Introduction
Gastric cancer (GC) is a highly lethal disease among
cancers worldwide. Diagnosis of GC, however, is usually confusing
and misleading due to atypical symptoms or unintelligible
complaints. As we all know, various metabolic processes proceed in
the stomach. Liver biomarkers in serum therefore have been
introduced into diagnosis of GC recent years. For example, low
serum pepsinogen levels imply the presence of atrophic gastritis, a
premonition of intestinal type GC. Nevertheless the threshold
values of serum pepsinogen levels were difficult to be determined,
due to various factors (1). Thus,
it is important to seek more metabolism-related genes to
investigate GC through metabolic perspective. To assess crucial
genes relevant to GC metabolism, we analyzed publicly available
RNA-Seq datasets, composed of the comprehensive transcriptome
profiles of 22 GC tissues and their non-cancerous counterpart
samples, and performed an integrative analysis across different GC
gene expression datasets. First, we identified a series of
differentially expressed genes (DGEs) in tumor tissues. Then, we
verified these genes by analyzing four independent microarray
datasets from distinct microarray platforms using GC samples from
different regions and ethnicities. Third, we performed gene
function enrichment analysis on DEGs. As a result, we found that
gastric lipase (LIPF), an enzyme which correlated with the
digestion of dietary triglycerides in the gastrointestinal tract,
was consistently downregulated in each dataset. Lipid metabolism is
an established hallmark of cancer. Ether lipid levels have been
shown to be elevated in tumors (2).
Thus, our research may provide a novel significant perspective to
investigate the relationship between lipid metabolic disturbance
and GC progenesis.
Materials and methods
RNA-Seq datasets of GC patients
With the aim of obtaining an integrative series of
DEGs in the stomach of GC patients compared with normal control
tissues, we evaluated SRA-formatted files of RNA-Seq data from the
Gene Expression Omnibus (GEO) database (3) under the accession number of SRP049809.
It consisted of 22 separate samples derived from GC tissues and
their matched non-cancerous samples, studied by the researchers in
the National Cancer Center of Korea (NCCK) and the University of
Texas MD Anderson Cancer Center (TMDACC). The dataset contained
transcriptome of the gastric tissues in different locations
isolated from eleven male and eleven female GC patients. In these
experiments, total RNA was purified for library preparation
(paired-end, 50 and 35 nt), template bead preparation and SOLiD
v4.0 sequencing, following standard protocols provided by Life
Technologies (Carlsbad, CA, USA).
After removing poly-A tails and low quality reads
from the original data, we mapped short read data on the human
genome reference sequence hg38 by using TopHat v2.0.14 (4). The number of mapped reads was
calculated with HTSeq (5).
Expression levels were evaluated using edgeR package (6). Significance of DEGs was expressed as
Q-value, representing FDR-adjusted P<0.05. Criteria of
determining DGEs was set to two times fold changes.
Microarray datasets of GC patients
We investigated four distinct microarray datasets of
GC retrieved from GEO dataset under accession nos. GSE13911,
GSE19826, GSE29272, and GSE33335.
The GSE13911 dataset contained transcriptome of
resected stomach tissues studied on an Affymetrix Human Genome U133
Plus 2.0 Array containing 47,000 transcripts (Affymetrix, Inc.),
and the data were normalized by the Robust Multichip Average
algorithm. The samples were collected by the researchers in IRBM
(Merck Research Laboratories, Italy). There were 38 GCs and normal
control samples with or without microsatellite instability (MSI)
which presented mismatch repair (MMR) inactivation or activation
(age, 73.94±7.21; males, 15; females, 23). This dataset contained 6
patients with diffuse GC (age, 72.83±7.14; males, 1; females, 5),
26 patients with intestinal GC (age, 74.5±7.44; males, 12; females,
14), 4 patients with mixed GC (age, 68.6±6.4; males, 3; females, 1)
and 2 patients with uncharacterized GC.
The GSE19826 dataset contained 27 transcriptome of
GC samples studied on an Affymetrix Human Genome U133 Plus 2.0
Array with the same normalized method as GSE13911 above. The
samples were collected by the researchers in the First People's
Hospital affiliated to Shanghai Jiao Tong University, China. This
dataset contained 24 GCs and 3 normal control samples from patients
with GC. There were three replicates in each histological stage.
Other details, such as age and gender, were unavailable.
The GSE29272 dataset contained 268 samples of paired
adjacent non-tumor tissues and cardia/non-cardia GC tissues. There
were 62 patients with cardia GC and 72 patients with non-cardia GC.
Samples were analyzed using the Affymetrix U133A Array. The cardia
gastric dataset consisted of 1 male patient in stage I (age, 48),
56 patients in stage III (age, 58.73±10.29; males, 39; females,
17), 5 patients in stage IV (age, 59.8±4.97; males, 5; females, 0).
The non-cardia gastric dataset contained 4 patients in stage I
(age, 55±10.10; males, 3; females, 1), 4 patients in stage II (age,
62±5.29; males, 4; females, 0), 59 patients in stage III (age,
54.71±10.07; males, 48; females, 11), 4 patients in stage IV (age,
51±16.02; males, 2; females, 2).
The GSE33335 dataset contained 50 samples of paired
adjacent non-cancerous tissues and GC tissues. There were pairs of
adjacent tissues and tumor tissues from 25 patients. These samples
were analyzed with Affymetrix Human Exon 1.0 ST Array which
contained 0.3 million probes. The dataset consisted of 7 patients
in stage I (age, 67.43±11.15; males, 4; females, 3), 6 patients in
stage II (age, 59.50±13.50; males, 4; females, 2), 6 patients in
stage IV (age, 71.33±9.11; males, 4; females, 2) and a patient
unclassified.
To assess the statistically significant DGEs between
tumor and normal or adjacent non-tumor groups, we performed a
paired t-test in LIMMA package (7–9).
Gene function enrichment analysis
Gene IDs of DEGs were submitted into the Functional
Annotation Tool of Database for Annotation, Visualization and
Integrated Discovery (DAVID) v6.7. David educes enriched gene
ontology (GO) terms in the series of DEGs. The Fisher's exact test
was employed to evaluate statistical significance. The significant
threshold was set to P<0.05 after Benjamini's correction. Then
we searched the potential pathway related to GO terms enrichment to
find the possible downstream or upstream genes.
Immunohistochemistry experiments
GC tissue sections containing HStm-Ade180Sur-02 (90
cancer cases) were provided by Outdo Biotech (Shanghai, China).
Experiments were authorized by the Ethics Committee of Jinan
central Hospital affiliated to Shandong University conforming with
the Helsinki Declaration. Histological parameters were ascertained
according to the criteria of the World Health Organization. Tumor
lymph node metastasis classification by the Current International
Union Against Cancer was employed to determine the pathologic
stages.
Ninety gastric tumor tissue samples and their
counterpart normal tissue samples were deparaffinized in xylene,
sequentially antigen were retrieved with citrate buffer solution
(Wuhan Boster Biological Technology, Ltd., Wuhan, Hubei, China).
Endogenous peroxidase activity was inactivated by 0.3%
H2O2 for 15 min at room temperature. The
sections were blocked with low lental serum for 30 min and
subsequently incubated with primary antibodies [LIPF: Abcam,
Cambridge, MA, USA; diacylglycerol kinase α (DGKA): Proteintech
Group, Inc., Rosemont, IL, USA]. Incubation of secondary antibodies
was performed with kits (ZSGB-BIO, Beijing, China). Antibody
staining was visualized with DAB (D-5637; Sigma) and hematoxylin
counterstain. Sections with 5% labeled cells were scored as 0; with
5–25% labeled cells as 1; with 26–50% labeled cells as 2; with
50–80% labeled cells as 3; and with >80% labeled cells as 4. The
staining intensity was scored similarly, with 0 indicating negative
staining, 0.5 weakly positive, 1 moderately positive and 2 strongly
positive. The scores for the percentage of positive tumor cells and
staining intensity were multiplied to generate an immunoreactive
score for each specimen. Samples with scores ≥3 were considered as
high expression while those with scores <3 were considered as
low expression.
Statistical analysis
The correlation of LIPF and its potential relevant
genes, DGKA clinicopathological characteristics were evaluated by
the Spearman's rank test and χ2 test. Expression level
of these biomarkers was investigated through Mann-Whitney test.
Kaplan-Meier survival curves were calculated using the log-rank
test. Multivariate Cox regression analysis was employed to
investigate the potential prognostic factors for survival in
patients with GC. P<0.05 was considered statistically
significant. The statistical calculation was performed in R
language package.
Results
RNA-Seq and microarray data analysis of
GC tissues
Based on RNA-Seq data analyzed with TopHat and
HTSeq, we studied transcriptome of the stomach samples from
resected tumor and normal tissues collected by NCCA and TMDACC.
Among them, we identified a series of DEGs that satisfied Q-value
(FDR-corrected P-value) <0.05 and |logFC| >2 times, when
compared between GC and normal groups. Sequentially, we divided
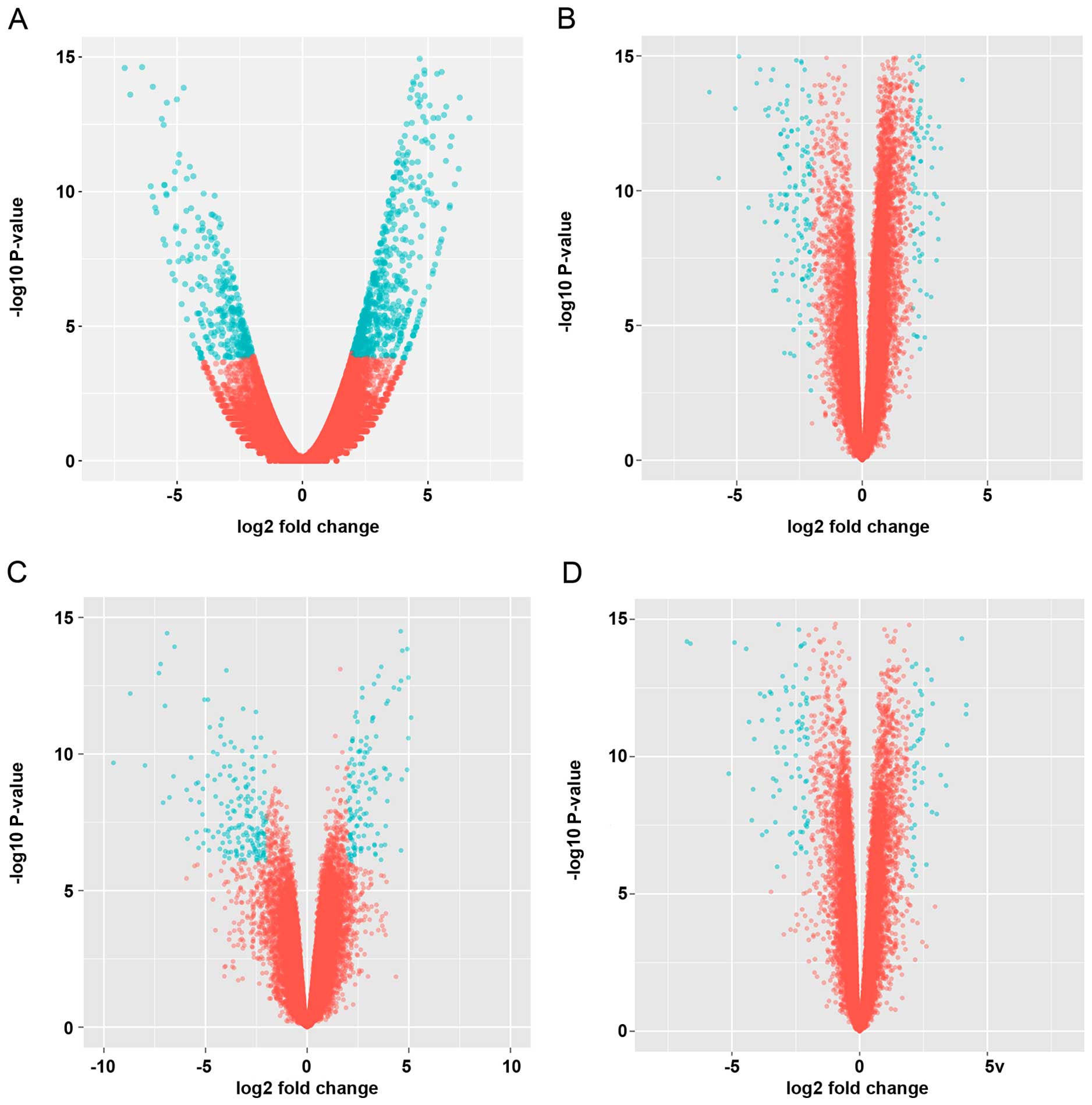
these DEG into 476 upregulated genes and 222 downregulated genes in
GC (Fig. 1A). Tumor progression
always correlated with overexpression of oncogenes and inactivation
of anti-oncogenes. Downregulating of anti-oncogenes may play a
crucial role in tumorigenesis (10).
Previous research was mainly focused on upregulated
genes, lacking of deep data evaluations on download genes. DEGs
were assessed and refined to obtain a core set which reduced
redundancies, according to Q-values, fold change values and
functions. Obviously, the expression of LIPF, an enzyme involved in
the digestion of dietary triglycerides in the gastrointestinal
tract, and responsible for 30% of fat digestion processes occurring
in human, was greatly reduced at fold changes of −2.55
(Q=1.62E-04).
To confirm the results of RNA-Seq data analysis, we
studied four microarray datasets of GC tissues numbered GSE13911,
GSE19826, GSE29272 and GSE33335. First, GSE13911, composed of
transcriptome of GC tissues with and without MSI isolated in
European patients, were investigated. We identified a series of 260
DEGs, including 166 downregulated and 94 upregulated genes. Because
previous research was mainly focused on upregulated, instead of
downregulated, genes we intended to obtain a deep panorama of
downregulated gene networks. Conspicuously, expression of LIPF
decreased at fold change equal to −6.09 times in gastric tumor
tissues (P=6.05E-12). In contrast, other lipase did not show
significant expression difference (P>0.05) between GC and NC in
this dataset.
Sequentially, we analyzed datasets of transcriptome
isolated from adjacent normal/tumor-matched gastric tissues in
Asian patients (GSE19826 on Human Genome U133 Plus 2.0 Array;
GSE29272 on Human Genome U133A Array and GSE33335 on Human Exon 1.0
ST Array). From the GSE19826 dataset, 378 DEGs were downregulated
in the gastric tumor tissues with the |logFC| >2 times of GC
versus NC. The expression of LIPF decreased at fold change of
−10.47 times (P=1.15E-13). From the GSE29272 dataset 75
downregulated DEGs were assessed in the gastric cardia and
non-cardiac adenocarcinomas from Chinese patients. Analogically,
the expression of LIPF decreased in GC at fold change of −5.23
times (P=4.01E-52). From the GSE33335 dataset, we identified 161
DEGs which constituted 107 downregulated and 54 upregulated genes.
The expression of LIPF decreased in GC at fold change equal to
−7.86 times (P=3.22E-17). The outline of the spread of DEGs in each
dataset is shown in Fig. 1. We
combined RNA-Seq and microarray data to select a core set of 373
DEGs. Taken together, from these transcriptome data, the expression
of LIPF was significantly decreased in the GC tissues in spite of
different location in the stomach, microarray platforms, or
ethnicity of the samples.
Top rank DEGs which possessed statistical
significance are listed in Table I.
The average logFC values were calculated to obtain a comprehensive
assessment on distinct datasets.
 | Table ITop rank differentially expressed
genes among distinct datasets. |
Table I
Top rank differentially expressed
genes among distinct datasets.
| Gene ID | Gene name | Full name | Average logFC |
|---|
| 2694 | GIF | Gastric intrinsic
factor | −6.586 |
| 496 | ATP4B | ATPase,
H+/K+ exchanging, β polypeptide | −6.54044 |
| 8513 |
LIPF | Gastric lipase | −6.438 |
| 56287 | GKN1 | Gastrokine 1 | −5.722 |
| 495 | ATP4A | ATPase,
H+/K+ exchanging, α polypeptide | −5.114 |
| 2104 | ESRRG | Estrogen-related
receptor γ | −4.65 |
| 643834 | PGA3 | Pepsinogen 3, group I
(pepsinogen A) | −4.426 |
| 10690 | FUT9 | Fucosyltransferase 9
(α1,3-fucosyltransferase) | −4.406 |
| 9992 | KCNE2 | Potassium
voltage-gated channel, Isk-related family, member 2 | −4.372 |
| 5225 | PGC | Progastricsin
(pepsinogen C) | −4.362 |
| 27159 | CHIA | Chitinase,
acidic | −4.322 |
| 200504 | GKN2 | Gastrokine 2 | −4.318 |
| 57016 | AKR1B10 | Aldo-keto reductase
family 1, member B10 (aldose reductase) | −3.92 |
| 1358 | CPA2 | Carboxypeptidase A2
(pancreatic) | −3.818 |
| 3624 | INHBA | Inhibin βA | 4.235 |
| 1469 | CST1 | Cystatin SN | 3.69 |
| 4102 | MAGEA3 | Melanoma antigen
family A, 3 | 3.5775 |
| 6696 | SPP1 | Secreted
phosphoprotein 1 | 3.412 |
| 23213 | SULF1 | Sulfatase 1 | 3.078 |
Gene function annotation
DAVID annotation demonstrated that most
downregulated genes in gastric tumor tissues were enriched in GO
terms of ʻlipid homeostasis' (GO: 0055088, P=3.3E-2), ʻorganic
ether metabolic process' (GO: 0018904, P=1.8E-3), ʻlipid catabolic
process' (GO: 0016042, P=5.6E-5).
LIPF was related with the signal pathways
ʻglycerolipid metabolism' (hsa00561), ʻmetabolic pathways'
(hsa01100) and ʻfat digestion and absorption' (hsa04975) in KEGG
database. DGKA, a potential downstream gene of LIPF was reported to
play an important role in tumorigenesis (11). To explore the protein expression
levels of LIPF and DGKA in GC, we performed immunohistochemistry in
GC and adjacent non-tumor tissues and analyzed the correlation
between LIPF, DGKA expression and clinicophathological
characteristics of GC patients.
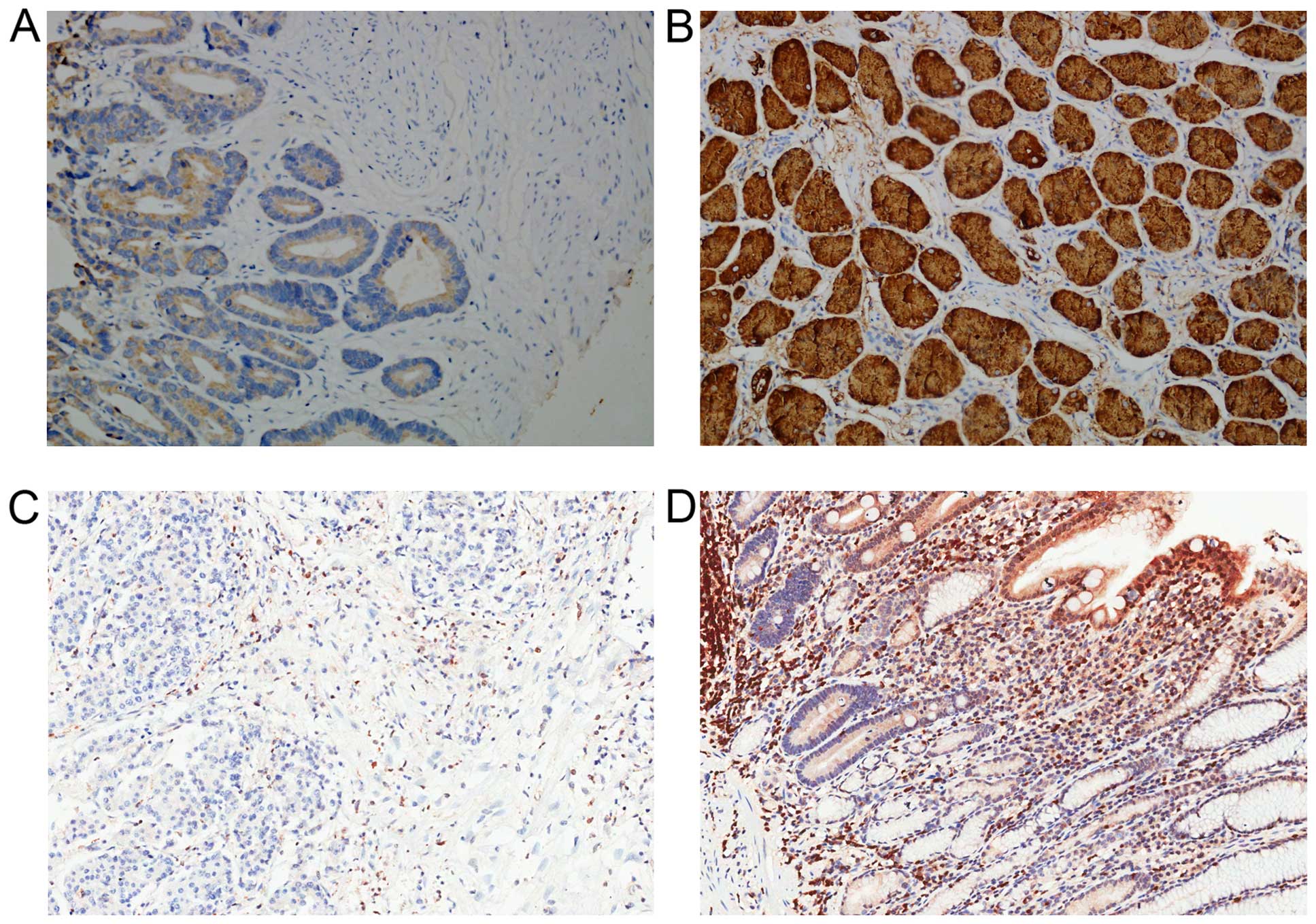
Results showed that LIPF protein was mainly located
in cell membranes and cytoplasm in gastric cells (Fig. 2). Among 90 GC and adjacent normal
tissues, LIPF was frequently expressed in normal tissues (94.4%,
85/90), while in GC tissues the positive percent is 59.1% (53/90).
A significant difference was observed between these two groups
(P<0.001). Additionally, higher level of LIPF expression was
detected in early stage of GC (stage I, II; mean, 2.000±0.1905,
N=32), while in advanced stage of GC the expression was lower
(stage III, IV; mean, 0.5517±0.1098, N=58) (Fig. 3A, Table
II). Analogical analysis was also performed on IHC results from
DGKA, and the results are listed in Table II. The positive rate of DGKA
decreased to 77.8% (70/90) in GC from 90% (81/90) in normal
tissues. There was a significant difference between GC and normal
tissues (P<0.05). Obvious disparity was found comparing early
stage (2.750±0.2490, N=32) and late stage (1.078±0.1056, N=58)
GC.
 | Table IICorrelation between expression of
LIPF, DGKA and clinicopathological features of GC patients. |
Table II
Correlation between expression of
LIPF, DGKA and clinicopathological features of GC patients.
| Characteristics | LIPF high
expression | LIPF low
expression | χ2 or
Fisher's exact test | P-value | DGKA high
expression | DGKA low
expression | χ2 or
Fisher's exact test | P-value |
|---|
| Age (years) | | | 2.18E-30 | 1.0 | | | 0.014 | 0.91 |
| <60 | 5 | 26 | | | 9 | 22 | | |
| ≥60 | 10 | 49 | | | 15 | 44 | | |
| Gender | | | | 1.0 | | | 2.94E-4 | 0.99 |
| Male | 10 | 52 | | | 16 | 46 | | |
| Female | 4 | 24 | | | 8 | 20 | | |
| Local invasion | | | 10.03 | 0.002 | | | | 0.0069 |
| T1+T2 | 6 | 5 | | | 7 | 4 | | |
| T3+T4 | 9 | 70 | | | 17 | 62 | | |
| Nodal status | | | 2.17 | 0.14 | | | 2.27 | 0.13 |
| Positive | 8 | 57 | | | 14 | 51 | | |
| Negative | 7 | 18 | | | 10 | 15 | | |
| Stage of
disease | | | 4.53 | 0.033 | | | 4.15 | 0.041 |
| I+II | 10 | 25 | | | 14 | 21 | | |
| III+IV | 5 | 50 | | | 10 | 45 | | |
| Distant
metastasis | | | | 1.0 | | | | 1.0 |
| M0 | 15 | 74 | | | 24 | 65 | | |
| M1 | 0 | 1 | | | 0 | 1 | | |
| Depth of tumor
invasion | | | 0.33 | 0.031 | | | 0.15 | 0.70 |
| Mucosa, submucosa,
muscularis propria, subserosa | 14 | 47 | | | 15 | 46 | | |
| Penetration of
serosa,adjacent structures | 1 | 28 | | | 9 | 20 | | |
Survival analysis
The 90 patients were predominantly males (68.9%)
with a performance status of 0 (females) or 1 (males). The median
age was 67 years (range, 42–83 years). A total of 35.5% of patients
belonged to early stage according to the AJCC classification
strategy (stage I and II) while the other patients belonged to late
stage (stage III and IV). Twelve percent of patients had low level
local invasion (T1 and T2) while the other 88% of patients had high
level local invasion (Table III).
Median follow-up time was 22 months (range from 1 to 78 months).
Among these patients, 56 (65.8%) died during the follow-up period.
In univariate analysis of prognostic value of clinical factors for
progression-free survival, poor prognosis was associated with lymph
node metastasis. Stage III and IV diseases were associated with a
poor prognosis compared with stage I and II diseases
(P=2.02×10−5). A better prognosis was associated with
high DGKA expression type as compared with low DGKA expression type
(Fig. 3B). A moderate increase in
LIPF expression was associated with a decreased risk but without
reaching statistical significance. Gender (P=0.69), age (P=0.40)
and local invasion status (P=0.13) did not reach a significance for
predicting prognostic value of clinical factors for
progression-free survival. In order to select the most appropriate
variables, a method following stepwise Akaike's Information
Criterion (AIC) was employed. A group of variables with the lowest
AIC value was chosen for subsequent multivariate Cox regression
analysis. In the multivariate Cox regression analysis, lymph node
metastasis (P=1.97×10−5) was also associated with a poor
prognosis. High expression of DGKA means a decrease of risk.
Compared to univariate analysis, the patient age (P=0.028) was
significantly associated with a low risk (Table IV).
 | Table IIIClinical characteristics of GC
tissues. |
Table III
Clinical characteristics of GC
tissues.
| Clinicopathological
features | No. | Percentage (%) |
|---|
| Age (years) |
| <60 | 31 | 34.4 |
| ≥60 | 59 | 65.6 |
| Gender |
| Male | 62 | 68.9 |
| Female | 28 | 31.1 |
| Tumor size
(cm) |
| <10 | 79 | 87.8 |
| ≥10 | 11 | 12.2 |
| Tumor site |
| Gastric
cardia | 11 | 12.2 |
| Body | 24 | 26.7 |
| Gastric
antrum | 38 | 42.2 |
| Gastric notch | 9 | 10 |
| Other | 8 | 8.8 |
| Tumor stage |
| I | 4 | 4.4 |
| II | 28 | 31.1 |
| III | 52 | 57.4 |
| IV | 1 | 1.1 |
| Follow-up time
(months) | 37 (1–78) | |
| Prognosis |
| Alive | 34 | 37.8 |
| Dead | 56 | 62.2 |
| Patients lived for
5 years | 31 | 34.4 |
| Nodal status |
| Positive | 65 | 72.2 |
| Negative | 25 | 27.8 |
| Pathological
type |
|
Adenocarcinoma | 90 | 100 |
 | Table IVMultivariate Cox regression analysis
of potential prognostic factors for survival in patients with
GC. |
Table IV
Multivariate Cox regression analysis
of potential prognostic factors for survival in patients with
GC.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender (male vs.
female) | 0.51–1.55 | 0.69 | | |
| Age (≥60 vs. <60
years) | 0.44–1.36 | 0.40 | 0.29–0.93 | 0.02 |
| LNM status (yes vs.
no) | 2.58–16.54 | 7.19E-05 | 1.39–15.51 | 0.01 |
| DGKA (high vs.
low) | 0.21–0.84 | 0.01 | 0.26–0.94 | 0.03 |
| LIPF (high vs.
low) | 0.15–0.95 | 0.07 | | |
| Tumor status (T3+T4
vs. T1+T2) | 0.76–7.79 | 0.13 | | |
| Stage (III+IV vs.
I+II) | 2.19–8.31 | 2.02E-05 | | |
Discussion
Recently, massive transcriptome studies of GC have
been published on GEO database. Analysis of these data will provide
important insights into the mechanisms underlying the progression
of GC, and give more hints to find novel diagnostic and therapeutic
methods for GC. We investigated the RNA-Seq datasets of GC
transcriptome derived from NCCK and TMDACC, and identified a core
set of 373 DEGs in GC vs. adjacent normal tissues. Then, we
combined four independent microarray datasets to filtrate these
DEGs into a core set. The downregulated genes in GC tissues,
related with digestion, lipid catabolic process, lipid binding,
lipase inhibitor activity, steroid binding by GO and pathway
analysis. Previous research focused on upregulated gene in GC
(12), however, downregulated genes
in this dataset demonstrated more tissue-specific patterns in GO
analysis. The downregulated genes in GC tissues were found to be
related with digestion, lipid catabolic process, lipid binding,
lipase inhibitor activity, steroid binding via GO and pathway
analysis. We compared the results of RNA-Seq data with four
different microarray datasets of GC tissues from different regions,
microarray platforms and ethnicities. Based on a comprehensive
assessment, we identified that LIPF was consistently downregulated
among these datasets.
LIPF, an enzyme involved in the digestion of dietary
triglycerides in the gastrointestinal tract, is responsible for 30%
of fat digestion processes occurring in human. It hydrolyzes the
ester bonds of triglycerides under acidic pH conditions. The
conserved lipase family is composed of a series of paralogs with
tissue-specificity, such as LIPA, LIPM. LIPF plays distinct roles
in neutral lipid metabolism. It is usually secreted by gastric
chief cells in the fundic mucosa of the stomach. The deficiency of
LIPF and its paralog, LIPA, is associated with Wolman disease and
cholesteryl ester storage disease (13). Recruitment of LIPF by free fatty
acid-rich particles inhibited triacylglycerol hydrolysis (14). LIPF was involved in two important
metabolic pathways, glycerolipid metabolism (KEGG: hsa00561) and
fat digestion and absorption (KEGG: ko04975). The downstream gene
of LIPF, DGKA, was reported to play a crucial role in secretion of
Fas ligand-containing exosomes (15). Study showed that exosomes release
was positively correlated with the invasiveness of ovarian cancer,
which brought potential for diagnosis in peripheral blood detection
(16). Different expression levels
of LIPF and its target genes in GC may suggest a novel perspective
upon relationship between tumor progression and lipid metabolic
disorder. These genes may provide metabolic insights for GC
investigation. Results of immunohistochemistry verified that the
expression of LIPF was significantly decreased and the expression
level was related to the pathological stage of GC. Compared with
early stage, samples of stage III and IV had a lower expression of
LIPF (Fig. 3A). We also
demonstrated that the local invasion, stage of disease, and depth
of tumor invasion correlated significantly with LIPF (P<0.05,
Table II). Besides, lipid
metabolism was involved in the initiation and/or progression of
cancer-associated cachexia (17).
In fatty acid metabolism of aggressive human cancer cells,
oxidative pathways were reduced, instead, pathways for generating
structural and signaling lipids were increased (18). Decreased LIPF level may play an
important role in this lipogenic conversion.
Diacylglycerol kinases (DGKs) are composed by 10
enzymes, which metabolize 1,2-diacylglycerol (DAG) to produce
phosphatidic acid (PA). DGKA is a key regulator of the polarized
secretion of exosomes, which had a particular lipid and protein
content (19). Takeishi et
al reported that DGKA enhanced hepatocellular carcinoma
progression through activating Ras-Raf-MEK-ERK pathway (11), playing a crucial role in dysfunction
of human tumor-infiltrating CD8+ T cells (20). DGKA was also considered as a
potential therapeutic target in glioblastoma cell lines, such as
U87MG and U251 (21). In addition,
downregulation of DGKA caused toxicity through key oncogenic
pathways, including mTOR-SREBP and Hif-1α. Moreover, overexpression
of mTOR and Hif-1α rescued the toxicity from DGKA knockdown and
inhibition.
We also compared the difference of DGKA expression
between GC and normal tissues. Expression level of DGK correlated
positively with LIPF in Spearman's rank test (P<0.05). Our
results showed that the difference of DGKA expression correlated
with local invasion, stage of disease (P<0.05).
Previous research indicated that high expression
level of DGKA was related to poor prognosis, in other words, low
expression was related to better prognosis (11). However, our results indicated an
alternative regulating pattern in GC. The Cox regression results
implied a protective effect of high DGKA rather than a poor risk
(HR, 0.19–0.79; 95% CI; P= 0.041). Analogically, the reversed
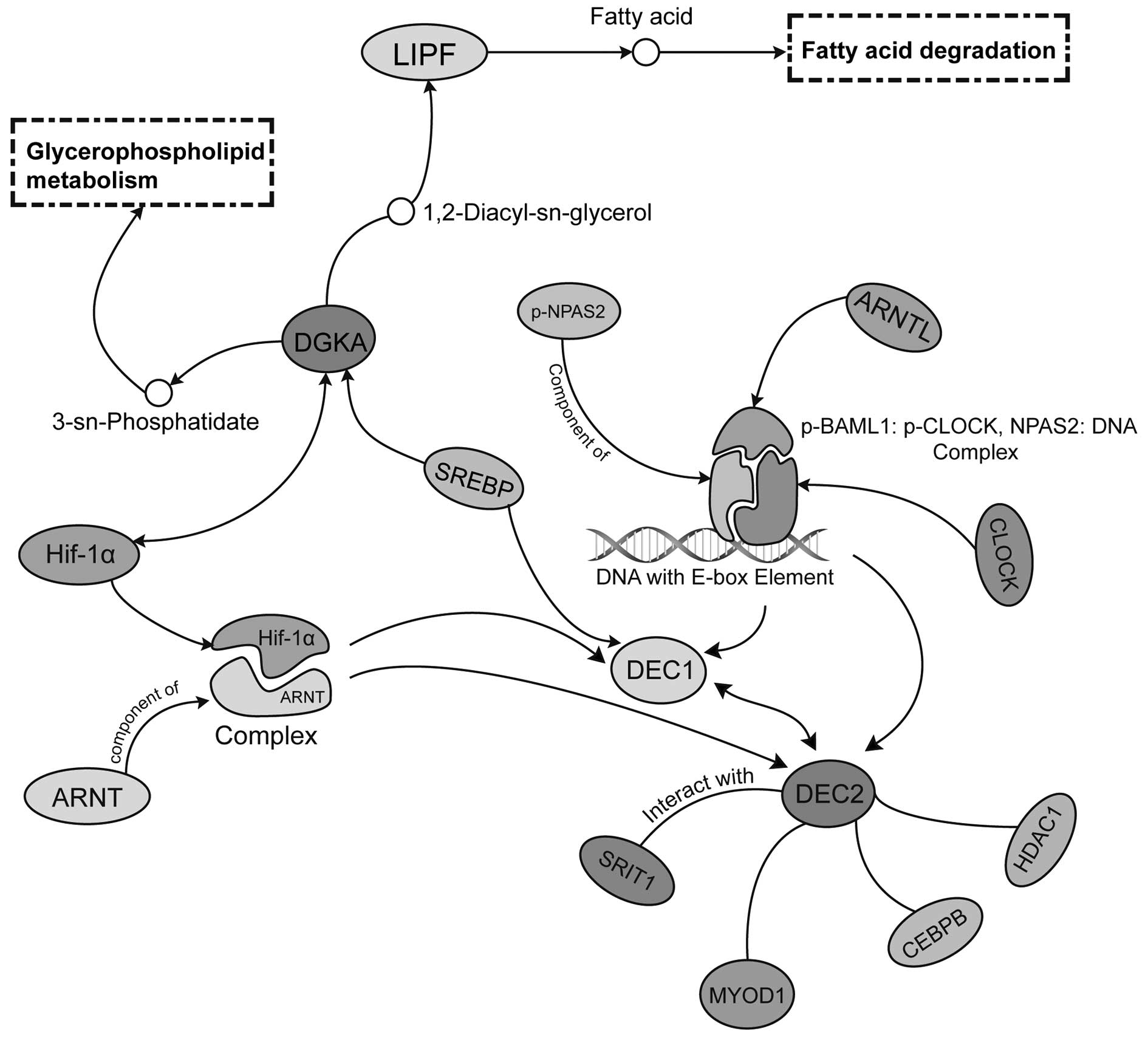
regulating pattern was also observed in DEC1, another important
regulating target of Hif-1α in GC (Fig.
4). Low DEC1 expression was associated with poor histological
differentiation and malignancy progression in hepatocellular
carcinoma, while high DEC1 expression was associated with poor
histological differentiation in gastric carcinoma and non-small
cell lung cancer (22–24). Interestingly, the regulating targets
of DGKA, Hif-1α and SERBP, also served as mediators of DEC1 and its
paralog, the DEC2. This interdependency implied a potential
regulating network, connecting glycerolipid metabolism with
hypoxia-dependent circadian clock derangement. The DGKA expression
demonstrated tissue-specificity, which indicated different
regulation mechanisms in various organisms.
In conclusion, bioinformatics data mining on RNA-Seq
and microarray datasets of GC indicated that LIPF was a
downregulated DEG in GC. IHC results confirmed that the
downregulation of LIPF and its target gene, DGKA, which might
reduce the lipid decompose metabolism, provided lipid resource to
the growth and division of cancer cells. Thus, we speculate that
LIPF might play a crucial role in the development of GC. Further
experiments need to be done to investigate the detail
mechanisms.
Acknowledgments
This study was financially supported by the National
Natural Science Foundation of China (NSFC nos. 81000869 and
81272588) and Project 973 (grant nos. 2012CB966503 and
2012CB966504).
References
|
1
|
Ohata H, Oka M, Yanaoka K, Shimizu Y,
Mukoubayashi C, Mugitani K, Iwane M, Nakamura H, Tamai H, Arii K,
et al: Gastric cancer screening of a high-risk population in Japan
using serum pepsinogen and barium digital radiography. Cancer Sci.
96:713–720. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Benjamin DI, Cozzo A, Ji X, Roberts LS,
Louie SM, Mulvihill MM, Luo K and Nomura DK: Ether lipid generating
enzyme AGPS alters the balance of structural and signaling lipids
to fuel cancer pathogenicity. Proc Natl Acad Sci USA.
110:14912–14917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets - update. Nucleic Acids Res. 41(D1): D991–D995.
2013. View Article : Google Scholar
|
|
4
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anders S, Pyl PT and Huber W: HTSeq - a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar
|
|
6
|
Robinson MD and Smyth GK: Moderated
statistical tests for assessing differences in tag abundance.
Bioinformatics. 23:2881–2887. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bolstad BM, Irizarry RA, Åstrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson TM and Attardi LD: Dissecting p53
tumor suppressor function in vivo through the analysis of
genetically modified mice. Cell Death Differ. 13:902–908. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takeishi K, Taketomi A, Shirabe K, Toshima
T, Motomura T, Ikegami T, Yoshizumi T, Sakane F and Maehara Y:
Diacylglycerol kinase alpha enhances hepatocellular carcinoma
progression by activation of Ras-Raf-MEK-ERK pathway. J Hepatol.
57:77–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang HR, Nam S, Kook MC, Kim KT, Liu X,
Yao H, Jung HR, Lemos R Jr, Seo HH, Park HS, et al: HNF4α is a
therapeutic target that links AMPK to WNT signalling in early-stage
gastric cancer. Gut. 65:19–32. 2016. View Article : Google Scholar
|
|
13
|
Tylki-Szymańska A and Jurecka A: Lysosomal
acid lipase deficiency: Wolman disease and cholesteryl ester
storage disease. Pril (Makedon Akad Nauk Umet Odd Med Nauki).
35:99–106. 2014.
|
|
14
|
Pafumi Y, Lairon D, de la Porte PL, Juhel
C, Storch J, Hamosh M and Armand M: Mechanisms of inhibition of
triacylglycerol hydrolysis by human gastric lipase. J Biol Chem.
277:28070–28079. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alonso R, Mazzeo C, Rodriguez MC, Marsh M,
Fraile-Ramos A, Calvo V, Avila-Flores A, Merida I and Izquierdo M:
Diacylglycerol kinase α regulates the formation and polarisation of
mature multivesicular bodies involved in the secretion of Fas
ligand-containing exosomes in T lymphocytes. Cell Death Differ.
18:1161–1173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi M, Salomon C, Tapia J, Illanes
SE, Mitchell MD and Rice GE: Ovarian cancer cell invasiveness is
associated with discordant exosomal sequestration of Let-7 miRNA
and miR-200. J Transl Med. 12:42014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Das SK and Hoefler G: The role of
triglyceride lipases in cancer associated cachexia. Trends Mol Med.
19:292–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Louie SM, Roberts LS, Mulvihill MM, Luo K
and Nomura DK: Cancer cells incorporate and remodel exogenous
palmitate into structural and oncogenic signaling lipids. Biochim
Biophys Acta. 1831:1566–1572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alonso R, Mazzeo C, Mérida I and Izquierdo
M: A new role of diacylglycerol kinase alpha on the secretion of
lethal exosomes bearing Fas ligand during activation-induced cell
death of T lymphocytes. Biochimie. 89:213–221. 2007. View Article : Google Scholar
|
|
20
|
Prinz PU, Mendler AN, Masouris I, Durner
L, Oberneder R and Noessner E: High DGK-α and disabled MAPK
pathways cause dysfunction of human tumor-infiltrating
CD8+ T cells that is reversible by pharmacologic
intervention. J Immunol. 188:5990–6000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dominguez CL, Floyd DH, Xiao A, Mullins
GR, Kefas BA, Xin W, Yacur MN, Abounader R, Lee JK, Wilson GM, et
al: Diacylglycerol kinase α is a critical signaling node and novel
therapeutic target in glioblastoma and other cancers. Cancer
Discov. 3:782–797. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jia YF, Xiao DJ, Ma XL, Song YY, Hu R,
Kong Y, Zheng Y, Han SY, Hong RL and Wang YS: Differentiated
embryonic chondrocyte-expressed gene 1 is associated with
hypoxia-inducible factor 1α and Ki67 in human gastric cancer. Diagn
Pathol. 8:372013. View Article : Google Scholar
|
|
23
|
Shi XH, Zheng Y, Sun Q, Cui J, Liu QH, Qü
F and Wang YS: DEC1 nuclear expression: A marker of differentiation
grade in hepatocellular carcinoma. World J Gastroenterol.
17:2037–2043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao
Y and Wang EH: The transcription factor DEC1
(BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in
non-small-cell lung cancer and inhibits the proliferation through
cyclin D1 in A549 and BE1 cells. Tumour Biol. 34:1641–1650. 2013.
View Article : Google Scholar : PubMed/NCBI
|