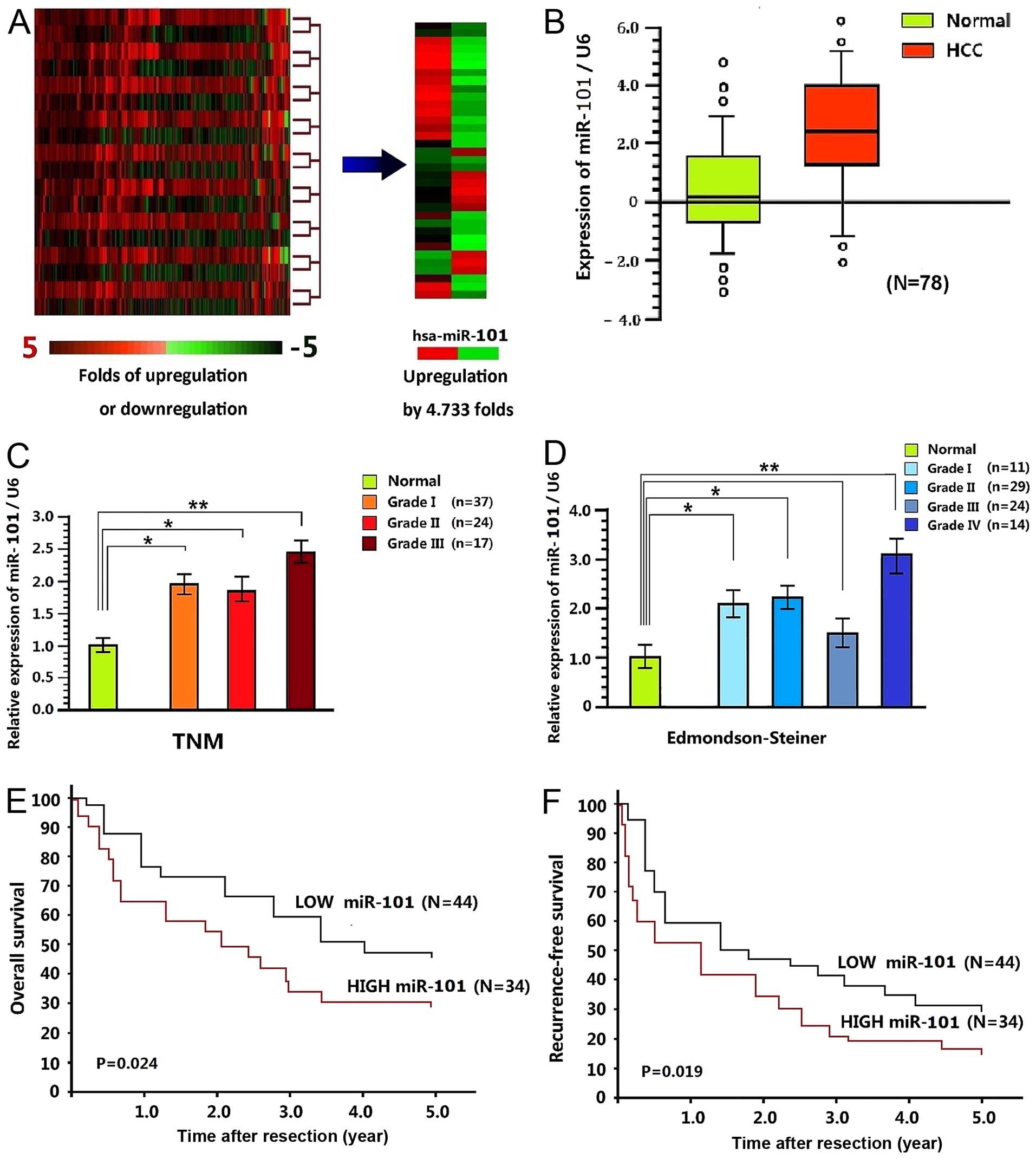
were screened by digital miRNA expression profiling (Table I). Obviously, the miR-101 expression
was significantly upregulated in tumor tissues (Fig. 1A), with the medians of paracancerous
and tumor tissues as 0.18 and 2.47, respectively (P<0.001)
(Fig. 1B). Of the 78 tumor tissues,
there were 37 cases of stage I, 24 cases of stage II and 17 cases
of stage III based on the TNM grading standard, with their miR-101
expression all higher than those in the corresponding paracancerous
tissues (miR-101 expressions were upregulated by 1.96-fold,
1.88-fold and 2.47-fold in the three stages, respectively).
Particularly, the expression levels in stage II and III differed
significantly between the two groups. According to the
Edmondson-Steiner grading standard, there were 11 cases of stage I,
29 cases of stage II, 24 cases of stage II and 14 cases of stage
IV, of which miR-101 expression was upregulated by 2.15-fold,
2.28-fold, 1.53-fold and 3.14-fold (Fig. 1D).
 | Table IDigital miRNA expression profiling
results. |
Table I
Digital miRNA expression profiling
results.
Upregulated miRNAs
| Downregulated
miRNAs
|
|---|
| miRNA | Difference
fold | q-value (%) | miRNA | Difference
fold | q-value (%) |
|---|
| hsa-miR-101 | 4.733 | 0 | hsa-miR-486 | 0.269 | 0 |
| hsa-miR-30e | 4.265 | 0 |
hsa-miRPlus-E1012 | 0.304 | 0 |
| hsa-miR-221 | 4.038 | 0 | hsa-miR-223 | 0.366 | 0 |
| hsa-miR-23a | 3.726 | 0 | hsa-miR-766 | 0.396 | 0 |
| hsa-miR-16 | 3.714 | 0 | hsa-miR-608 | 0.405 | 0 |
| hsa-miR-30a | 3.284 | 0 |
hsa-miRPlUS-El120 | 0.417 | 0 |
| hsa-miR-21 | 3.197 | 0 | hsa-miR-129 | 0.433 | 0 |
| hsa-miR-15a | 3.006 | 0 |
hsa-miRPlus-A1027 | 0.451 | 0 |
| hsa-miR-24 | 2.994 | 0 | hsa-miR-625 | 0.470 | 0 |
| hsa-miR-195 | 2.754 | 0 |
hsa-miRPlus-F1205 | 0.476 | 0 |
| hsa-miR-200a | 2.732 | 0 | hsa-miR-665 | 0.514 | 0 |
| hsa-miR-125a | 2.525 | 0 | hsa-miR-767-3p | 0.548 | 0 |
| hsa-miR-222 | 2.471 | 0 |
hsa-miRPlus-F1208 | 0.557 | 0 |
| hsa-miR-205 | 2.378 | 0 | hsa-miR-634 | 0.565 | 0 |
| hsa-miR-30b | 2.236 | 0 |
hsa-miRPlus-E124 | 0.594 | 0 |
| hsa-miR-378 | 1.941 | 0 |
hsa-miRPlus-18b | 0.608 | 0 |
| hsa-miR-27a | 1.879 | 0 | | | |
| hsa-miR-142-3p | 1.677 | 0 | | | |
| hsa-miR-497 | 1.524 | 0 | | | |
The correlations between miR-101 expression and
clinical pathological factors, including gender, age, cirrhosis
history, HBsAg state, serum AFP level, Child-Pugh grade, vascular
invasion status, tumor size, tumor number, tumor capsule, tumor
differentiation degree (Edmondson-Steiner grade) and TNM stage,
were analyzed. The 78 patients were divided into a low miR-101
expression group and a high miR-101 expression group based on the
average upregulation fold (2.47). First, univariate analysis and
log-rank test were performed. High miR-101 expression (P=0.019),
vascular invasion (P<0.001), tumor size ≥7 cm (P= 0.023), tumor
capsule (P= 0.035) and late stage (TNM II–III) (P=0.035) were the
risk factors of overall survival rate. Besides, high miR-101
expression (P=0.024), vascular invasion (P<0.001), tumor size ≥7
cm (P<0.001) and late stage (TNM II–III) (P=0.001) were the risk
factors of recurrence-free survival rate (Table II). Multivariate analysis showed
that high miR-101 level (P= 0.041) and vascular invasion (P=0.043)
were the risk factors of overall survival rate. High miR-101
expression level (P<0.001), AFP ≥200 µg/l (P=0.032) and
Edmondson-Steiner III–IV stages (P=0.033) were the risk factors of
recurrence-free survival rate (Table
II).
 | Table IIUnivariate and multivariate analyses
of factors affecting survival rates. |
Table II
Univariate and multivariate analyses
of factors affecting survival rates.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
RFS
| OS
| RFS
| OS
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| miRNA-101 (high vs.
low)a | 2.231
(1.234–3.876) | 0.019a | 2.274
(1.187–3.922) | 0.024a | 1.984
(1.121–4.258) | 0.041a | 3.354
(1.521–6.633) | <0.001a |
| Gender (M vs.
F) | 1.159
(0.379–3.894) | 0.633 | 1.154
(0.538–3.640) | 0.622 | – | – | – | – |
| Age (≥50 vs. <50
years) | 1.305
(0.478–2.321) | 0.421 | 0.589
(0.377–1.946) | 0.279 | – | – | – | – |
| Cirrhosis (yes vs.
no) | 0.698
(0.323–1.515) | 0.657 | 0.682
(0.214–1.663) | 0.536 | – | – | – | – |
| HBsAg (positive vs.
negative) | 2.305
(0.825–4.634) | 0.093 | 2.450
(0.998–4.547) | 0.055 | – | – | – | – |
| AFP (≥200 vs.
<200 µg/l) | 1.599
(0.842–3.362) | 0.062 | 1.687
(1.118–3.637) | 0.052 | 1.451
(0.628–3.255) | 0.223 | 2.124
(1.068–3.622) | 0.032a |
| Child-Pugh (B vs.
A) | 1.854
(0.685–4.212) | 0.143 | 1.573
(0.653–3.862) | 0.082 | – | – | – | – |
| Vascular invasion
(now vs. past) | 3.174
(1.450–5.691) | <0.001a | 2.356
(1.039–5.022) | <0.001a | 1.271
(0.674–3.389) | 0.043a | 1.328
(0.598–2.725) | 0.256 |
| Tumor size (≥7 vs.
<7 cm) | 2.743
(1.182–5.744) | 0.023a | 3.459
(1.547–5.364) | <0.001a | 1.635
(0.725–4.527) | 0.075 | 2.307
(1.126–3.549) | 0.065 |
| Tumor no. (many vs.
few) | 1.377
(0.684–2.771) | 0.165 | 1.741
(1.169–2.822) | 0.073 | – | – | 1.556
(0.578–3.642) | 0.210 |
| Tumor capsule (now
vs. past) | 1.215
(0.385–3.032) | 0.035a | 1.106
(0.493–2.548) | 0.106 | – | – | – | – |
| Edmondson-Steiner
(III–IV vs. I–II) | 1.476
(0.586–3.261) | 0.069 | 1.651
(0.985–3.459) | 0.083 | 1.446
(0.825–3.341) | 0.185 | 1.520
(1.039–3.472) | 0.033a |
| TNM stage (II–III
vs. I) | 2.238
(1.235–4.097) | 0.035a | 2.564
(1.087–4.218) | <0.001a | 1.206
(0.673–2.360) | 0.123 | 0.794
(0.255–2.306) | 0.184 |
Cox multiple regression analysis showed that high
miR-101 expression level was the independent prognostic factor of
overall and recurrence-free survival rates of HCC patients [OS,
(HR, 2.274; 95% CI, 1.187–3.922); RFS, (HR, 2.231; 95% CI,
1.234–3.876)]. The 1-, 2-, 3-, 4- and 5-year overall survival rates
of low miR-101 expression group were 77.2, 74.3, 60.2%, 51.9 and
48.2%, respectively, and those of high miR-101 expression group
were 64.7, 55.0, 34.1, 30.6 and 30.6%, respectively (P=0.024)
(Fig. 1E). The 1-, 2-, 3-, 4- and
5-year recurrence-free survival rates of low miR-221 expression
group were 59.7, 47.2, 41.0, 34.9 and 32.5%, and those of the high
expression group were 52.3, 34.4, 20.4, 19.6 and 17.7% (P=0.019)
(Fig. 1F).
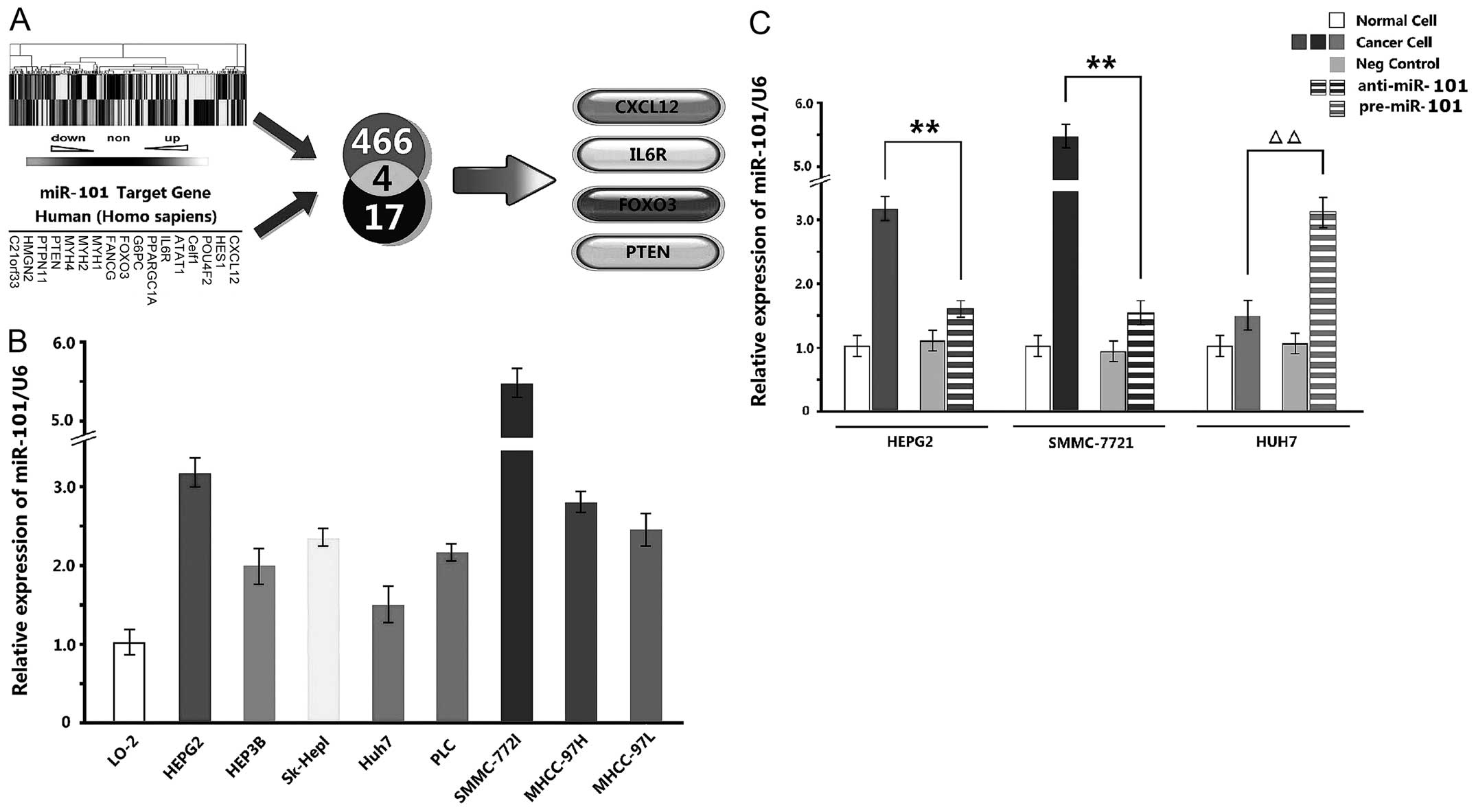
Screening of miR-101 target gene and
expression in different HCC cell lines
A total of 466 genes underwent significantly
different changes according to the results of digital gene
expression profiling, and CXCL12, HES1, POU4F2, Celf1, ATAT1, IL6R,
PPARGC1A, G6PC, FOXO3, FANCG, MYH1, MYH2, MYH4, PTEN, PTPN11, HMGN2
and C21orf33 were predicted as the target genes of miR-101 by
miRBase. Hence, CXCL12, IL6R, FOXO3 and PTEN were determined as the
desired genes based on the two tests (Fig. 2A).
qPCR detection exhibited that miR-101 expression
levels in 8 HCC cell lines were upregulated compared with that in
normal cell line L-02. Especially, the levels of HepG2 and
SMMC-7721 cells were upregulated maximally, while that of Huh7
cells was upregulated minimally (Fig.
2B). Therefore, miR-101 inhibitor was added in HepG2 and
SMMC-7721 cells, and miR-101 mimics were added in Huh7 cells.
Compared with NC and blank groups, the three kinds of cells
underwent significant changes after being transiently transfected
with inhibitor and mimics (P<0.01) (Fig. 2C).
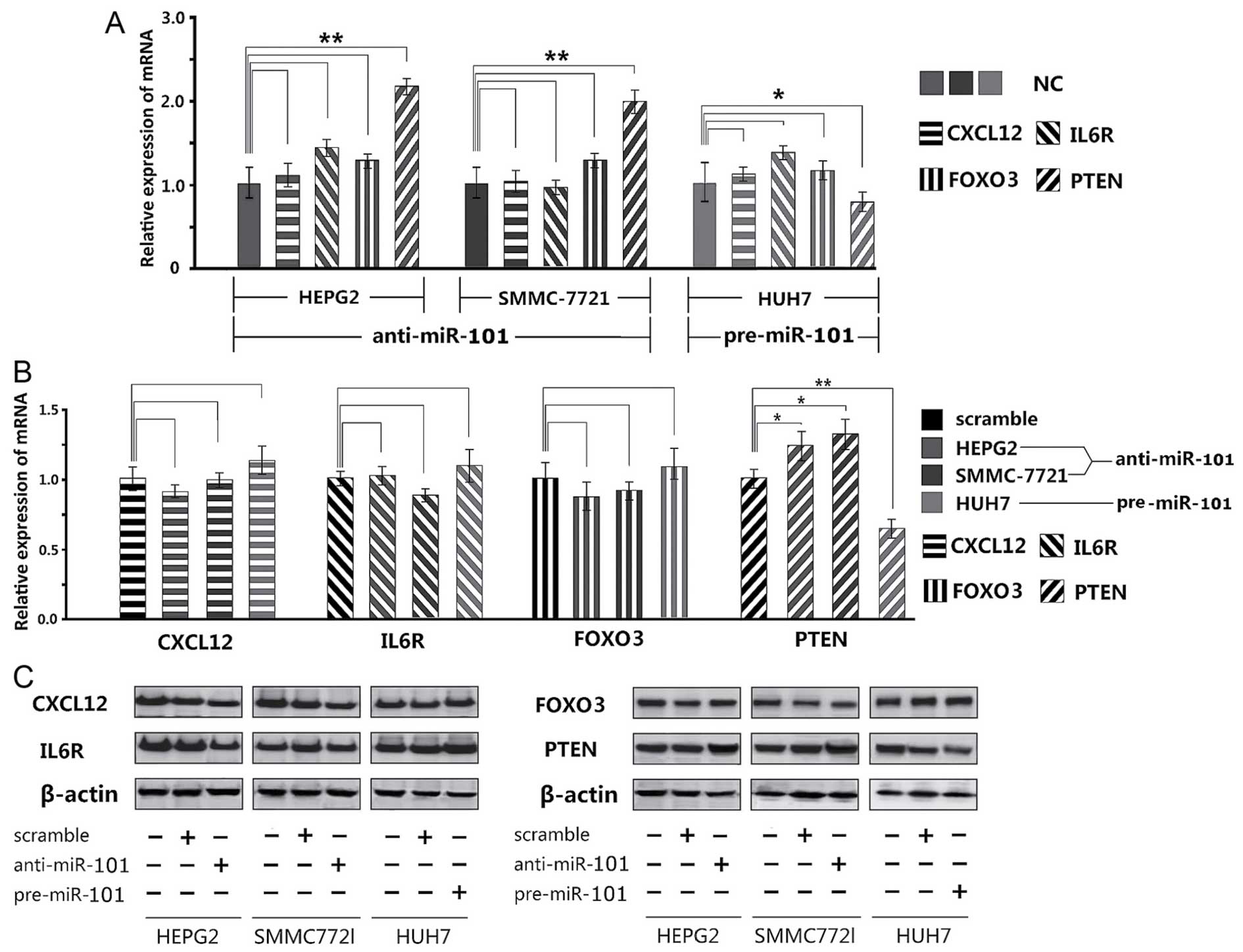
Determination of miR-101 desired genes by
qPCR and Western blotting
Of the 4 potential target genes, only PTEN gene was
expressed significantly differently, as suggested by qPCR results.
Adding miR-101 inhibitor significantly increased the PTEN
expressions in HepG2 and SMMC-7721 cells than that in the NC
control group, whereas adding miR-101 mimics led to significant
decrease in Huh7 cells (Fig. 3A).
In contrast, CXCL12, IL6R and FOXO3 did not experience the same
changes.
Western blotting also exhibited that only PTEN
protein was significantly upregulated (P<0.05) or downregulated
(P<0.01) (Fig. 3B and C).
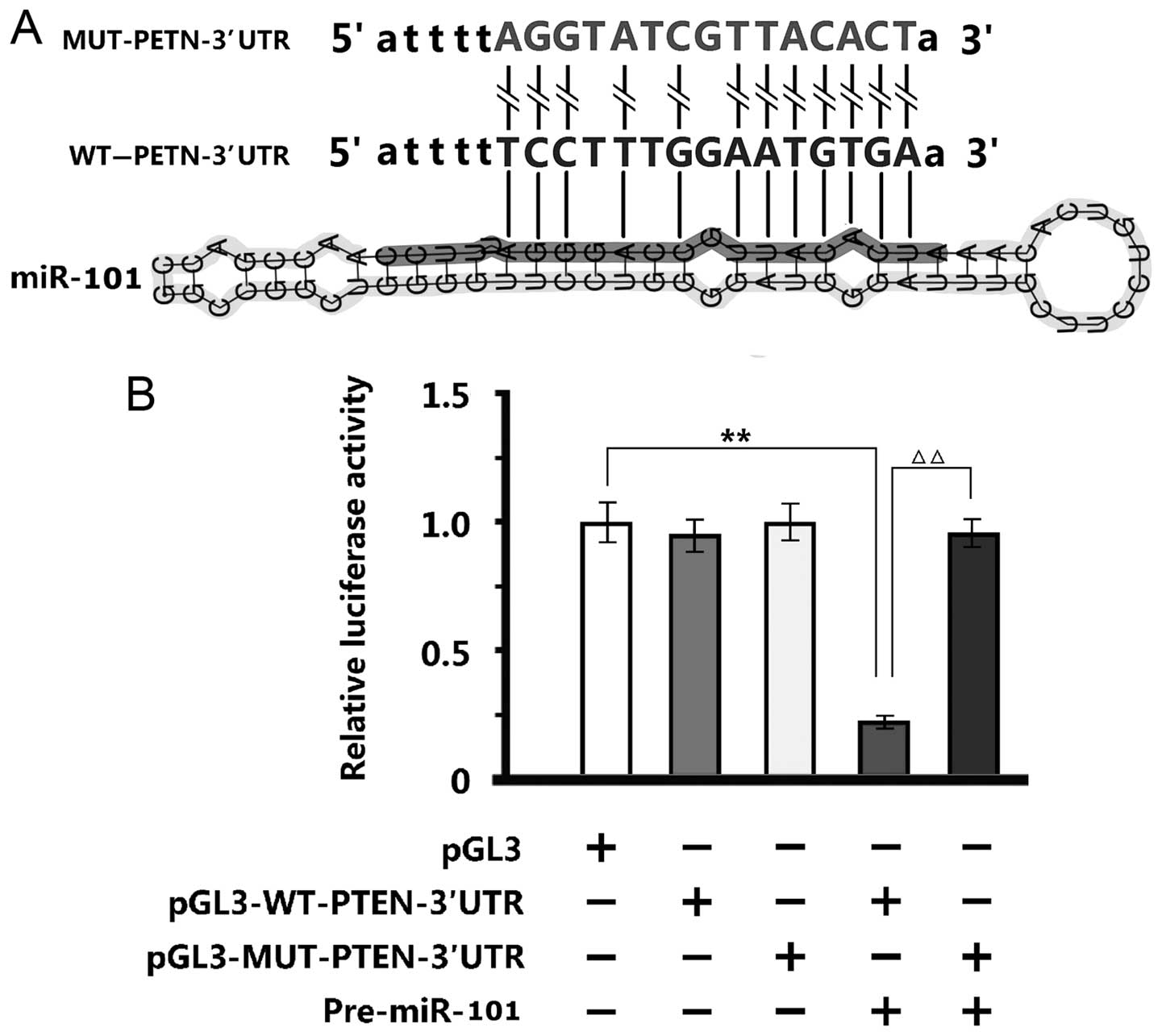
Verification of PTEN by luciferase
reporter assay
Twelve sequences of miR-101, which were identical to
the WT-PTEN-3′UTR end, were disrupted by the MUT-PTEN-3′UTR
(Fig. 4A). The luciferase reporter
assay of 293T cells showed that negative control
pGL3M-MUT-PTEN-3′UTR and pGL3M-WT-PTEN-3′UTR were similar to empty
plasmid pGL3M. Adding miR-101 mimics did not alter the activity of
MUT group significantly, but significantly reduced the fluorescent
intensity of WT group, indicating that miR-101 bound and changed
the specific sequences of WT-PTEN-3′UTR promoter (Fig. 4B).
Discussion
By using digital miRNA expression profiling, we
herein observed that miR-101 was significantly upregulated.
Besides, qPCR detection showed that miRNA expression in tumor
tissues was upregulated by over 2-fold. The expressions of miR-101
in SMMC-7721 and HepG2 cells were highly expressed, and that in
Huh7 cells was lowly expressed. Therefore, the three types of cells
were used (28).
Since miR-101 was ubiquitously highly expressed in
HCC cells, the results of digital miRNA expression profiling were
verified correctly. Moreover, correlation between miR-101
expression and clinical prognosis of HCC was also studied. Although
miR-101 has been correlated with the prognosis of malignant tumors
previously, it has rarely been correlated with that of HCC
(5). Upregulation of miR-101 was
the independent prognostic factor of overall and recurrence-free
survival rates. High miR-101 expression was correlated with poor
prognosis independent of tumor staging and other clinical
variables, suggesting that miR-101 is a promising molecular marker
for determining the prognosis of HCC. In this study, HCC
metastasis-associated miRNAs were correlated with recurrence and
survival rates, and the expression levels of most miRNAs were
correlated with survival time, being consistent with previous
studies (29–31).
Of the four candidate genes, only PTEN yielded
desirable results. Ever since 1997, PTEN, mutated in multiple
advanced cancers 1 (MMAC1) and TGF-β-regulated and epithelial
cell-enriched phosphatase (TEP1) were found (32,33)
which are actually the same gene that is referred to as PTEN now.
PTEN is one of the genes that are most prone to mutation, the
deletion and mutation of which are bound to induce malignant
glioma, prostate cancer and HCC (34,35).
As a highly conservative gene, PTEN affects many types of tumors by
bispecific lipase.
Anti-miR-101 regulates the transcription of PTEN
that influences the growth, apoptosis, proliferation and adhesion
of tumor cells by mainly regulating negatively the PDK/AKT
signaling pathway. Whereas, the PI3K/PKB/AKT signaling transduction
pathway mainly promotes cell proliferation (36). PTEN protein can antagonize the
effects of phosphatidylinositol PI3K by dephosphorylating the
substrate, and decreasing the contents of second messengers
phosphatidylinositol-3,4-diphosphate and phosphatidylinositol
(3,4,5)-triphosphate. Thus, the activity of
protein kinase can be maintained normal after negative regulation
of the PI3K/PKB/AKT signaling transduction pathway, thus regulating
cell proliferation, differentiation and apoptosis. Hence, this
pathway is able to suppress cancer (37), by which anti-miR-101 may exert
inhibitory effects. It has previously been reported that miR-101
may regulate cell proliferation, differentiation and apoptosis by
affecting mitogen-activated protein kinase (MAPK) via a certain
pathway (38).
As the first anti-oncogene with phosphatase
activity, PTEN inspires structural and functional studies for
potential malignant tumor treatment. Mutter et al diagnosed
early endometrial cancer by using PTEN as a marker (39). In 2011, RNA was associated with
cancer onset by manipulating the fate of molecules (12,13).
In case the dialogue between PTEN gene and miRNA is ruined,
unexpected disasters may occur. On the other hand, altering the
dialogue may be able to prevent and treat cancer (40). The RNA management network seems to
be applicable to non-protein-coding regions, thus playing an
essential role in maintaining normal muscle development.
Although the correlation between anti-miR-101 and
PTEN has been reported (40,41),
in vitro cell experiments and clinical studies have seldom
been referred simultaneously. We herein verified that high miR-101
expression may facilitate the proliferation of HCC cells and
miR-101 may participate in the onset of HCC as an oncogene.
Moreover, the impact of miR-101 on PTEN was also demonstrated,
being in accordance with the results of Karreth et al
(11). In conclusion, anti-miR-101
may function as a small-molecule antitumor agent for HCC, and this
study provides evidence for clarifying the molecular mechanism of
HCC onset.
References
|
1
|
Fang M, Zhao YP, Zhou FG, Lu LG, Qi P,
Wang H, Zhou K, Sun SH, Chen CY and Gao CF: N-glycan based models
improve diagnostic efficacies in hepatitis B virus-related
hepatocellular carcinoma. Int J Cancer. 127:148–159. 2010.
View Article : Google Scholar
|
|
2
|
Pang RW, Joh JW, Johnson PJ, Monden M,
Pawlik TM and Poon RT: Biology of hepatocellular carcinoma. Ann
Surg Oncol. 15:962–971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao Q, Ren P, Li Y, Peng B, Dai L, Lei N,
Yao W, Zhao G, Li L and Zhang J: Autoantibodies against
glucose-regulated protein 78 as serological diagnostic biomarkers
in hepatocellular carcinoma. Int J Oncol. 41:1061–1067.
2012.PubMed/NCBI
|
|
4
|
Wang L, Li B, Li L and Wang T:
MicroRNA-497 suppresses proliferation and induces apoptosis in
prostate cancer cells. Asian Pac J Cancer Prev. 14:3499–3502. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saumet A, Vetter G, Bouttier M, Antoine E,
Roubert C, Orsetti B, Theillet C and Lecellier CH: Estrogen and
retinoic acid antagonistically regulate several microRNA genes to
control aerobic glycolysis in breast cancer cells. Mol Biosyst.
8:3242–3253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Varambally S, Cao Q, Mani RS, Shankar S,
Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al:
Genomic loss of microRNA-101 leads to overexpression of histone
methyltransferase EZH2 in cancer. Science. 322:1695–1699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saito A, Suzuki HI, Horie M, Ohshima M,
Morishita Y, Abiko Y and Nagase T: An integrated expression
profiling reveals target genes of TGF-β and TNF-α possibly mediated
by microRNAs in lung cancer cells. PLoS One. 8:e565872013.
View Article : Google Scholar
|
|
9
|
Piepoli A, Tavano F, Copetti M, Mazza T,
Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia
G, et al: Mirna expression profiles identify drivers in colorectal
and pancreatic cancers. PLoS One. 7:e336632012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fulci V, Chiaretti S, Goldoni M, Azzalin
G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F,
Messina M, et al: Quantitative technologies establish a novel
microRNA profile of chronic lymphocytic leukemia. Blood.
109:4944–4951. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Karreth FA, Tay Y, Perna D, Ala U, Tan SM,
Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al:
In vivo identification of tumor-suppressive PTEN ceRNAs in an
oncogenic BRAF-induced mouse model of melanoma. Cell. 147:382–395.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee SS, Shin HS, Kim HJ, Lee SJ, Lee HS,
Hyun KH, Kim YH, Kwon BW, Han JH, Choi H, et al: Analysis of
prognostic factors and 5-year survival rate in patients with
hepatocellular carcinoma: A single-center experience. Korean J
Hepatol. 18:48–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ang SF, Tan SH, Toh HC, Poon DY, Ong SY,
Foo KF and Choo SP: Activity of thalidomide and capecitabine in
patients with advanced hepatocellular carcinoma. Am J Clin Oncol.
35:222–227. 2012. View Article : Google Scholar
|
|
15
|
Zhou L, Rui JA, Wang SB, Chen SG and Qu Q:
Risk factors of poor prognosis and portal vein tumor thrombosis
after curative resection of solitary hepatocellular carcinoma.
Hepatobiliary Pancreat Dis Int. 12:68–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Deng ZS, Liao MM, Wang N, Zhang
XQ, Yu HY and Zhang YD: Tumor associated glycoprotein-72 is a novel
marker for poor survival in hepatocellular carcinoma. Pathol Oncol
Res. 18:911–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang W, Sun T, Cao J, Liu F, Tian Y and
Zhu W: Downregulation of miR-210 expression inhibits proliferation,
induces apoptosis and enhances radiosensitivity in hypoxic human
hepatoma cells in vitro. Exp Cell Res. 318:944–954. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo J, Yao F, Lou Y, Xu C, Xiao B, Zhou W,
Chen J, Hu Y and Liu Z: Detecting carcinoma cells in peripheral
blood of patients with hepatocellular carcinoma by immunomagnetic
beads and rt-PCR. J Clin Gastroenterol. 41:783–788. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng X, Pan C, Diao Y, You Y, Yang C and
Hu Z: Development of microsatellite markers by transcriptome
sequencing in two species of Amorphophallus (Araceae). BMC
Genomics. 14:4902013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia H, Zhao C, Hou L, Li A, Zhao S, Bi Y,
An J, Zhao Y, Wan S and Wang X: Transcriptome profiling of peanut
gynophores revealed global reprogramming of gene expression during
early pod development in darkness. BMC Genomics. 14:5172013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gorur A, Balci Fidanci S, Dogruer Unal N,
Ayaz L, Akbayir S, Yildirim Yaroglu H, Dirlik M, Serin MS and Tamer
L: Determination of plasma microRNA for early detection of gastric
cancer. Mol Biol Rep. 40:2091–2096. 2013. View Article : Google Scholar
|
|
22
|
Burgucu D, Guney K, Sahinturk D, Ozbudak
IH, Ozel D, Ozbilim G and Yavuzer U: Tbx3 represses PTEN and is
over-expressed in head and neck squamous cell carcinoma. BMC
Cancer. 12:4812012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ekizoglu S, Dalay N, Karaman E, Akdeniz D,
Ozaydin A and Buyru N: LKB1 downregulation may be independent of
promoter methylation or FOXO3 expression in head and neck cancer.
Transl Res. 162:122–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Oliveira KB, Guembarovski RL,
Guembarovski AM, da Silva do Amaral Herrera AC, Sobrinho WJ, Ariza
CB and Watanabe MA: CXCL12, CXCR4 and IFNγ genes expression:
Implications for proinflammatory microenvironment of breast cancer.
Clin Exp Med. 13:211–219. 2013. View Article : Google Scholar
|
|
25
|
Tsukamoto K, Ohta N, Shirai Y and Emi M: A
highly polymorphic CA repeat marker at the human interleukin 6
receptor (IL6R) locus. J Hum Genet. 43:289–290. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fulzele A, Malgundkar SA, Govekar RB,
Patil A, Kane SV, Chaturvedi P, D'Cruz AK and Zingde SM: Proteomic
profile of keratins in cancer of the gingivo buccal complex:
Consolidating insights for clinical applications. J Proteomics.
91:242–258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dou LP, Li YH, Wang LL and Yu L: HOXA9 is
direct target of miR-196a. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
27:166–169. 2011.In Chinese. PubMed/NCBI
|
|
28
|
Wang YP, Tang XJ, Chen XH, Che GW, Zhu DX,
Sun ZL and Zhou QH: Study on cloning of hTERT promoter and its
targeting transcriptional activities in telomerase-positive lung
cancer cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 37:497–501.
2006.In Chinese. PubMed/NCBI
|
|
29
|
Zhi Q, Zhu J, Guo X, He S, Xue X, Zhou J,
Hu B, Li H, Chen S, Zhao H, et al: Metastasis-related miR-185 is a
potential prognostic biomarker for hepatocellular carcinoma in
early stage. Biomed Pharmacother. 67:393–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katayama Y, Maeda M, Miyaguchi K, Nemoto
S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S and Tanaka H:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncol Lett. 4:817–823.
2012.PubMed/NCBI
|
|
31
|
Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH,
Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, et al: Association
study of microRNA polymorphisms with hepatocellular carcinoma in
Korean population. Gene. 504:92–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Furnari FB, Lin H, Huang HS and Cavenee
WK: Growth suppression of glioma cells by PTEN requires a
functional phosphatase catalytic domain. Proc Natl Acad Sci USA.
94:12479–12484. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dahia PL, Marsh DJ, Zheng Z, Zedenius J,
Komminoth P, Frisk T, Wallin G, Parsons R, Longy M, Larsson C, et
al: Somatic deletions and mutations in the Cowden disease gene,
PTEN, in sporadic thyroid tumors. Cancer Res. 57:4710–4713.
1997.PubMed/NCBI
|
|
34
|
Inaba N, Kimura M, Fujioka K, Ikeda K,
Somura H, Akiyoshi K, Inoue Y, Nomura M, Saito Y, Saito H, et al:
The effect of PTEN on proliferation and drug-, and radiosensitivity
in malignant glioma cells. Anticancer Res. 31:1653–1658.
2011.PubMed/NCBI
|
|
35
|
Barbosa M, Henrique M, Pinto-Basto J,
Claes K and Soares G: Prostate cancer in Cowden syndrome: Somatic
loss and germline mutation of the PTEN gene. Cancer Genet.
204:224–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ogita S and Lorusso P: Targeting
phosphatidylinositol 3 kinase (PI3K)-Akt beyond rapalogs. Target
Oncol. 6:103–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karra L, Shushan A, Ben-Meir A, Rojansky
N, Klein BY, Shveiky D, Levitzki R and Ben-Bassat H: Changes
related to phosphatidylinositol 3-kinase/Akt signaling in
leiomyomas: Possible involvement of glycogen synthase kinase 3alpha
and cyclin D2 in the pathophysiology. Fertil Steril. 93:2646–2651.
2010. View Article : Google Scholar
|
|
38
|
Kim HA, Kim KJ, Seo KH, Lee HK and Im SY:
PTEN/MAPK pathways play a key role in platelet-activating
factor-induced experimental pulmonary tumor metastasis. FEBS Lett.
586:4296–4302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mutter GL, Lin MC, Fitzgerald JT, Kum JB,
Baak JP, Lees JA, Weng LP and Eng C: Altered PTEN expression as a
diagnostic marker for the earliest endometrial precancers. J Natl
Cancer Inst. 92:924–930. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang
B, Min L and Liu W: miRNA-200c increases the sensitivity of breast
cancer cells to doxorubicin through the suppression of
E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 7:1579–1584.
2013.PubMed/NCBI
|
|
41
|
Xiong X, Ren HZ, Li MH, Mei JH, Wen JF and
Zheng CL: Down-regulated miRNA-214 induces a cell cycle G1 arrest
in gastric cancer cells by up-regulating the PTEN protein. Pathol
Oncol Res. 17:931–937. 2011. View Article : Google Scholar : PubMed/NCBI
|